Abstract
Alternative splicing is an important source of protein diversity, and is an established but not yet fully understood mechanism for gene regulation in higher eukaryotes. Its regulation is governed by a variety of mechanisms, including variation in the expression levels of splicing factors engaged in spliceosome formation. SRp55 is one of the most ubiquitous splicing factors and one that can be up-regulated by DNA damage in the absence of p53, and we had previously found that depletion of its activity increased resistance to DNA damage in p53-dependant manner. To assess its influence on the splicing patterns of genes involved in apoptosis, we performed splice-specific microarray analysis of cells treated with siRNA specific for this gene. This analysis, backed by RT-PCR verification, identified three genes, KSR1, ZAK and mda7/IL24, which are sensitive to SRp55 depletion. We also analyzed the splice patterns of apoptosis-related genes in p53-deficient U2OS cells following treatment with the genotoxic drug mitomycin C. This analysis revealed that DNA damage resulted in changes in splicing activity that modified the splicing pattern of Fas, a key proapoptotic, p53-inducible death receptor. Interestingly, this modification led to an enrichment of the antiapoptotic soluble Fas isoform, and this secreted isoform was detected in the media surrounding cells subjected to DNA damage. These findings show that modulation of splicing activity in p53-deficient cells during the early response to sub-lethal DNA damage results in a change in the splicing of target genes, thus modifying the cellular response to genotoxic agents.
Keywords: Alternative splicing, Splice-specific microarray, p53, Fas, mda7/IL24
1. Introduction
Genome-wide analyses of human mRNA transcripts have shown that more than two thirds of multiexon genes have alternatively spliced isoforms (Johnson et al., 2003), and it is generally acknowledged that alternative splicing (AS) is a constitutive part of gene regulation in higher eukaryotes. However, in contrast to the well-established mechanisms of transcriptional regulation, molecular mechanisms controlling AS events are poorly understood.
AS patterns are known to be highly tissue- and cell-type specific, implying that splicing regulation is under tight control. Splice site selection is controlled by formation of the spliceosome, a riboprotein complex that consists of a diverse array of factors including members of the SR and hnRNP families of proteins (Matlin et al., 2005). The decision by which the splice site is chosen is believed to be regulated by combinatorial control of the effective concentration and activity of splicing factors assembled within the spliceosome (Stamm, 2002). Alterations in splicing can therefore be achieved by changing the expression levels, intracellular localization, and posttranslational modifications of the constituent splicing factors.
The importance of AS in regulating gene expression is demonstrated by a growing number of diseases caused by abnormal splicing patterns, including such well-studied pathologies as cystic fibrosis and spinal muscular atrophy (for review, see (Novoyatleva et al., 2006)). Studies of AS in cancer cells have also revealed that regulation of splicing is significantly altered during tumorigenesis, and expression of certain cancer-specific isoforms correlates with cancer progression (reviewed by (Skotheim and Nees, 2007)). Although the connection between disregulation of splicing activity and tumorigenesis is clearly established, the mechanisms governing this process have not yet been identified.
One of the reasons for our poor understanding of AS regulation is the absence of a reliable model system that is able to modify splicing in response to external stimuli. There are a few examples showing that cellular stress, including temperature, osmotic shock and DNA damage by UVC irradiation, can change the splicing patterns of certain genes (Stamm, 2002). However, the precise regulatory mechanism of these splicing modifications is not known.
Recently, we found that immediately following the application of a sub-lethal concentration of a DNA damaging agent, p53-deficient cells activate AS activity by transiently increasing expression of a number of SR proteins, including SRp55 (Filippov et al., 2007). Discovery of this transient splicing activation prompted us to search for possible gene targets of this increased alternative splicing. Initially, we tested genes known for their sensitivity to splicing factors that belong to the SR protein family. This approach allowed us to identify the CD44 receptor as a target of increased splicing activity in p53 deficient cells, and to demonstrate that increased expression of SRp55 was necessary for its AS. To perform a more thorough and unbiased search for the targets of changed splicing activity following DNA damage, we decided to analyze changes in RNA splicing using a splicing microarray approach in two complementary comparisons. In the first, we compared the splice patterns of apoptosis-related genes in cells with and without a knock-down of SRp55 splicing factor activity, as SRp55 had been identified as a factor whose expression and activity increased following DNA damage. Microarray analysis followed by RT-PCR validation revealed that at least three apoptosis-related genes require SRp55 activity for proper splicing. In the second, we compared the splice patterns of apoptosis-related genes in cells treated and not treated with the DNA damaging agent mitomycin C. In this set of experiments, we found that p53-deficient cells modulate their response to DNA damage by shifting the splice pattern of the Fas gene such that the soluble, pro-survival variant is favored over the membrane-bound, pro-apoptotic isoform. Together, these observations demonstrate that cells can respond to external stressors by directly regulating the expression of specific splicing factors, thereby changing the splicing profile of target genes.
2. Materials and Methods
2.1. Cell lines and cell culture
U2OS cells, derived from a human osteosarcoma, were obtained from the ATCC (Manassas, VA). They were cultured in McCoy’s 5A medium (Invitrogen, Carlsbad, CA) supplemented to contain 10% fetal bovine serum (Invitrogen, Carlsbad, CA), penicillin (100 µg/ml) and streptomycin (100 µg/ml) (Sigma, St. Louis, MO). Construction of the pHA-E6S and pHA-E6AS plasmids, which respectively contain either the sense or the antisense versions of HA-tagged E6 under the control of the CMV promoter, and establishment of the U2OSE64b and U2OSE6AS cell lines by stable transfection with these plasmids have been described previously (Filippova et al., 2002). Construction and characterization of the Tet-Off U2OSE6tet24 cell line, in which E6 expression is regulated by the presence or absence of doxycycline, is described in (Filippova et al., 2004).
2.2 Cell Survival and p53 analysis
Cell survival analysis was performed using the MTT test (Filippova et al., 2002), and the p53 ELISA was conducted as described previously (Filippova and Duerksen-Hughes, 2003).
2.3. Semi-quantitative RT-PCR
Reverse transcriptase – polymerase chain reaction (RT-PCR) analysis was used to estimate transcript levels in total RNA samples isolated from cell cultures. 10 µg of total RNA was used as a template for cDNA synthesis with SuperScript III reverse transcriptase and an oligo(dT) primer according to the manufacturer’s instructions (Invitrogene, Carlsbad, CA). The cDNA obtained was normalized by PCR with primers specific to cofilin1 (CFL1). Serial dilutions of the normalized cDNA samples were used to find the linear range of amplification for gene-specific DNA fragments. PCR was performed at 94°C for 30 s, at 57°C for 30 s, and at 72°C for 1 min for 35 cycles using Taq DNA polymerase and the recommended protocol (NEB, Beverly, MA). The following sets of primers were used for RT-PCR analysis to detect expression of the ATF3 gene: ATGATGCTTCAACACCCAGGC and TTAGCTCTGCAATGTTCCTTC; CASP gene: GAAGAAAGAGCTGAACATTCTGAA and GAGATGATGGAAAGCAGTGAAT; CFL1 gene; CCTTCCCAAACTGCTTTTGAT and CTGGTCCTGCTTCCATGAGTA; Fas isoforms: GACATGGCTTAGAAGTGGAAA and TTAGTGTCATGACTCCAGCAA; KSR1-1 isoform: CTGGAGAAACTTCCCAAGCTGAAC and AAAGAAGTAGCCTCGAGAGAGACG ; KSR1-2 isoform: CTGGAGAAACTTCCCAAGCTGAAC and CCAACAGTCTGAATCTAGGACGTG ; CUTL1 gene: TAACTCTTACAGCTTTGCCTTG and GGAATCCAAACTAGTGTGTTTAGA; ZAK (α isoform): CTTTAAGGAGCAGGAGCTTAAA and ATCTGCATCCCAGAATGCAT ; ZAK (β isoform): CTTTAAGGAGCAGGAGCTTAAA and TGAGTCCTTAATTAGTGGTGGG; SFRS6 gene: CAGGTCGAGTTCCAGAGATTA and TCAAACTGCAATTTCAACTCA; and mda7/IL24 gene: CAGCCCTCAAGCATCACTTACA and TGCTCTCCGGAATAGCAGAAAC.
2.4. siRNA inhibition of SFRS6 gene expression
Cells were transfected with pre-designed small interfering (si) RNAs for the human SFRS6 gene obtained from Santa Cruz Biotechnology (sc-4418), or with control siRNA (Santa Cruz Biotechnology, Santa Cruz, CA) using the siRNA transfection kit as recommended by the manufacturer (Santa Cruz Biotechnology, Santa Cruz, CA). To obtain different levels of silencing of the SFRS6 gene, two concentrations of siRNA were used: 0.5 µg and 0.25 µg per 6-well plate. After 48 hrs of incubation, the silencing effect was evaluated by RT-PCR analysis as described above.
2.5. RNA extraction and splice array data collection
Total RNA was extracted using Trizol Reagent (Invitrogen, Carlsbad, CA) and additionally purified using the RNeasy kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. RNA samples for the siRNA inhibtion experiment were analyzed by RT-PCR to verify successful silencing of SFRS6 gene expression prior to splice array analysis. The splice array analysis was performed at the facilities of ExonHit Therapeutics (ExonHit Therapeutics, Gaitherburg, MD) using the Human Apoptosis SpliceArrays and employing a technical dye-swap.
2.6. Splice array data analysis
Initial data analyses were performed using the SpliceAnalysis Tool supplied by ExonHit Therapeutics as well as with Bioconductor. The Bioconductor version used included the limmaGUI software package for differential expression analysis as developed for two-color microarrays (Wettenhall and Smyth, 2004). The splice events that contained probes with the highest statistical B values were further analyzed to determine whether the ratios of probe hybridization outputs for probes specific to a splice variant were in agreement with the changes of probes specific for the reference RNA transcript, as previously described (Heinzen et al., 2007).
2.7. Collection of the cell media, soluble Fas ELISA, immunoprecipitation and Western blot analysis
Confluent cells were exposed to 2 µg/ml mitomycin C (Roche, Indianapolis, IN) for 5 hrs, then the medium was collected and cleared by centrifugation. The concentration of soluble Fas in media from treated and untreated cell cultures was estimated using a sCD95(APO1/Fas) ELISA kit (Cell Sciences, Canton, Massachusetts). To perform immunoprecipitations, the media were first concentrated from 4 ml to 1 ml using Centricon centrifugal filtration through a YM-3 membrane (Millipore, Billerica, MA). Concentrated media were incubated with agarose-conjugated Fas monoclonal antibodies (sc-8009, Santa Cruz Biotechnology, Santa Cruz, CA) for 16 hrs at 4°C and washed 3 times with the washing buffer (50 mM Tris-HCl, pH 7.5, 500 mM NaCl, 01% NP-40, 1mM EDTA, 1 mM DTT). The precipitates were fractionated by 10% SDS-PAGE and immunoblotting was performed as described previously (Filippova et al., 2005). Fas protein was detected using rabbit polyclonal Fas antibodies (sc-714, Santa Cruz Biotechnology, Santa Cruz, CA).
3. Results
3.1. The SRp55 contribution to the DNA damage response is p53-dependant
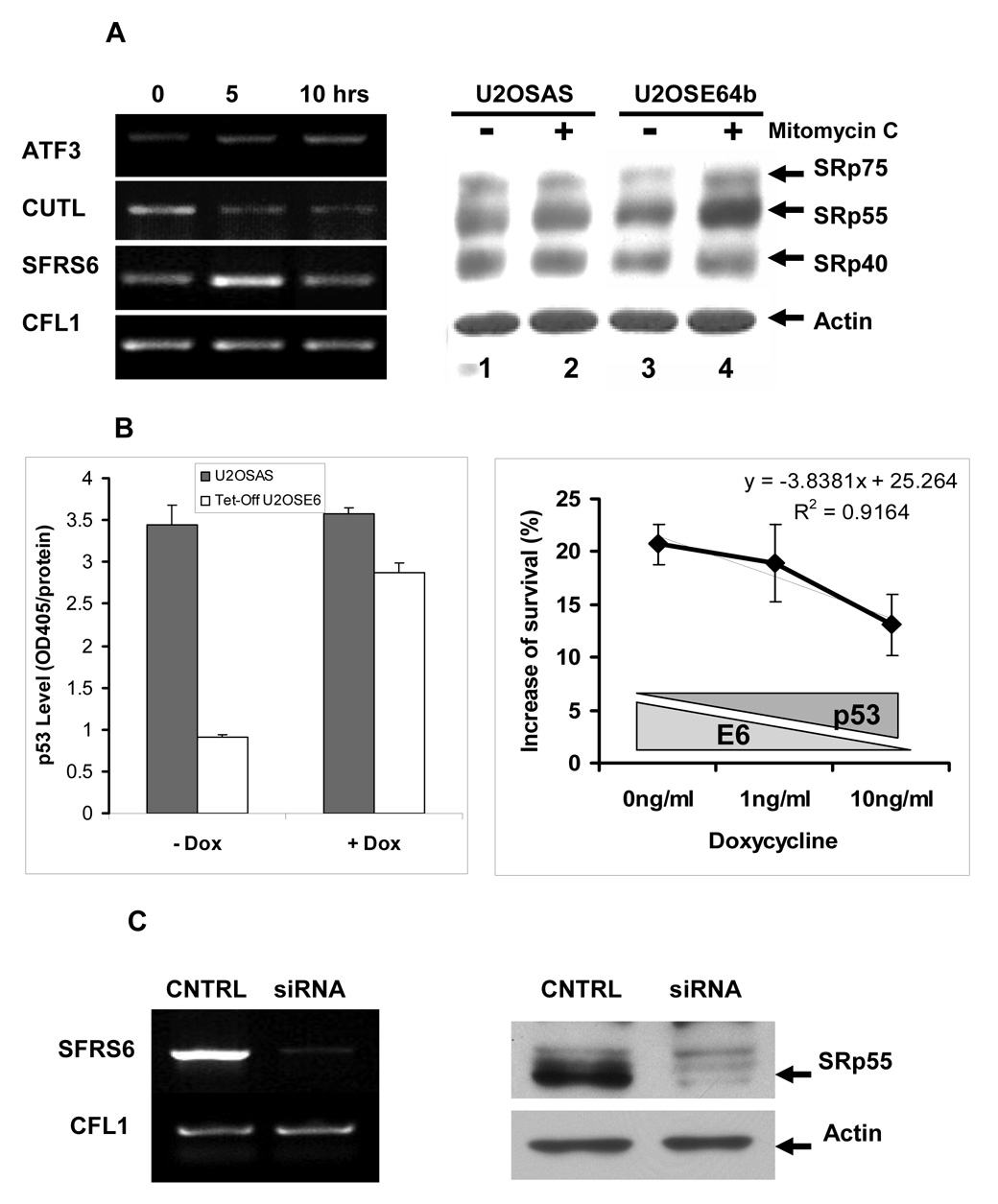
As we have previously shown, DNA damage activates AS activity in the HPV16 E6-expressing U2OS cell line U2OSE64b, with this activity peaking 5 hrs following drug treatment. Furthermore, we found that this activation depends on p53, which is efficiently degraded in the presence of E6 (Filippov et al., 2007). The activation of AS activity was due, at least in part, to an up-regulation in transcription of the SFRS6 gene, leading to an increase in protein expression of the SRp55 splicing factor encoded by that gene. Semi-quantitative RT-PCR showed that the increase in SFRS6 gene expression is transient during DNA damage caused by mitomycin C treatment, such that after the observed increase at 5 hrs, expression returns to its basal level by 10 hrs. This is in contrast to the continued up-regulation of the transcriptional factor ATF3, as well as the down-regulation of the transcriptional factor CUTL in the same RNA samples (Figure 1A, left panel). The up-regulation of SRp55 following DNA damage in cells lacking p53 can also be demonstrated at the protein level (Figure 1A, right panel).
Figure 1. Characterization of RNA samples subjected to SpliceArray™ analysis and effect of SRp55 silencing on cell survival.
A. Left panel: Level of ATF3, CUTL, SFRS6, and CFL1 transcripts in U2OSE64b cells treated with 2 µg/ml mitomycin C for 5 and 10 hrs as determined by RT-PCR analysis. Right panel: The SRp55 protein encoded by the SFRS6 gene has elevated expression levels in U2OSE64b cells after incubation with mitomycin C (2 µg/ml) for 5 hrs, but not in the control U2OSAS cells (right panel). Lanes 1 and 3 show lysates of untreated U2OSAS and U2OSE64b cells, and lanes 2 and 4 show lysates of U2OSAS and U2OSE64b cells treated with 2 µg/ml mitomycin C for 5 hrs, respectively. The levels of splicing factors were determined by immunoblotting with monoclonal antibody 1H4 (ATCC, Manassas, VA). β-Actin antibodies were used for re-blotting to verify uniformity of sample loading. B. Left panel: Depletion of doxycycline (−Dox) in the medium effectively decreases p53 levels in the Tet-Off U2OSE6tet24 cell line but not in control U2OSAS cells. U2OSAS or Tet-Off U2OSE6 cells, grown in the presence or absence of 10 ng/ml Dox, were lysed and the lysates analyzed for p53 by ELISA. Right panel: Sensitivity of SRp55-depleted cells to mitomycin C treatment depends on the level of p53. U2OStetE624 cells, maintained in the presence of three concentrations of doxycycline, were transfected with SRp55 or scrambled siRNAs, then subjected to DNA damage by treatment with 4 µg/ml mitomycin C for 24 hrs. Viable cells were quantified using the MTT assay, and the differences between SRp55 silenced and control cells were compared by subtracting the number of viable cells in control scrambled siRNA samples from the number of surviving cells transfected with SRp55 siRNA. Means from triplicate measurements are shown, with the error bars representing the standard deviation. C. Left panel: Treatment of U2OS cells with SFRS6-specific siRNA effectively decreases the SFRS6 mRNA level. The level of SFRS6 mRNA was determined by RT-PCR and normalized against expression levels of the CFL1 gene. Right panel: Inhibition by siRNA leads to a decrease in the level of the SRp55 protein. Immunoblot of protein lysates isolated from control U2OS cells (CNTRL) and U2OS cells transfected with SFRS6 siRNA (siRNA). The β-actin antibodies were used to verify uniformity of sample loading.
These observations, as well as our previous observation that knock-down of SFRS6 (encoding SRp55) by siRNA in cells lacking p53 led to increased viability following DNA damage (Filippov et al., 2007), led to the prediction that the ability of SRp55 to affect cell viability following DNA damage would be dependant on the presence, and possibly the amount, of p53. To test this prediction, we employed an approach utilizing siRNA-mediated SRp55 silencing (Filippov et al., 2007) in cells where the levels of E6 expression could be controlled by changing the level of doxycycline present in the media, the Tet-Off U2OSE6 cells (Filippova et al., 2004). To confirm that the level of biologically active E6 could indeed be controlled by the level of doxycycline, we compared p53 levels by ELISA in mitomycin C-treated control U2OSAS cells that do not express E6, as well as in Tet-Off U2OSE6 cells either expressing E6 at a high level (−Dox) or when E6 expression is inhibited by the antibiotic (+Dox). As expected, a high level of E6 expression caused rapid p53 degradation, while repression of E6 expression by Dox almost completely restored p53 to the level observed in the control U2OSAS cells (Figure 1B, left panel).
We then asked whether manipulating the level of p53 in Tet-Off U2OSE6 cells would affect the ability of SRp55 to modulate biological activities such as cell survival following DNA damage. To address this question, Tet-Off U2OSE6 cells were transfected with either SRp55-specific or control scrambled siRNA and incubated in the presence of three different concentrations of Dox. After 24 hr treatment with mitomycin C, these cells were evaluated for viability using the MTT assay. Comparison of survival rates of Tet-Off U2OSE6 cells with depleted SRp55 expression due to siRNA inhibition versus the control cells showed a clear dependency on the concentration of Dox in medium, such that the ability of siRNA directed against SFRS6 to impact survival was greatest in the absence of p53 (Fig. 1B, right panel). This demonstrates that in the absence of p53, the activity of at least one splicing factor, SRp55, is capable of interfering with the cellular response to mitomycin C treatment. The most plausible mechanism for this interference may be an alteration in the splicing patterns of target genes.
3.2. Splice array analysis identified changes in splicing events
To identify candidate target genes, we compared splicing events in apoptosis-related genes in two pairs of RNA samples using ExonHit SpliceArray™ microarrays. In the first analysis, we monitored changes in the splicing of apoptosis-related genes in U2OS cells that were sensitive to depletion of SRp55 splicing factor activity. U2OS cells were transfected with siRNA directed against SFRS6 mRNA, encoding the SRp55 protein, or with control scrambled siRNA oligonucleotides. These cells were then examined for the resulting down-regulation of the SFRS6 message and SRp55 protein (Figure 1C), and analyzed for their expression of splice variants of apoptosis-related human genes using ExonHit splice arrays. In the second comparison, we compared RNA isolated from U2OSE64b cells treated with 2 µg/ml mitomycin C for 5 hrs with RNA isolated from untreated cells. This analysis allowed us to identify candidate genes with splice patterns that could be affected by increased AS activity induced by DNA damage.
The raw hybridization data were then processed using tools developed by Bioconductor for the analysis of two-color microarrays in order to yield lists of probes with the highest statistical B values. These B-statistics evaluate the probability that sequences hybridizing to a particular probe are differentially expressed. Since for each of the two experiments, siRNA silencing and mitomycin C treatment, only one technical repeat was subjected to splice array hybridization, the B values for differential hybridization of array probes were considerably low, and were considered only as an indication that hybridization to a probe might reflect splicing changes. Next, we analyzed selected splicing events in which probes with the highest B values were involved. Each splicing event is represented on the array by up to six probes that can quantify the presence of purely exonic sequences as well as exon-exon junctions in analyzed RNA samples. In situations where an actual shift in the splicing patterns have occurred, an increase in the binding of probes for a particular splice variant should be accompanied by an increase in the ratio for the binding of its specific probe(s), as well as a decrease in the binding for probes specific to the reference transcript (Heinzen et al., 2007). Using this rationale, we selected four candidate splicing events, designed primers specific for these events and performed subsequent RT-PCR analysis.
3.3. Depletion of SRp55 levels change the isoform ratios for the KSR1 and ZAK kinases
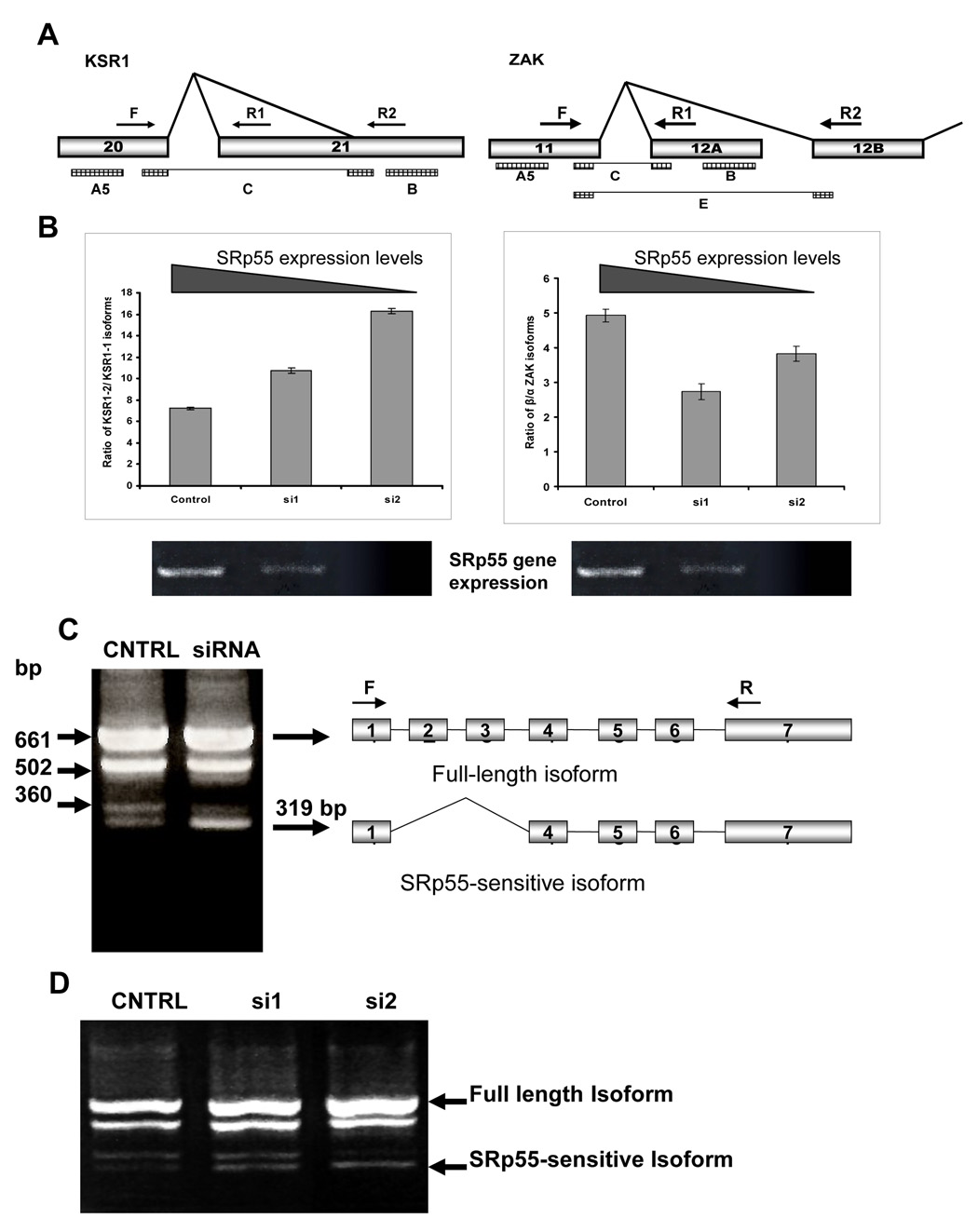
The effectiveness of the approach we used for verification of the splice array data is supported by the analysis of two splicing events. In the first splicing event, microarray hybridization showed coordinated changes in the hybridization of three probes specific for AS of the last exon of the KSR1 gene, encoding the Kinase Suppressor of Ras1. As shown on the scheme, these three probes, A5, B and C, monitor splicing of an alternative acceptor site located within exon 21 of the KSR1 gene (Figure 2A, left panel). While the 5’ probe A5 and the 3’ probe B should be present in both isoforms, probe C represents the junction of exon 20 with the alternative acceptor site of exon 21.
Figure 2. Splicing patterns of KSR1, ZAK and mda7/IL24 genes are sensitive to SRp55 expression.
A. Schemes of the regions of the KSR1 and ZAK gene structures involved in the tested AS events. Arrows F, R1 and R2 show the primers used for RT-PCR analyses for each of these genes. Rectangles A5, B, C, and E show localization of the SpliceArray™ probes. B. Left panel: Increased expression of the KSR1 isoform with truncated exon 21 (KSR1-2) in SRp55-depleted U2OS cells. The graph shows quantification of the ratio between KSR1 isoforms, as recognized by primers F+R1 (KSR1-1) and F+R2 (KSR1-2) in U2OS cells with different levels of SRp55 expression. Right panel: Changes in the splicing of the ZAK gene in SRp55-depleted U2OS cells. The graph shows quantification of the ratio between the two ZAK isoforms, as recognized by primers F+R1 (β isoform) and F+R2 (α isoform) in U2OS cells with different levels of SRp55 expression. Two lower panels show RT-PCR analysis of SFRS6 expression in these RNA samples isolated from U2OS cells treated with scrambled siRNA (Control), 0.25 µg or 0.5 µg SRFS6 specific siRNA (si1 and si2, respectively). C. Silencing of SRp55 expression by specific siRNA up-regulates the abundance of the isoform represented by a 319 bp band in RT-PCR analysis using the primers designated by arrows F and R. D. The amount of the SRp55-sensitive isoform is proportional to the levels of silencing of SRp55. U2OS cells were treated with scrambled siRNA (CNTRL), 0.25 µg or 0.5 µg SRFS6 specific siRNA (si1 and si2, respectively) to decrease SRp55 expression gradually. Isolated RNA was subjected to RT-PCR analysis.
The second example shows AS of the ZAK gene, a member of the MAPKKK family of signal transduction kinases. In one alternative variant of the gene, exon 11 is spliced to alternative exon 12A, which can be monitored by exon specific probe B and junction-specific probe C (Figure 2A, right panel). This splice variant is a truncated version of the gene that produces its β isoform (Gotoh et al., 2001). Probe E for this splice event is specific for the junction of exon 11 to exon 12B that represents the α isoform of the gene, while probe A5 is common for both of the isoforms (Figure 2A, right panel).
To examine these two splicing events, qRT-PCR analysis was performed using primers able to distinguish between the isoforms of the KSR1 and ZAK genes affected by these splicing events. The levels of these isoforms were measured in samples of RNA isolated from cells transfected with scrambled siRNA (Control), 0.25 µg of siRNA specific for SRp55 (si1), and 0.5 µg of siRNA specific for SRp55 (si2). This allowed us to manipulate the degree of gene silencing, as shown in Figure 2B (lower panels). Calculation of the ratios between the two KSR1 isoforms in cells with different levels of SRp55 silencing showed that reduced activity of the splicing factor led to a dose-dependent increase of the KSR1 isoform with the truncated exon 21 (Figure 2B, left panel). With regards to the ZAK gene, the qRT-PCR data showed that its expression in control cells is predominantly of the β isoform. Quantification of the ZAK isoforms in the silenced and untreated RNA samples did show some increase in the α splice variant in SRp55 depleted cells (Figure 2B, right panel). However, we did not observe a strong correlation between the degree of SRp55 silencing and changes in the isoform ratio. This observation may be an indication that regulation of this splicing event is quite complex and that other factors may be involved.
3.4. Inclusion of exons 2 and 3 of mRNA from mda7/IL24 requires SRp55 activity
The mda7/IL24 gene encodes a cytokine in the IL10 family that has been demonstrated to be a potent tumor suppressor (Gupta et al., 2006). This gene consists of seven exons and currently has two isoforms deposited in the RefSeq database. Two microarray probes for this gene showed a significantly changed signal ratio in the siSRp55 experiment. RT-PCR expression analysis of this gene in U2OS cells using a primer set that covers the entire transcript showed that its pre-mRNA undergoes extensive and varied splicing in U2OS cells, yielding at least 4 distinguishable isoforms (Figure 2C). Comparison of the splicing patterns of mda7/IL24 transcripts in control and siRNA treated cells confirmed the splice microarray data, indicating that regulation of splicing for at least one isoform of this gene is sensitive to silencing of the SRp55 splicing factor. Depletion of SRp55 splicing activity resulted in a substantial increase of the isoform represented in RT-PCR by a 319 bp fragment (Figure 2C). Moreover, we have found that the increase in this isoform is proportional to the decrease in the level of SRp55 in treated cells (Figure 2D). Sequencing of this fragment revealed that this is the annotated isoform 2 that lacks exons 2 and 3.
3.5 Mitomycin C treatment of E6 expressing cells induces expression of soluble Fas
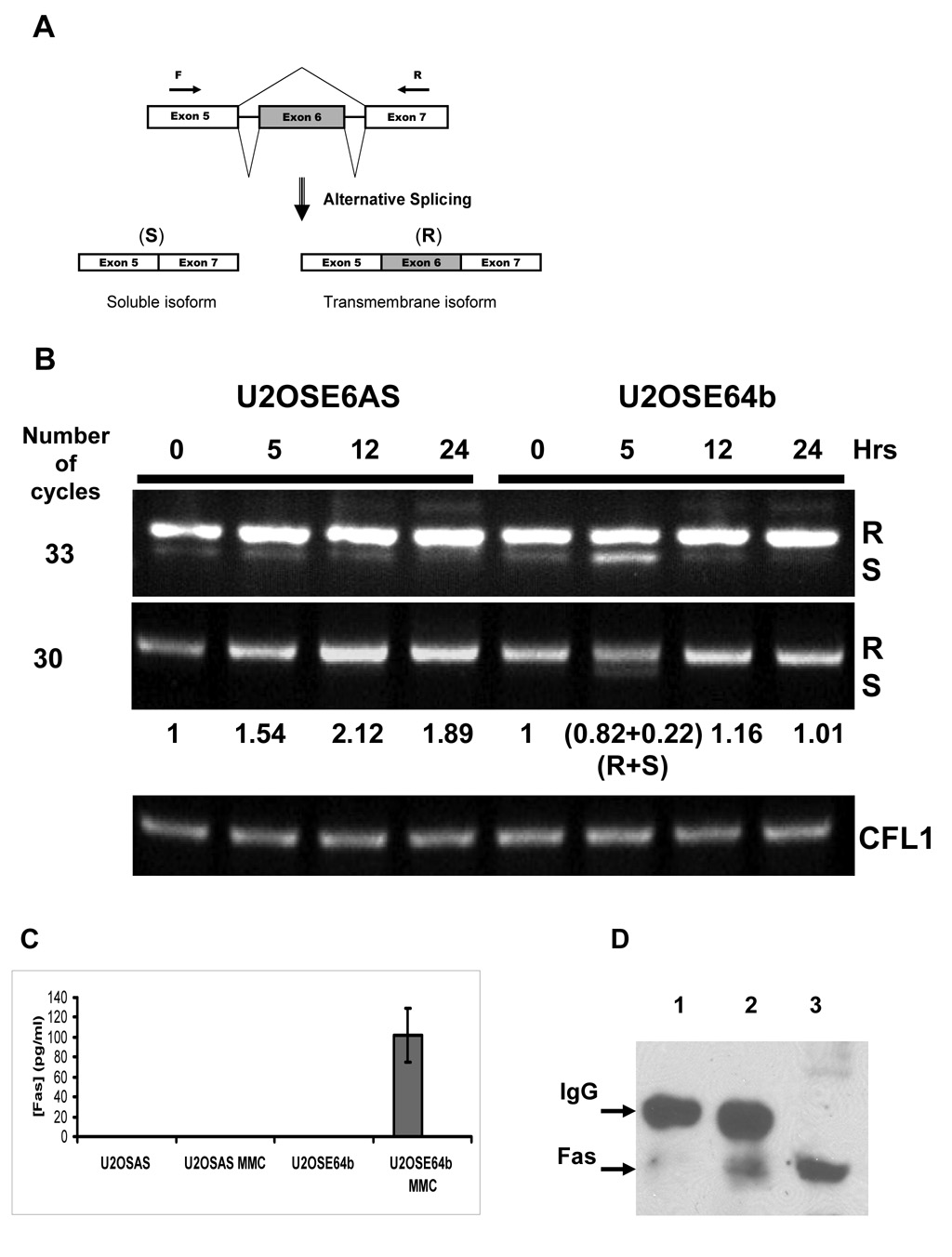
The microarray hybridization of RNA samples isolated from U2OSE64b cells treated with 2 µg/ml mitomycin C for 5 hours produced considerably fewer probes with significant changes in the signal probe ratio as compared to the siRNA silencing experiment. One of the probes from this list belonged to a sequence specific to the Fas receptor. Fas (CD95) receptor is a member of the TNF-Receptor superfamily that is activated by binding to Fas ligand. It then transduces a pro-apoptotic signal into the cell by formation of a death-inducing signaling complex (DISC) (Thorburn, 2004). To monitor splicing of the Fas gene, we used primers designed to distinguish between the soluble and membrane-bound forms of the message (Figure 3A). In addition to the expected membrane-bound form, we found detectable amounts of the soluble Fas (sFas) isoform in both untreated U2OSE6AS and U2OSE64b cells under conditions where a high number of amplification cycles were used (Figure 3B, upper panel). These results also demonstrated that there is an increased amount of the sFas isoform following five hours of mitomycin C treatment in the U2OSE64b cells. To further explore this, semi-quantitative RT-PCR using fewer replication cycles was performed (Figure 3B, lower panel). In this experiment, we were able to detect this transcript variant only following 5 hrs of treatment, and only in the U2OSE64b cells. Finally, following 5 hrs of treatment of the U2OSE64b cells, the combined density of both bands, representing the membrane and soluble variants of the gene, was approximately equal (1.04) to the density of the Fas gene in untreated cells, and its expression was split between these isoform in a 4:1 ratio (Figure 3B). This suggests that the gene itself has not been up-regulated, although the regulation of its splicing pattern has changed.
Figure 3. Mitomycin C treatment induces up-regulation of Fas gene expression in U2OSE6AS cells and changes its splicing pattern in U2OSE64b cells.
A. Scheme of alternative splicing of the Fas gene. Skipping exon 6, which codes for the transmembrane domain of the receptor, leads to expression of the soluble isoform (S), while retention of the exon produces membrane bound receptor (R). Two arrows (F and R) show the primers used to distinguish between these isoforms. B. Time course of Fas gene expression during mitomycin C (2 µg/ml) treatment by RT-PCR analysis. The CFL1 gene was used as a control. The ratio of the RT-PCR Fas R isoform bands to control (0 hrs of drug treatment) is shown between the panels, with the density of the S isoform for the U2OSE64b cells after 5 hrs of treatment noted separately. C. Detection of soluble Fas in cell culture media. The concentration of soluble Fas protein secreted into the media was estimated by ELISA in media collected from U2OSE6AS and U2OSE64b cells untreated and treated with mitomycin C (MMC) (2 µg/ml) for 5 hrs. Means from triplicate measurements are shown, with the error bars representing the standard deviation. D. Immunoprecipitation analysis of media collected from treated and untreated U2OSE64b cell cultures. Lane 1 – Fas immunoprecipitation of medium from untreated U2OSE64b cells. Lane 2 - Fas immunoprecipitation of medium from U2OSE64b cells treated with 2 µg/ml mitomycin C for 5 hrs. Lane 3 – U2OSE64b lysate.
This increase in message coding for the sFas isoform was accompanied by the secretion of this form of the protein into the cell medium, as detected by ELISA. Both the p53-expressing U2OSE6AS control cells and the p53-deficient U2OSE64b cells were treated with 2 µg/ml mitomycin C for 5 hours. The media were collected and subjected to ELISA analysis for the presence of sFas protein. Media from the control treated and untreated cells as well as the untreated U2OSE64b cells did not contain detectable amounts of sFas protein in any of the analyzed samples. The only samples that contained detectable levels (the minimum detectable dose is 47 pg/ml) of the protein were from media collected from mitomycin C treated U2OSE64b cells (Figure 3C). To confirm the release of sFas into the medium by mitomycin C treated U2OSE64b cells, we immunoprecipitated this protein from the collected media using agarose conjugated Fas antibodies, and then performed an immunoblot analysis. This analysis showed that while medium from untreated cells contained a trace amount of Fas, the media collected from the drug-treated cells contained a much higher level of this isoform of this protein (Figure 3D).
4. Discussion
Our discovery that AS activity is activated in p53-deficient cells treated with a sub-lethal dose of a DNA damaging agent prompted us to search for genes that could be affected by changes in splicing regulation. A search for possible targets among genes that were known to be sensitive to the splicing activity of SR proteins allowed us to identify such a target gene of this splicing activation, the CD44 receptor (Filippov et al., 2007). Identification of such a target gene encouraged us to search for other possible targets using a more systematic approach. Currently, genome-wide analysis of AS using microarray technology is considered the most comprehensive approach for identification and quantification of splice events. Splicing microarray technology enables the acquisition of a high resolution, exon-level snapshot of the population of RNA transcripts. While it is potentially a very powerful tool for transcriptional studies, splice microarray technology is still under development and a number of challenges remain.
In this report, we describe our use of splice microarray technology to analyze splice events in two cellular systems. In the first system, splicing changes were analyzed in cells where the splicing factor SRp55 had been depleted by siRNA. In the second system, we identified splicing changes that occurred in p53-deficient cells that were treated with a sub-lethal dose of a DNA damaging agent. The results obtained in both experiments provided partial, rather than complete data on global splice changes for several reasons. First, the number of genes tested was only 609, as the SpliceArray chips we used covered a pool that contained only apoptosis-related genes. Secondly, only one technical repeat was used for each experiment, which reduced the quality of statistical data obtained. Third, the tools used for statistical processing of hybridization data were somewhat limited and are still under development. Further development of microarray technology will certainly provide a more comprehensive picture of splicing changes in both tested systems. However, despite these limitations, we were able to identify several splicing targets using this approach.
SRp55 belongs to the family of arginine/serine-rich splicing factors and is involved in both constitutive and alternative splicing. Though its role in constitutive splicing is believed to be redundant, its function was found to be essential for Drosophila development, since its mutation is lethal (Hoffman and Lis, 2000). A global analysis of all annotated splicing events that generate the 8315 alternatively spliced RNAs in Drosophila revealed that silencing of SRp55 by siRNA directly affected 107 events (Blanchette et al., 2005). This confirmed that while it may be redundant for constitutive splicing, Drosophila SRp55 is required for regulation of several splicing events. Our observation that depletion of SRp55 activity increases cellular resistance to DNA damage in a p53-dependant manner (Figure 1B, right panel) suggests that in humans this splicing factor may be required for splicing regulation of some genes engaged in the p53-independent response to DNA damage. To further examine this possibility, we compared normal gene splice patterns in human osteosarcoma U2OS cells with the patterns seen in cells depleted of SRp55 activity by siRNA. Analysis of the microarray data, coupled with RT-PCR validation, showed that of the 609 genes present on the chip, at least three: KSR1, ZAK and mda7/IL24, are sensitive to SRp55 activity during their splicing.
Microarray data pointed to possible SRp55 sensitive modifications in the splicing of two kinases: KSR1 and ZAK (Figure 2A). RT-PCR analysis of these events presented compelling data that SRp55 participates in regulation of splicing of the last exon of the KSR1 gene. Depletion of this splicing factor caused a decrease in utilization of the full-length exon 21, making the splice acceptor site located within exon 21 more preferable (Figure 2B). RT-PCR data also showed that depletion of SRp55 caused less dramatic changes in the regulation of the ratio between the α and β ZAK isoforms. Though siRNA silencing led to an increase of the α isoform by as much as 2-fold, the absence of a direct correlation between SRp55 activity and the isoform ratio may be due to the involvement of other splicing factors in the regulation of this splicing event. Interestingly, while alteration of KSR1 splicing affects only the 3’-UTR region of the gene, changes in ZAK splicing directly affect the coding region, potentially modifying one or more functions of the protein. RT-PCR analysis also confirmed SRp55 involvement in splicing of the mda7/IL24 gene (Figure 2C, D). In this case, splicing factor depletion led to the generation of isoform 2, in which exons 2 and 3 have been excluded. Identification of these SRp55 sensitive splicing events indicates that, as in the case of its Drosophila homologue, human SRp55 is required for certain splicing events, and other splicing factors cannot completely substitute for its activity.
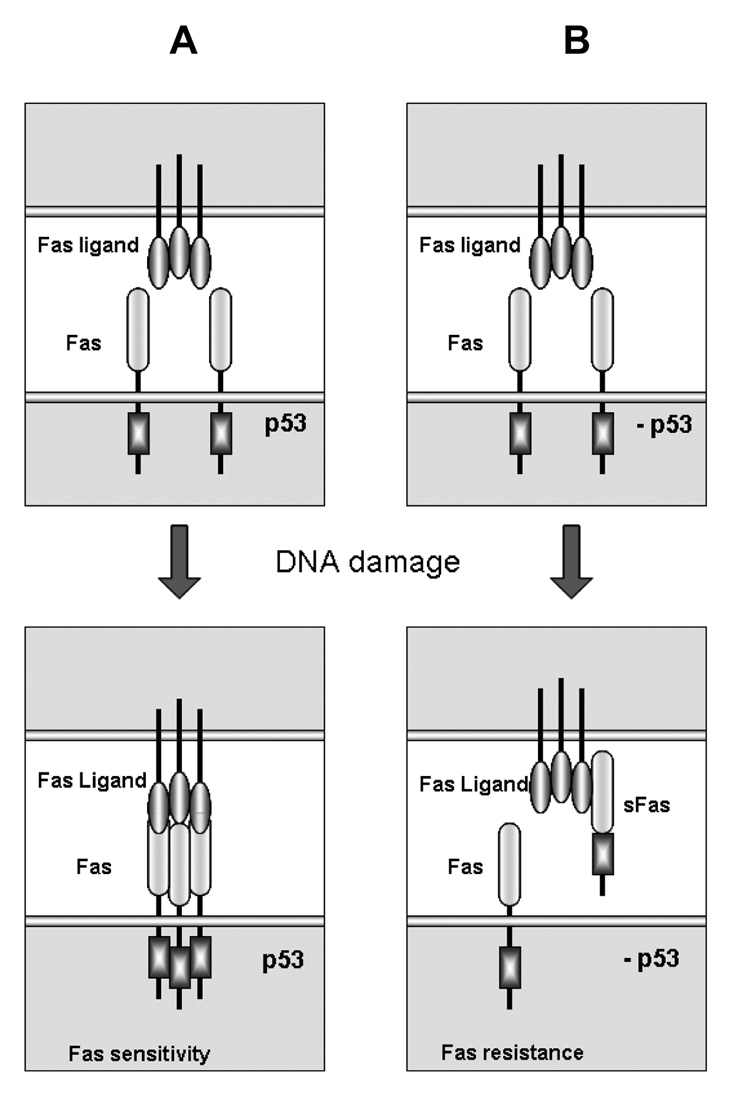
Microarray analysis of splicing in p53-deficient U2OSE64b cells treated with the DNA damaging drug mitomycin C identified changes in splicing of the Fas gene. The Fas receptor is one of the key inducers of apoptosis in response to a variety of stresses, including DNA damage (Sharma et al., 2000). It encodes a transmembrane member of the TNF receptor superfamily that effectively mediates apoptosis after binding to its ligand (FasL) (Krammer, 2000). Fas can be up-regulated by p53, and it is generally believed that this p53-dependent induction during the DNA damage response sensitizes cells to Fas mediated apoptosis and toxicity (Friesen et al., 1996). Consistent with this idea, we also observed an up-regulation of Fas in p53-expressing U2OSE6AS cells (Figure 3B). However, in the absence of p53, rather than an overall up-regulation of Fas expression, we observed increased expression of the soluble Fas isoform (Figure 3B). Exclusion of exon 6, which encodes the transmembrane region of the receptor molecule, produces a soluble isoform of the Fas gene that is unable to transmit the apoptotic signal (Figure 3A) (Cheng et al., 1994). Instead, this soluble isoform binds to ligand in the surrounding medium, thus preventing the ligand from binding membrane-bound Fas and protecting cells from apoptosis. This increased level of sFas RNA transcript was also accompanied by the appearance of a detectable amount of soluble Fas protein in the medium of U2OSE64b cells treated with mitomycin C (Figure 3C and 3D). Based on these data, one may propose a model of how cells modify their response to DNA damage by modulation of splicing regulation. Activation of the p53 pathway following DNA damage increases the number of Fas molecules on the cell surface, thus making cells more susceptible to Fas-mediated apoptosis (Figure 4A, left side). In the absence of the proper p53 response to DNA damage, however, cells not only fail to up-regulate Fas receptor expression, but also produce an increased amount of its soluble form by modulation of splicing activity such that exclusion of exon 6 is favored. This soluble version of the receptor is released from the cells and is able to bind to Fas ligand, thus reducing engagement of membrane-bound Fas and the consequent triggering of Fas-mediated apoptosis (Figure 4B, right side).
Figure 4. Regulation of Fas expression differs in p53-deficient cells during the early response to DNA damage.
A. A scheme of the p53-dependent response to DNA damage. p53 induces expression of the Fas gene, leading to an increased amount of Fas receptor molecules on the cellular surface, thus making the cell more sensitive to FasL-mediated apoptosis. B. A scheme of the early response to DNA damage in the absence of p53. In p53-deficient cells, the total expression of the Fas gene does not change and activation of AS leads to increased expression of the soluble isoform of the receptor. This results in a decreased amount of Fas receptor on the cellular surface, accompanied by the appearance of the soluble anti-apoptotic isoform in the intercellular space, and resulting in protection of the cell from FasL-mediated apoptosis.
By monitoring Fas gene expression, we also showed that induction of sFas synthesis is temporary and occurs only within the first few hours of the response to mild DNA damage (Figure 3B). This induction coincides with the transient increase of splicing activity in the p53-deficient cells. Moreover, we have previously shown that disruption of splicing activity in these cells by siRNA silencing of SRp55 expression postponed apoptosis, indicating that the observed increase of splicing activity is functionally important (Filippov et al., 2007).
Identification of the CD44 and Fas genes, coding for receptor molecules with known functions, as targets for AS following DNA damage sheds light on how activation of splicing activity may change the cellular response to DNA damage. The glycoprotein receptor, CD44, known to be involved in cell-cell interactions, and its numerous functionally distinct isoforms are closely connected with apoptosis and tumorigenesis (Marhaba and Zoller, 2004). The Fas receptor, a powerful inducer of apoptosis, also exists as a soluble splice variant. Increased expression of its soluble, anti-apopotic splice isoform, in cells lacking a proper p53 response to DNA damage indicates that these cells are able to engage a protective mechanism by modification of splicing activity. This observation may represent one of the mechanisms by which p53 deficient cells become resistant to apoptosis. Further analysis of genes targeted by AS activity is needed to determine the full extent of how alterations of splicing activity can modulate cellular responses.
ACKNOWLEDGEMENTS
This work was supported by NCI Grant R01 CA095461 from the National Institutes of Health. We would also like to thank Richard Einstein and Weiyin Zhou (ExonHit Therapeutics) for their suggestions and comments.
Abbreviations
- AS
alternative splicing
- bp
base pair
- CMV promoter
promoter of cytomegalovirus immediate-early gene
- ELISA
enzyme-linked immunosorbent assay
- HPV16 E6
human papilloma virus type 16 E6 oncogene
- kDa
kilodalton
- MTT
Thiazolyl Blue Tetrazolium Bromide
- qRT-PCR
quantitative RT-PCR
- RT-PCR
reverse transcription- polymerase chain reaction
- siRNA
small interfering RNA
- Tet-Off cell line
mammalian cell line in which expression of the gene of interest can be regulated by tetracycline or doxycyline
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Blanchette M, Green RE, Brenner SE, Rio DC. Global analysis of positive and negative pre-mRNA splicing regulators in Drosophila. Genes Dev. 2005;19:1306–1314. doi: 10.1101/gad.1314205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Zhou T, Liu C, Shapiro JP, Bruaer MJ, Kiefer MC, Barr PJ, Mountz JD. Protection from Fas-mediated apoptosis by a soluble form of the Fas molecule. Science. 1994;263:1759–1762. doi: 10.1126/science.7510905. [DOI] [PubMed] [Google Scholar]
- Filippov V, Filippova M, Duerksen-Hughes PJ. The early response to DNA damage can lead to activation of alternative splicing activity resulting in CD44 splice pattern changes. Cancer Res. 2007;67:7621–7630. doi: 10.1158/0008-5472.CAN-07-0145. [DOI] [PubMed] [Google Scholar]
- Filippova M, Brown-Bryan TA, Casiano CA, Duerksen-Hughes PJ. The human papillomavirus 16 E6 protein can render cells either sensitive or resistant to TNF: Effect of dose. Cell Death Differ. 2005;12:1622–1635. doi: 10.1038/sj.cdd.4401678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippova M, Duerksen-Hughes PJ. Inorganic and dimethylated arsenic species induce cellular p53. Chem. Res. Toxicol. 2003;16:423–431. doi: 10.1021/tx025606a. [DOI] [PubMed] [Google Scholar]
- Filippova M, Parkhurst L, Duerksen-Hughes PJ. The human papillomavirus 16 E6 protein binds to Fas-associated death domain and protects cells from Fas-triggered apoptosis. J. Biol. Chem. 2004;279:25729–25744. doi: 10.1074/jbc.M401172200. [DOI] [PubMed] [Google Scholar]
- Filippova M, Song H, Connolly JL, Dermody TS, Duerksen-Hughes PJ. The human papillomavirus 16 E6 protein binds to tumor necrosis factor (TNF) R1 and protects cells from TNF-induced apoptosis. J. Biol. Chem. 2002;277:21730–21739. doi: 10.1074/jbc.M200113200. [DOI] [PubMed] [Google Scholar]
- Friesen C, Herr I, Krammer PH, Debatin KM. Involvement of the CD95 (APO-1/FAS) receptor/ligand system in drug-induced apoptosis in leukemia cells. Nat. Med. 1996;2:574–577. doi: 10.1038/nm0596-574. [DOI] [PubMed] [Google Scholar]
- Gotoh I, Adachi M, Nishida E. Identification and characterization of a novel MAP kinase kinase kinase, MLTK. J. Biol. Chem. 2001;276:4276–4286. doi: 10.1074/jbc.M008595200. [DOI] [PubMed] [Google Scholar]
- Gupta P, Su ZZ, Lebedeva IV, Sarkar D, Sauane M, Emdad L, Bachelor MA, Grant S, Curiel DT, Dent P, Fisher PB. mda-7/IL-24: multifunctional cancer-specific apoptosis-inducing cytokine. Pharmacol. Ther. 2006;111:596–628. doi: 10.1016/j.pharmthera.2005.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzen EL, Yoon W, Weale ME, Sen A, Wood NW, Burke JR, Welsh-Bohmer KA, Hulette CM, Sisodiya SM, Goldstein DB. Alternative ion channel splicing in mesial temporal lobe epilepsy and Alzheimer's disease. Genome Biol. 2007;8:R32. doi: 10.1186/gb-2007-8-3-r32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman BE, Lis JT. Pre-mRNA splicing by the essential Drosophila protein B52: tissue and target specificity. Mol. Cell Biol. 2000;20:181–186. doi: 10.1128/mcb.20.1.181-186.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, Shoemaker DD. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- Krammer PH. CD95's deadly mission in the immune system. Nature. 2000;407:789–795. doi: 10.1038/35037728. [DOI] [PubMed] [Google Scholar]
- Marhaba R, Zoller M. CD44 in cancer progression: adhesion, migration and growth regulation. J. Mol. Histol. 2004;35:211–231. doi: 10.1023/b:hijo.0000032354.94213.69. [DOI] [PubMed] [Google Scholar]
- Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat. Rev. Mol. Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- Novoyatleva T, Tang Y, Rafalska I, Stamm S. Pre-mRNA missplicing as a cause of human disease. Prog. Mol. Subcell. Biol. 2006;44:27–46. doi: 10.1007/978-3-540-34449-0_2. [DOI] [PubMed] [Google Scholar]
- Sharma K, Wang RX, Zhang LY, Yin DL, Luo XY, Solomon JC, Jiang RF, Markos K, Davidson W, Scott DW, Shi YF. Death the Fas way: regulation and pathophysiology of CD95 and its ligand. Pharmacol. Ther. 2000;88:333–347. doi: 10.1016/s0163-7258(00)00096-6. [DOI] [PubMed] [Google Scholar]
- Skotheim RI, Nees M. Alternative splicing in cancer: noise, functional, or systematic? Int. J. Biochem. Cell Biol. 2007;39:1432–1449. doi: 10.1016/j.biocel.2007.02.016. [DOI] [PubMed] [Google Scholar]
- Stamm S. Signals and their transduction pathways regulating alternative splicing: a new dimension of the human genome. Hum. Mol. Genet. 2002;11:2409–2416. doi: 10.1093/hmg/11.20.2409. [DOI] [PubMed] [Google Scholar]
- Thorburn A. Death receptor-induced cell killing. Cell Signal. 2004;16:139–144. doi: 10.1016/j.cellsig.2003.08.007. [DOI] [PubMed] [Google Scholar]
- Wettenhall JM, Smyth GK. limmaGUI: a graphical user interface for linear modeling of microarray data. Bioinformatics. 2004;20:3705–3706. doi: 10.1093/bioinformatics/bth449. [DOI] [PubMed] [Google Scholar]