INTRODUCTION
The actin cytoskeleton of Saccharomyces cerevisiae is composed of a single, conventional actin isoform that is 86% identical to mammalian actins in addition to a battery of similarly conserved associated proteins. This conservation of components and their functions has led yeast geneticists and cell biologists to use the experimental power of yeast toward the study of the regulation of actin cytoskeleton assembly and the basic, housekeeping functions of the actin cytoskeleton (for a comprehensive review, see Botstein et al., 1997). Important to these studies has been the visualization of the actin cytoskeleton through the cell cycle and how mutations in regulators and components of the yeast cytoskeleton affect this organization.
The first images of the yeast actin cytoskeleton showed that it consists of two filament-based structures: the actin cortical patch and the actin cables (Adams and Pringle, 1984; Kilmartin and Adams, 1984). The actin cortical patches show a polarized distribution that changes during the cell cycle: first they appear at the incipient bud site, suggesting a role in bud emergence; soon thereafter they are also found within the growing bud, indicating a role in bud growth; and late in the cell cycle they reorganize into two rings in the neck, where they are believed to be involved in septation and cytokinesis. By electron microscopy, the actin cortical patches have been shown to be invaginations of the plasma membrane around which actin filaments and actin-associated proteins are organized (Mulholland et al., 1994). Recently it has been shown that subsets of actin cortical patches can move at speeds of up to 1 μm/s (Doyle and Botstein, 1996; Waddle et al., 1996). The actin cables, which consist of bundled actin filaments, were observed to generally run along the long axis of budding cells. This organization fits well with the understanding that actin is involved in polarized cell growth, dynamic reorganization of the cell cortex, membrane trafficking at the cell cortex, and organelle segregation at cell division.
Because certain aspects of the organization of the yeast actin cytoskeleton cannot be well addressed or documented by conventional two-dimensional microscopy, I have investigated the use of a DeltaVision deconvolution microscope for visualization of the yeast actin cytoskeleton in three dimensions. This instrument digitally captures focal sections in the Z plane and mathematically removes out-of-focus light by examining neighboring focal sections, from each section in a process called iterative deconvolution (Agard et al., 1989; Scalettar et al., 1996). The clarified focal sections can then be assembled to produce high-resolution three-dimensional images.
I have found that this technology is extremely powerful for the study of yeast cell anatomy. With respect to the organization of the actin cytoskeleton, I have confirmed that the actin cables (like the patches) are cortical in their arrangement and can be observed to attach both at their ends and laterally to the cortical patches. In some cases, multiple cables can be seen to attach to single cortical patches. In addition, I have observed the earliest stages of bud emergence and have found that buds first emerge through a ring of cortical patches, and only after this stage do the cortical patches migrate into the bud. In my strain background (S288C), I do not observe a polarized distribution of cortical patches in the bud at any time during bud growth.
MATERIALS AND METHODS
Rhodamine-Phalloidin Staining
Yeast strain FY23x86 (ura3-52/ura3-52 leu2Δ1/leu2Δ1 trp1Δ63/TRP1 HIS3/his3Δ200) was grown in 25 ml of YPD at 30°C to low log (5 × 106/ml) and fixed in the medium with 4% formaldehyde (Polysciences, Warrington, PA) for 10 min. The cells were resuspended in PBS plus 4% formaldehyde and incubated for 1 h at 25°C on a roller drum. The cells were washed twice with PBS and resuspended in 500 μl of PBS. To 100 μl of cell suspension was added 10 μl of rhodamine-phalloidin (Molecular Probes, Eugene, OR) dissolved in methanol according to the manufacturer’s instructions. The cells were incubated in the dark for 1 h, washed five times with 1 ml of PBS, and finally suspended in 100 μl of mount solution (90% glycerol, 0.1× PBS, 92.5 μM p-phenylenediamine [Sigma, St. Louis, MO], pH adjusted to >8 with 0.5 M sodium carbonate, pH 9.0). Cells were stored at −20°C until use.
Image Capture and Analysis
Rhodamine-phalloidin–stained cells (2.5 μl) were spotted on a dust-free slide and covered with a 22 × 22-mm coverslip. The cells were visualized on an integrated DeltaVision system (Applied Precision, Issaquah WA) including a Nikon (Garden City, NY) E6 inverted microscope, with a 100× (numerical aperture, 1.4) objective. Images were captured with a Princeton Instruments (Trenton, NJ) cooled, charge-coupled device camera. thirty-five to 40 optical sections were captured at 0.2-μm intervals, and out-of-focus light was removed by iterative deconvolution (Agard et al., 1989; Scalettar et al., 1996) on a Silicon Graphics (Mountain View, CA) Iris workstation. The deconvolved images were transferred to an Apple (Cupertino, CA) Macintosh computer, and projections were generated with NIH Image 1.60 software (National Institutes of Health, Bethesda, MD). Stereo pairs were generated from 5° rotations of the image projections.
RESULTS
Diploid strain FY23x86 was stained with rhodamine-conjugated phalloidin and visualized by fluorescence microscopy on an integrated DeltaVision microscopy workstation. Optical sections were taken at 0.2-μm increments, allowing us to obtain 35–40 optical sections per cell. The sections were then mathematically deconvolved, and three-dimensional projections were generated. The projections can be viewed in three dimensions as movies or as stereo pairs, or they can be collapsed into two dimensions. Figure 1 shows a representative sample of collapsed images of the actin cytoskeleton from cells at distinct points in the cell cycle. Even though represented in two dimensions, these images are far superior to those obtainable by conventional microscopy in resolution, contrast, and detail.
Figure 1.
The yeast actin cytoskeleton through the cell cycle. Diploid yeast cells were stained with rhodamine-phalloidin and imaged with a DeltaVision workstation, and the deconvolved optical sections were collapsed into two dimensions.
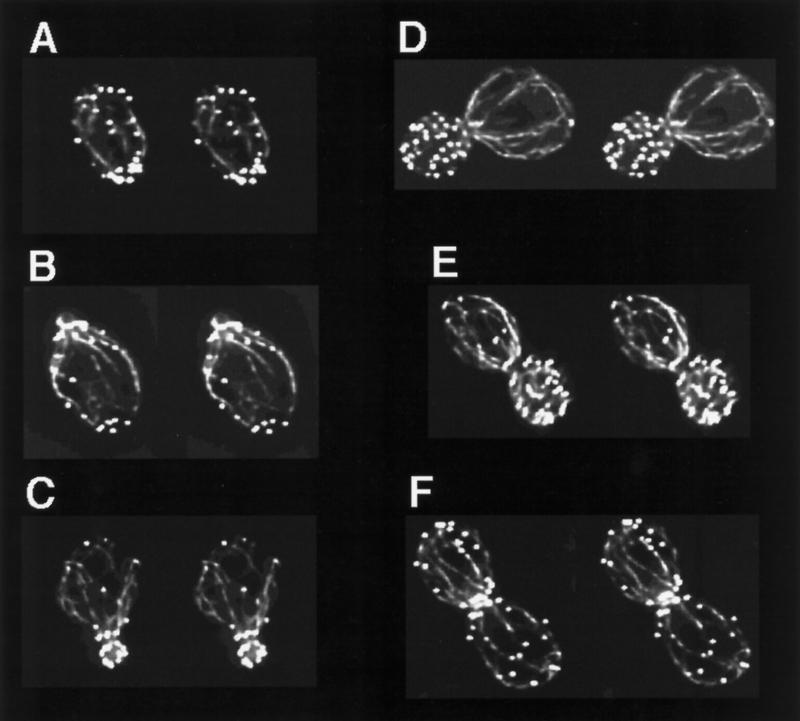
Figure 2 shows stereo pairs generated from the projections of the images displayed in Figure 1. The cells are arranged in their presumed order in the cell cycle, beginning in Figure 2A with cells at start; Figure 2B shows a cell at the earliest stages of bud emergence; Figure 2C shows a cell soon after bud emergence; Figure 2, D and E, shows cells preparing for cytokinesis; and Figure 2F shows a cell at or soon after cytokinesis. A number of features of the yeast actin cytoskeleton that were previously unappreciated and/or unresolved can be clearly seen in these stereo pairs and in the accompanying on-line version of this article, which contains movies showing rotations of these images.
Figure 2.
Stereo pair images of the yeast actin cytoskeleton through the cell cycle. Projections generated from deconvolved optical sections of yeast cells stained with rhodamine-phalloidin were rotated 5° to generate stereo pairs. Cells are displayed by their presumed order in the cell cycle beginning with start (A) and ending with the completion of cytokinesis (F).
The Actin Cables Are Almost Exclusively Located at the Cortex of the Cell.
This can be appreciated in many of the images but is perhaps most dramatic in the cell displayed in Figure 2D.
The Actin Cables Interact with the Actin Cortical Patches.
Some but not all cables can be seen to contact cortical patches within the mother cell. These interactions can be observed at the apparent termini of cables, and in fact multiple cables sometimes appear to terminate at a single cortical patch (Figure 2E, see top right corner of the mother cell). Single cables can also be seen to interact laterally with cortical patches, as if the cortical patches may be partly responsible for the cortical organization of the cables. A particularly dramatic example of this can be seen at the right side of the mother cell in Figure 2B, where a single cable appears to form lateral contacts with several different cortical patches. I have been unable to visualize attachment of cables to patches in the daughter cells, possibly because they are obscured by the strong fluorescence emitted by the densely packed cortical patches found within the buds. Although I have seen cables extend from the mother cell body into the bud, frequently cables that originate in the mother appear to terminate in the neck region (see Figure 2D). It is possible that because of the large amount of out-of-focus light (from the strongly fluorescent cortical patches), the deconvolution procedure has artificially eliminated some of the cables in the bud. Alternatively, it may be correct that cables in the mother do not generally extend into the bud. Further experimentation will be needed to resolve this issue.
At Bud Emergence, the Bud Does Not Contain Cortical Patches.
In several cells, I was able to observe the earliest stages of bud emergence. In these cells the bud appears to be extruded through the ring of cortical patches, and at this point the bud contains no cortical patches. In the cell shown in Figure 2B, I have captured the first cortical patch migrating into a bud that is otherwise devoid of cortical patches. Very soon after this stage (Figure 2C) the bud becomes well occupied with cortical patches, and at no time do I observe, in my strain background, a polarized distribution of cortical patches in the bud.
DISCUSSION
I have shown that the application of focal sectioning in combination with iterative deconvolution can lead to a much more detailed view of yeast cell anatomy than can be achieved through conventional fluorescence microscopy. Even confocal microscopes are unable to obtain thin enough optical sections to be useful for studies of yeast cells (my unpublished observations). In this case I examined the organization of the actin cytoskeleton during the budding cycle with a DeltaVision deconvolution microscope. The yeast actin cytoskeleton is a good test case for use of this instrument because its visualization presents technical difficulties. For example, the small diameter of the actin cables can make them difficult to resolve, and the relatively stronger fluorescence of the cortical patches can make simultaneous photography of both patches and cables problematic. I expect that this technology will allow for much more detailed studies of the organization of yeast and the relative spatial relationships between proteins of interest in vivo, in short, making an experimental organism touted for its genetic power more attractive for cell biological analysis.
My analysis of the yeast actin cytoskeleton pointed out features of its structure that were previously undocumented and/or underappreciated. I show here that the actin cables, like the patches, are located at the cortex of the cell and can be observed to interact with the cortical patches. This suggests that the patches may facilitate anchoring of the cables to the cortex and/or may be the sites of formation of the actin cables. Furthermore, the cables may be involved in the observed motility of the cortical patches (Doyle and Botstein, 1996; Waddle et al., 1996), possibly serving as tracks on which the patches move. If so, movement of the cortical patches on cables might be catalyzed by myosin motors or possibly through treadmilling of actin filaments within the cables. On the other hand, not all actin patches in a given cell are moving at one time, and perhaps attachment to cables serves to restrict the movement of subsets of cortical patches.
These images of the actin cytoskeleton also present a clearer picture of the initial stages of bud formation. At earliest times I can observe bud extrusion through a ring of cortical patches, and at this time the bud does not appear to contain cortical patches. Soon thereafter cortical patches migrate into or are formed in the bud in a nonpolarized distribution and remain nonpolarized, in the bud, until the cytoskeleton reorients for cytokinesis. It has become a common misconception that there is a shift from polarized bud growth to isotropic bud growth, which is accompanied by a similar shift in the arrangement of cortical patches within the bud. It was previously shown that yeast strains with elongated cell morphologies can display a polarization of patches in the bud (Kilmartin and Adams, 1984), but the results presented here clearly show that in the common laboratory strain S288C this is not true. Although it is true that some mutations (such as in CDC4) and pseudohyphal growth lead to hyperpolarization of both actin cortical patches and bud growth (Adams and Pringle, 1984; Cali et al., 1998), perhaps in these situations the cells are activating alternative cell polarity programs such as those used during mating or unipolar growth as opposed to the normal budding program.
Supplementary Material
ACKNOWLEDGMENTS
I thank David Botstein for suggesting the use of a DeltaVision microscope and making the instrument available for this study, Susan Palmieri for technical assistance, and Craig Cummings for help making the Quicktime movies. At the time this work was performed, D.C.A. was a SmithKline Beecham Pharmaceuticals Fellow of the Life Sciences Research Foundation.
Footnotes
REFERENCES
- Adams A, Pringle J. Relationship of actin and tubulin distribution to bud growth in wild-type and morphogenetic-mutant Saccharomyces cerevisiae. J Cell Biol. 1984;98:934–945. doi: 10.1083/jcb.98.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agard DA, Hiraoka Y, Shaw P, Sedat JW. Fluorescence microscopy in three dimensions. Methods Cell Biol. 1989;30:353–377. doi: 10.1016/s0091-679x(08)60986-3. [DOI] [PubMed] [Google Scholar]
- Botstein D, Amberg DC, Huffaker T, Mulholland J, Adams A, Drubin D. The genetics of the yeast cytoskeleton. In: Broach JR, Pringle JR, editors. The Molecular and Cellular Biology of the Yeast Saccharomyces. Cell Cycle and Cell Biology. E.W. Jones, New York: Cold Spring Harbor Laboratory Press; 1997. pp. 1–90. [Google Scholar]
- Cali BM, Doyle TC, Botstein D, Fink GR. Multiple functions of actin during filamentous growth of Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:1873–1889. doi: 10.1091/mbc.9.7.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle T, Botstein D. Movement of yeast cortical actin cytoskeleton visualized in vivo. Proc Natl Acad Sci USA. 1996;93:3886–3891. doi: 10.1073/pnas.93.9.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin J, Adams A. Structural rearrangements of tubulin and actin during the cell cycle of Saccharomyces cerevisiae. J Cell Biol. 1984;98:922–933. doi: 10.1083/jcb.98.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulholland J, Preuss D, Moon A, Wong A, Drubin D, Botstein D. Ultrastructure of the yeast actin cytoskeleton and its association with the plasma membrane. J Cell Biol. 1994;125:381–391. doi: 10.1083/jcb.125.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scalettar BA, Swedlow JR, Sedat JW, Agard DA. Dispersion, aberration and deconvolution in multi-wavelength fluorescence images. J Microsc. 1996;182:50–60. doi: 10.1046/j.1365-2818.1996.122402.x. [DOI] [PubMed] [Google Scholar]
- Waddle JA, Karpova TS, Waterston RH, Cooper JA. Movement of cortical actin patches in yeast. J Cell Biol. 1996;132:861–870. doi: 10.1083/jcb.132.5.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.