Abstract
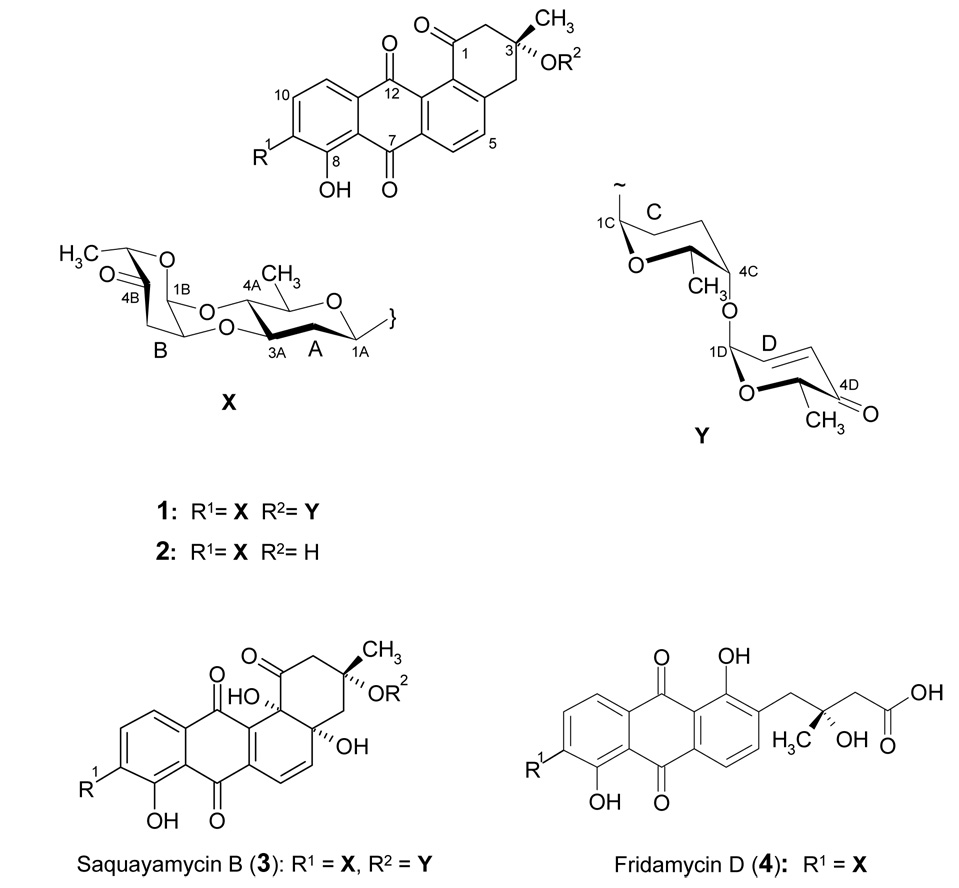
Two new anticancer antibiotics of the angucycline class, moromycins A and B (1, 2), along with the known microbial metabolites saquayamycin B (3) and fridamycin D (4) were isolated from the ethyl acetate extract of a culture broth of the terrestrial Streptomyces sp. KY002. The structures consist of a tetrangomycin core, and various C- and O-glycosidically linked deoxysugars. The chemical structures of the new secondary metabolites were elucidated by 1D and 2D NMR and by mass spectrometry. Moromycin B (2) showed significant cytotoxicity against H-460 human lung cancer and MCF-7 human breast cancer cells.
Angucyclines are a large group of naturally occurring quinone-saccharides isolated from the culture broths of different microorganisms.1–3 The angucycline group of compounds exhibits a broad range of biological activities, such as anticancer, antibacterial, antiviral, enzyme inhibitory and inhibition of platelet aggregation.1–4 Their characteristic polyketide derived tetracyclic benz[a]anthracenone (angucyclinone) frame was first found in tetrangomycin,5 isolated from the culture broth of Streptomyces rimosus. Since then, an increasing number of derivatives with different activities have been isolated from natural sources, predominantly from streptomycetes,1, 2, 6–11 many of these with saccharide residues but only a few with a C-glycosidically bound sugar moiety. This subgroup includes the saquayamycins A~F12, 13 and Z,14 frigocyclinone,15 and the most recently reported marmycins A and B.16 For our projects studying and modifying the biosyntheses of unusual angucyclinone-derived natural products we were interested in finding new angucyclines with unusual sugar moieties and/or saccharide structures that suggest interesting sugar moiety-modifying enzymes.
The Southern Appalachian Mountains are an ancient, topographically diverse mountain range that provides a wide array of habitats and microhabitats for both flora and fauna which are endowed with a high level of biodiversity unparalleled in the temperate zone. For example, the Southern Appalachian Mountains house nearly 10% of global salamander diversity (~ 55 species, 21 of these are known from nowhere else, overall 134 reptiles and amphibians), more than 460 different arachnids, a record diversity of over 230 millipedes, 255 birds, 78 mammals and more than 2,000 species of vascular plants.17–21 Microorganisms and fungi are less well explored. Nearly 2,300 species of fungi have been identified, but it is estimated that as many as 20,000 exist. Although nothing is known about the numbers and diversity of microorganisms of this region, we assumed it would be worthy to look into Appalachian microbes, because they likely yield a rich diversity and novel microbial natural products with exciting biological activities. From a soil sample collected in the Kentucky foothills of the Appalachian Mountains near Morehead (Cave Run Lake) were isolated two Streptomyces species. HPLC-MS screening of the metabolites from one of the isolate of Streptomyces sp. KY002 revealed several peaks with a quinone-type chromophore, and an MS fragmentation pattern that suggested the existence of deoxysugar residues. We herein describe fermentation, isolation, structure elucidation and biological activities of two new C-glycosyl-angucyclines designated as moromycin A (1) and B (2).
Results and Discussion
Six days-grown agar cultures of Streptomyces sp. KY002 served to inoculate 50 of 250 mL Erlenmeyer flasks each containing 100 mL of soy peptone/glucose (SG) medium. Fermentation of the strain was carried out at 28 °C for 3 days while shaking at 250 rpm. After centrifugation, extraction and evaporation, working up of the crude extract resulted in the isolation of two new metabolites, named moromycin A (1, yield: 2 mg/L) and B (2, yield: 1mg/L), along with the two known compounds saquayamycin B12, 13 (3, yield: 4 mg/L) and fridamycin D22–25 (4, yield: 1.5 mg/L).
Fridamycin D (4) and saquayamycin B (3) were isolated as yellow solids with molecular weights of 596 and 820, respectively, based on the negative ion mode of the APCI mass spectrum. From their deduced molecular formulae and their NMR data, both compounds were identified rapidly as known angucycline group antibiotics. Also compound 1 appeared in isolated form as a yellow solid. The UV spectrum (maxima were visible at 200, 265 and 405 nm) revealed a hydroxyl-anthraquinone chromophore. The ESI-MS of 1 displayed in the positive ion mode a mass of m/z = 787.3 [M+H]+, and of m/z = 785.3 [M-H]− in the negative mode suggesting a molecular weight of 786 Da, and from the HRFAB MS a molecular formula of C43H46O14 (m/z 786.2887 found, calcd 786.2882) was confirmed. The 1H NMR of 1 showed one signal at δ = 12.75 which could be assigned to a chelated peri-hydroxy group and two sets of aromatic signals each with ortho-couplings at δ 8.34 (7.8 Hz), 7.96 (7.9 Hz), 7.80 (7.8 Hz) and 7.63 (7.9 Hz). Additionally, an olefinic AB system at δ 7.03 (10.0 Hz) and 6.01 (10.0 Hz) was observed. Between δ 5.26 ~ 2.41, several signals typical for deoxysugars were visible. Furthermore, four methyl doublets at δ 1.34, 1.27, 1.20 and 1.12 and one methyl singlet at δ1.53 were detected. The 13C NMR spectrum of 1 showed five carbonyl signals, three ketone groups (δ 207.7, 196.6 and 195.5), two of these being α,β-unsaturated, and a typical quinone system (δ 188.2 and 182.4). In the sp2-region, twelve signals corresponding to two isolated aromatic system were visible. Overall, the 13C NMR data of the polyketide derived non-sugar moiety was in good agreement with tetrangomycin.5, 6, 26 In the sugar region, the 13C NMR spectrum showed three acetal carbon signals (δ 95.0, 91.3 and 91.4) and four methyl groups (δ 16.9, 16.6, 15.6 and 14.5) pointing to four deoxysugars, one of which needed to be C-glycosidically linked.
Compound 2 was obtained as a yellow solid as well. It gave a quasi-molecular ion at m/z 561.2 [M-H]−1 by (−)-APCIMS. The HRFAB-MS of 2 showed a molecular ion at m/z 562.1844 consistent with the molecular formula C31H30O10 (calcd 562.1839). Again, the UV spectrum recorded in MeOH (maxima at 270 and 415 nm) suggested a hydroxy-anthraquinone chromophore. The 1H NMR data of 2 showed a close similarity with those of compound 1. Identical signals for one chelated hydroxy group at δ 12.75, and four ortho-coupled aromatic proton signals were found. However, in the sugar region, less signals were observed, and the spectrum showed only one anomeric signal at δ 5.25. The 13C NMR spectra of 2 revealed all 31 carbons, 19 of them again belonging to a tetracyclic tetrangomycin moiety. The presence of only one acetal carbon signal (δ 91.5), but two methyl doublets (δ 17.3, 16.0) and overall 12 sugar carbons gave evidence for two deoxysugar moieties, again one C-glycosidically bound.
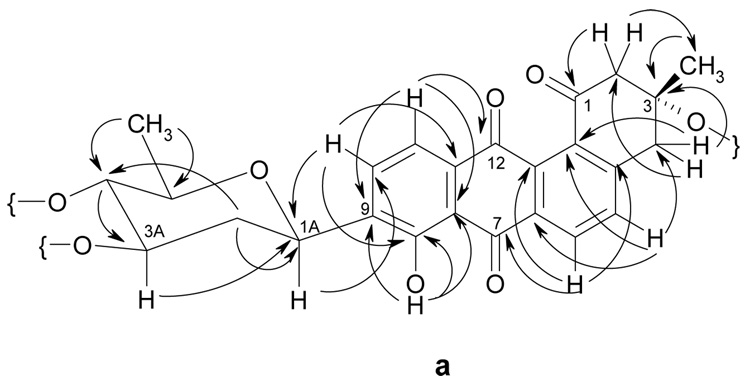
Comprehensive analysis of 2D NMR spectra (H,H COSY, HSQC, HMBC and ROESY) of both new compounds confirmed the existence of a tetrangomycin core. The data indicated for both compounds a β-C-glycosidic linkage (H-1A shows a 11.0 Hz transaxial coupling to Ha-2A) of one of the sugars to the 9-position of this tetrangomycin core. The linkage position was directly deducible from 3JC-H coupling observed between H-1A and C-9 as well as between H-9 and C-1A in the HMBC spectrum (Figure 2). From the analysis of the H,H COSY spectrum and the coupling pattern of each H-signal the C-glycosidic moiety in both compounds 1 and 2 could be deduced as D-olivose. The D-configuration was assumed considering the β-linkage and the fact that so far all glycosyltransferases (GTs) found in Streptomyces secondary metabolites belong to the GT-1 family of inverting enzymes.27–29 The three other O-glycosidically linked sugars in 1 were identified in the same manner as α-l-cinerulose, α-l-rhodinose and α-l-aculose (Figure 3 and Figure 4 show key HMBC and ROESY correlations), and by comparison with previous literature.12, 13 All of these sugars show only small couplings to their anomeric protons. The spin system of each sugar unit was also investigated further by 1H-1H DQFCOSY.
Figure 2.
HMBC correlations in the aglycon moieties of moromycins A (1) and B (2).
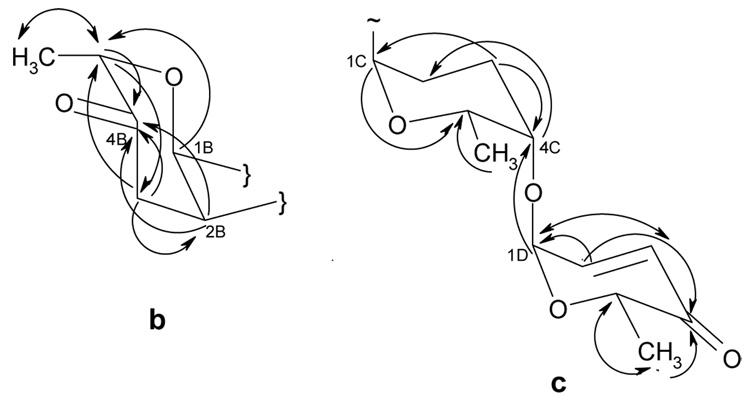
Figure 3.
HMBC correlations in the O-glycosidic sugar moieties of moromycins A (1, fragment b and c) and B (2, fragment b), b = α-l-cinerulosyl, c = α-l-aculosyl-(1–4)- α-l-rhodinosyl.
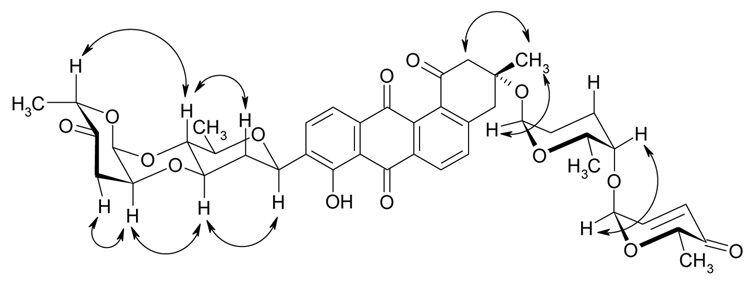
Figure 4.
Selected ROESY couplings for moromycin A (1).
The connection of the moromycin aglycon fragment a (this includes the C-glycosidic moiety, as shown in Figure 1) with the O-glycosidically linked sugar residues in compounds 1 and 2 was deduced by the HMBC and ROESY couplings (Figure 3, Figure 4). In the HMBC spectrum of 1, the proton signals at δ 3.90 (H-3A) and 3.58 (H-4A) were coupled to the carbon signals at δ 91.3 71.1 (C-2B) and (C-1B), respectively, which shows the connection of the l-cinerulose unit (fragment b) to both the 3A- and 4A-oxygens of the β-d-olivose of fragment a. Comparison with compound 2 showed the same α-l-cinerulosyl moiety connected both the 3- and 4-positions of the C-glycosidically linked d-olivose. The remaining two sugars (fragment c) of compound 1 were found to be linked as a disaccharide to 3-O of the tetrangomycin core (H-1C, belonging to the l-rhodinose building block, at δ 5.25 coupled to C-3 at δ 76.2, by HMBC). Also the connectivity between the two sugars of this disaccharidal moiety of 1, l-aculose and l-rhodinose (fragment c), was confirmed by HMBC couplings (the anomeric proton of the L-aculose at δ 5.26, H-1D coupled to δ 76.4, C-4C). Thus, moromycin A (1) contains two disaccharide chains, and α-l-cinerulosyl-(1→4, 2→3)-β-l-olivosyl chain, C-glycosidically linked to C-9 of a tetrangomycin core, and an α-l-aculosyl-(1→4)-α-l-rhodinosyl chain, linked O-glycosidically to C-3 of the tetrangomycin polyketide core. In comparison, moromycin B (2) lacks the O-glycosidic disaccharide. The overall structures of 1 and 2 were also confirmed by comparison of the NMR data with those of the saquayamycins B and B1.13.
Figure 1.
Compounds 1, 2 and saquayamycin B were assessed for their antitumor activities against a human lung (H-460) and a human breast cancer cell line (MCF-7, Table 2). Interestingly, the antitumor activity of moromycin B (2) with only one disaccharide chain is superior compared to moromycin A (1) and saquayamycin B, while the toxicity (LC50) is similar for both moromycins Moromycin A (1) appears less active than saquayamycin B (3). This difference must be caused by the angucyclic core moiety, which is less aromatic in the saquayamycins due to tertiary OH residues in 4a- and 12b-positions. The two compounds differ only in this respect. The results are consistent with those reported by Zeeck et al., who reported that any change in the angucyclic core of the saquayamycins decreased their cytotoxic activity.30.
Table 2.
Cytotoxicity assay of moromycins A (1) and B (2) in comparison with saquayamycin B (3) against human lung H-460 and human breast MCF-7 cancer cell lines (given are GI50, TGI and LC50 values in µmol). The data represent the average of three measurements.
| H-460 | MCF-7 | |||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 1 | 2 | 3 | |
| GI50 | 18.7 | 4.1 | 12.2 | 25.1 | 5.6 | 15.2 |
| TGI | 51.2 | 14.1 | 26.5 | 38.9 | 9.2 | 21.3 |
| LC50 | 69.1 | 52.3 | 34.3 | 55.0 | 16.3 | 28.1 |
GI50= 50% inhibition of cell growth; TGI=total growth inhibition; LC50=50% lethal concentration.
Overall, our search for new angucyclines in the foothills of the Southern Appalachian mountains yielded the terrestrial Streptomyces sp. KY002 strain, which produced two new angucyclines with interesting sugar moieties for further exploration. Particularly interesting, although not novel but so far not studied, are the disaccharide chains with the two 4-keto deoxysugars l-aculose and l-cinerulose.12, 31 The connection of C-2 of cinerulose with 3A-O of the preceding d-olivose unit might be established due to the acidic environment during fermentation. Such an acid-induced ether bridge formation has been reported previously in aclacinomycin B31 and in other saquayamycins12 with apparent negative effect on the biological activity. Aclacinomycin B (ID50 0.24 µg/mL) was found to be half as active against cultured l 1210 leukemia cells than aclacinomycin A (ID50 0.12 µg/mL), but exhibited roughly twice the toxicity in mice assays.31 In contrast, presence of the ether bridge did not reveal any significant difference in biological activity between saquayamycins A and B, which showed similar activity against murine P388 leukemia cells.12 Obviously, the bridge formation has two consequences, (i) it forces the l-cinerulose sugar into an unusual 4C1 conformation for an l-sugar with the 5-methyl group in the axial position, and it (ii) takes away the typical flexibility of the saccharide chain for a relatively rigid structural tricyclic side chain. The involved glycosyltransferases (GTs) are also interesting. While the C-GT resembles UrdGT232–35 and is no longer unique, GTs able to transfer ketosugars are rare, and new in our armamentarium for combinatorial biosynthetic approaches. It will be interesting to study these unique enzymes with respect of their regio-, stereo- and substrate specificity. They may become valuable resources for generating novel angucyclines-C-glycosides through combinatorial methods.
Experimental Section
General Experimental Procedures
Structure Elucidation
Optical rotations were recorded using a Perkin-Elmer 241 spectrophotometer. The UV spectra were recorded on a Varian model CARY50 spectrophotometer, and IR spectra were obtained from a pure sample of KBr disks in a Bio-Rad model FTS3000MX FT-IR spectrometer. All NMR data were recorded in d6-acetone using Varian Inova spectrometer equipped with a 9.4 T magnet. The ESI mass spectra (ESI-MS) were acquired using a Finnigan LCQ mass spectrometer. The high resolution fast atom bombardment mass spectrometry (FAB) spectra were acquired using a model VG70SQ double focusing magnetic sector MS instrument.
HPLC-MS
Analysis was performed on a Waters Alliance 2695 system, equipped with a Waters 2996 photodiode array detector and a Micromass ZQ2000 mass spectrometer by using an APCI probe (solvent A: 0.1% formic acid in H2O; solvent B: CH3CN; flow rate = 0.5 mL/min; 0–10 min 75% A and 25% B to 100% B, linear gradient, 10–12 min 100% B, 12–14 min 100% B to 75% A and 25% B (linear gradient), 14–15 min 75% A and 25% B). Column: Waters Symmetry C-18, 4.6 × 50 mm, particle size 5 mm. The column temperature was 23°C, and the UV detection wavelength was 452 nm.
SG medium
Glucose (20 g), soytone (10 g), CaCO3 (2 g) and CoCl2 (1 mg) were dissolved in 1000 mL of tap H2O. The medium was adjusted to pH 7.2 with 2 N NaOH and then sterilized for 33 min at 121°C.
M2 agar
Glucose (4 g), malt extract (10 g), yeast extract (4 g) and agar (15.0 g) were dissolved in 1 l of tap H2O then sterilized for 33 min at 121°C. The medium was adjusted to pH 7.2 with 2 M NaOH before sterilization.
Isolation and Phylogenetic Identification of Streptomyces sp. KY002
The soil sample was collected from Cave Run Lake site (Eastern Kentucky, USA). An aliquot of soil (2 g) was suspended in 20 mL sterile H2O, and the suspension was heated to 80 °C for 30 min. The supernatant was diluted serially (10−1, 10−2, 10−3) with sterile H2O and 100 µ1 sample was spread on oatmeal agar plates supplemented with nalidixic acid (1 mg) and cycloheximide (10 mg). Bacterial colonies were visible after 6 days of incubation of the culture plates at 30°C. Each colony was streaked on M2-agar plate. Morphological and physiological studies to identify the genus of the strain were carried out following a standard protocol (see supplementary material).36, 37.
Genomic DNA was isolated from 50 mL culture of the strain in TSB-sucrose medium (20 g/L, tryptic soy broth; 103 g/L, sucrose) following a standard protocol.38 A set of degenerate primers (16S-F: 5’-TCACGGAGAGTTTGATCCTG-3’ and 16S-R1: 5’-GCGGCTGCTGGCACGTAGTT-3’) were designed from the consensus 16S rRNA gene sequences of various Streptomyces species. Advantage®-GC-2 PCR kit (Clonetech Laboratories, Inc.) was used to amplify 16S rRNA gene sequence in 50 µL reaction mixture composed of 40ng genomic DNA, 1 nM of each primer, 200 µM of each dNTP. Thus obtained PCR product was cloned in to pGEM®-TEasy vector (Promega). Three different clones were sequenced to verify that sequencing errors had not been incorporated. Homology search (BLAST search) in the database revealed that the amplified nucleotide sequence (511 bp) has highest identity (99%) to the 16S rRNA gene sequence of Streptomyces sp. 1A01511. The deduced 16S rRNA nucleotide sequence has been deposited in the NCBI nucleotide database with an accession no AM922111.
Fermentation of the Isolate Streptomyces sp. KY002
Streptomyces sp. KY002 was inoculated from its soil culture on three M2 agar plates. After incubation for 72 h at 28 °C the well-developed colonies were used to inoculate fifty of 250 cm3 Erlenmeyer flasks each containing 100 mL of SG-Medium. The flasks were incubated with 220 rpm for 3 days at 28 °C.
Extraction and Isolation of Compounds 1~4
The fermentation broth was harvested after 3 days and centrifugation at 4200 rpm for 30 min. The resulting mycelial cake was extracted three times with acetone. After removal of acetone, the aqueous solution was extracted three times with EtOAc. The EtOAc-soluble portion was concentrated under reduced pressure. Also the culture broth was extracted three times with EtOAc. As the LC/MS of both extracts from culture filtrate and mycelia showed the same composition, they were combined and concentrated under reduced pressure. The crude extract was subjected to Sephadex LH-20 (column, 4 × 120 cm, MeOH) resulting in three fractions. The middle fraction containing compounds 1~4 (LC/MS) was purified by preparative HPLC (column: mBondapak C18 radial compression cartridge, PrepPak cartridge, 19 K 150 mm, Waters; eluent: CH3CN and H2O (gradient from 35% to 100% CH3CN in 30 min, flow rate 10 mL/min). Compounds 1 (7.0 mg), 2 (2.8 mg), 3 (12.0 mg) and 4(4.0 mg) were eluted at retention times of 15, 17, 13 and 12 min, respectively. Due to the lack of compounds of interest (monitored by LC/MS), the rest of the fractions from the Sephadex chromatography were not further analyzed.
Moromycin A (1)
yellow solid (7.0 mg); [α]D 25 +11.1 (c 0.27, MeOH); UV (MeOH) λmax (log ε) 405 nm (3.57), 265 nm (4.35); IR (KBr) νmax 3421, 2960, 1700, 1437, 1341, 1275, 1065 cm−1; 1H NMR (400 MHz, acetone-d6) and 13C NMR (100.6 MHz, acetone-d6), see Table 1; (−)-APCI-MS m/z 785 [M-H]−; ESI-MS m/z 785.3 [M-H]−, (+)-ESI-MS m/z 787.3 [M+H]+; HRFAB m/z 786.2887 (calcd for C43H46O14, 786.2882).
Table 1.
13C and 1H NMR data for compounds 1 and 2, δ in ppm (quantity, multiplicity and J/Hz).
| Moromycin A (1) | Moromycin B (2) | |||
|---|---|---|---|---|
| Position | δCa | δHb | δCa | δHb |
| 1 | 195.5 | - | 196.0 | - |
| 2 | 51.6 | 2.91 (1Ha, d, 16.5); 3.31 (1He, dd, 16.5, 1.5) | 53.5 | 3.08 (1H, d, 17.1); 3.11 (1H, dd, 17.1, 1.5) |
| 3 | 76.2 | - | 72.0 | - |
| 4 | 41.3 | 3.23 (1Ha, d, 14.9); 3.44 (1He, dd, 14.9, 1.5) | 44.1 | 3.30 (1H, d, 14.9); 3.32 (1H, dd, 14.9, 1.5) |
| 4a | 148.4 | - | 149.4 | - |
| 5 | 134.4 | 7.80 (1H, d, 7.8) | 134.2 | 7.75 (1H, d, 7.9) |
| 6 | 128.8 | 8.34 (1H, d, 7.8) | 128.7 | 8.31 (1H, d, 7.9) |
| 6a | 133.4 | - | 133.6 | - |
| 7 | 188.2 | - | 188.3 | - |
| 7a | 115.1 | - | 115.3 | - |
| 8 | 157.9 | - | 157.8 | - |
| 8-OH | - | 12.75 (1H, br s) | - | 12.75 (1H, br s) |
| 9 | 136.3 | - | 136.3 | - |
| 10 | 136.4 | 7.96 (1H, d, 7.9) | 136.4 | 7.96 (1H, d, 7.8) |
| 11 | 118.5 | 7.63 (1H, d, 7.9) | 118.7 | 7.62 (1H, d, 7.8) |
| 11a | 135.8 | - | 135.6 | - |
| 12 | 182.4 | - | 182.6 | - |
| 12a | 133.6 | - | 133.7 | - |
| 12b | 133.7 | - | 133.8 | - |
| 13 | 25.2 | 1.53 (3H, s) | 28.8 | 1.48 (3H, s) |
| 1A | 71.3 | 5.05 (1H, dd, 11.0,1.6) | 71.4 | 5.06 (1H, dd, 11.0,1.6) |
| 2A | 36.6 | 1.59 (1Ha, ddd, 13.0, 11.0, 3.7); 2.41 (1He, ddd, 13.0, 3.8,1.1) | 36.9 | 1.64 (1Ha, ddd, 13.0, 11.0, 3.7); 2.47 (1He, ddd, 13.0, 3.8, 1.2) |
| 3A | 76.5 | 3.90 (1H, ddd, 13.0, 9.0, 3.8) | 76.7 | 3.90 (1H, m) |
| 4A | 74.1 | 3.58 (1H, dd, 9.1, 9.0) | 74.0 | 3.58 (1H, dd, 9.0, 9.0) |
| 5A | 74.2 | 3.60 (1H, dq, 9.0, 6.1) | 74.5 | 3.62 (1H, dq, 9.0, 5.8) |
| 6A | 16.9 | 1.34 (3H, d, 6.1) | 17.3 | 1.34 (3H, d, 5.8) |
| 1B | 91.3 | 5.13 (1H, d, 1.2) | 91.5 | 5.25 (1H, d, 2.9) |
| 2B | 71.1 | 4.43 (1H, ddd, 2.9, 2.9, 2.9) | 71.6 | 4.44 (1H, ddd, 3.0, 3.0, 3.0) |
| 3B | 39.7 | 2.55 (1Ha, dd, 12.9, 3.0) ; 2.84(1He, dd, 12.9, 3.0) | 40.0 | 2.54 (1 Ha, dd, 12.9, 3.0); 2.81(1 He, dd, 12.9, 3.0) |
| 4B | 207.7 | - | 207.6 | - |
| 5B | 77.3 | 4.79 (1H, q, 6.2) | 77.6 | 4.79 (1H, q, 6.2) |
| 6B | 16.6 | 1.27 (3H, d, 6.8) | 16.0 | 1.26 (3H, d, 6.2) |
| 1C | 91.4 | 5.25 (1H, d, 2.1) | - | - |
| 2C | 25.1 | 1.72 (1Ha, m), 1.76 (1He, m) | - | - |
| 3C | 24.0 | 1.80 (1Ha, m),1.82 (1He, m) | - | - |
| 4C | 76.4 | 3.63 (1H, m) | - | - |
| 5C | 66.2 | 3.97 (1H, dq, 9.1, 6.8) | - | - |
| 6C | 15.6 | 1.12 (3H, d, 6.8) | - | - |
| 1D | 95.0 | 5.26 (1H, d, 2.1) | - | - |
| 2D | 144.2 | 7.03 (1H, dd, 10.0, 2.8) | - | - |
| 3D | 126.2 | 6.01 (1H, d, 10.0) | - | - |
| 4D | 196.4 | - | - | - |
| 5D | 70.0 | 4.48 (1H, q, 6.2) | - | - |
| 6D | 14.5 | 1.20 (3H, d, 6.2) | - | - |
d6-acetone, 100.6 MHz.
d6-acetone, 400 MHz.
Moromycin B (2)
yellow solid (2.80 mg); [α]D 25 +44.0 (c 0.25, MeOH); UV (MeOH) λmax (logε) 415 nm (3.36), 270 nm (4.02); IR (KBr) νmax 3420, 2929, 1647, 1458, 1383, 1066 cm−1; 1H NMR (400 MHz, acetone-d6) and 13C NMR (100.6 MHz, acetone-d6), see Table 1; (−)-APCI-MS m/z 561 [M-H]−, (+)-APCI-MS m/z 563 [M+H]+; HRFAB m/z 562.1844 (calcd for C31H30O10, 562.1833).
Cytotoxicity Assays
In vitro cytotoxicity against human lung H-460 and human breast MCF-7 cell lines were assessed using the sulforhodamine B (SRB) assay, after 48 h exposure to the drugs.39, 40.
Supplementary Material
Taxonomy of strain Streptomyces sp. KY 002 (cultural characteristics, physiological properties, electron-microscopic photograph). This material is available free of charge via Internet at http://pubs.acs.org.
Acknowledgments
This work was supported by a grant from the National Institutes of Health (CA 102102) to J.R. The NMR core center of the University of Kentucky is acknowledged for the use of their spectrometers. We also thank J. Goodmann (University of Kentucky Mass Spectrometry Facility) for the mass spectra, and V. R. Adams and C. Mattingly for the cytotoxicity assays.
References and Notes
- 1.Rohr J, Thiericke R. Nat. Prod. Rep. 1992;9:103–137. doi: 10.1039/np9920900103. and references therein. [DOI] [PubMed] [Google Scholar]
- 2.Krohn K, Rohr J. Topics Curr. Chem. 1997;188:127–195. and references therein. [Google Scholar]
- 3.Thomson RH. Naturally Occurring Quinones. 4th ed. London: Blackie Academic & Professional; 1996. p. 519. and references therein. [Google Scholar]
- 4.Oka M, Kamei H, Hamagishi Y, Tomita K, Miyaki T, Konishi M, Oki T. J. Antibiot. 1990;43:967–976. doi: 10.7164/antibiotics.43.967. [DOI] [PubMed] [Google Scholar]
- 5.Dann M, Lefemine DV, Barbatschi F, Shu P, Kunstmann MP, Mitscher LA, Bohonos N. Antimicrob. Agents Chemother. (Bethesda) 1965;5:832–835. [PubMed] [Google Scholar]
- 6.Abdelfattah M, Maskey RP, Asolkar RN, Grün-Wollny I, Laatsch H. J. Antibiot. 2003;56:539–542. doi: 10.7164/antibiotics.56.539. [DOI] [PubMed] [Google Scholar]
- 7.Taniguchi M, Nagai K, Watanabe M, Niimura N, Suzuki K, Tanaka A. J. Antibiot. 2002;55:30–35. doi: 10.7164/antibiotics.55.30. [DOI] [PubMed] [Google Scholar]
- 8.Méndez C, E K, Lipata F, Lombó F, Cotham W, Walla M, Bearden DW, Braña AF, Salas JA, Rohr J. J. Nat. Prod. 2002;65:779–782. doi: 10.1021/np010555n. [DOI] [PubMed] [Google Scholar]
- 9.Puder C, Zeeck A, Beil W. J. Antibiot. 2000;53:329–336. doi: 10.7164/antibiotics.53.329. [DOI] [PubMed] [Google Scholar]
- 10.Schimana J, Fiedler HP, Groth I, Süssmuth R, Beil W, Walker M, Zeeck A. J. Antibiot. 2000;53:779–787. doi: 10.7164/antibiotics.53.779. [DOI] [PubMed] [Google Scholar]
- 11.Sun CH, Wang Y, Wang Z, Zhou JQ, Jin WZ, You XF, Gao H, Zhao LX, Si SY, Li X. J. Antibiot. 2007;60:211–215. doi: 10.1038/ja.2007.25. [DOI] [PubMed] [Google Scholar]
- 12.Uchida T, Imoto M, Watanabe Y, Miura K, Dobashi T, Matsuda N, Sawa T, Naganawa H, Hamada M, Takeuchi T. J. Antibiot. 1985;38:1171–1181. doi: 10.7164/antibiotics.38.1171. [DOI] [PubMed] [Google Scholar]
- 13.Sekizawa R, Iinuma H, Naganawa H, Hamada M, Takeuchi T, Yamaizumi J, Umezawa K. J. Antibiot. 1996;49:487–490. doi: 10.7164/antibiotics.49.487. [DOI] [PubMed] [Google Scholar]
- 14.Ströch K, Zeeck A, Antal N, Fiedler HP. J. Antibiot. 2005;58:103–110. doi: 10.1038/ja.2005.13. [DOI] [PubMed] [Google Scholar]
- 15.Bruntner C, Binder T, Pathom-aree W, Goodfellow M, Bull AT, Potterat O, Puder C, Horer S, Schmid A, Bolek W, Wagner K, Mihm G, Fiedler HP. J. Antibiot. 2005;58:346–349. doi: 10.1038/ja.2005.43. [DOI] [PubMed] [Google Scholar]
- 16.Martin GDA, Tan LT, Jensen PR, Dimayuga RE, Fairchild CR, Raventos-Suarez C, Fenical W. J. Nat. Prod. 2007;70:1406–1409. doi: 10.1021/np060621r. [DOI] [PubMed] [Google Scholar]
- 17.Petranka JW. Salamanders of the United States and Canada. Washington, DC: Smithonian Institute Press; 1998. [Google Scholar]
- 18.Bernardo J, Ossola RJ, Spotila J, Crandall KA. Biol. Lett. 2007;3:695–698. doi: 10.1098/rsbl.2007.0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bernardo J, Spotila JR. Biol. Lett. 2006;2:135–139. doi: 10.1098/rsbl.2005.0417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neves RJ, Bogan AE, Williams JD, Ahlstedt SA, Hartfield W. Aquatic fauna in peril: the southeastern perspective. Decatur, Ga: Lenz Design and Communications; 1997. p. 554. Special Publication No. 1. Southeastern Research Institute ed. [Google Scholar]
- 21.Stein BA, Kutner LS, Adams JS. Precious Heritage: The Status of Biodiversity in the United States. Oxford, UK: Oxford University Press; 2000. For further information on life-form diversity see also the following web sites http://www.samab.org; http://www.sabionline.org/sabihome.html; http://www.wcu.edu/hbs/biodiversity.htm. [Google Scholar]
- 22.Kricke P. Ph.D. thesis. Georg-August-Universität Göttingen: Göttingen; 1984. [Google Scholar]
- 23.Krohn K, Baltus W. Tetrahedron. 1988;44:49–54. [Google Scholar]
- 24.Ueberbacher BJ, Osprian I, Mayer SF, Faber K. Eur. J. Org. Chem. 2005:1266–1270. [Google Scholar]
- 25.Maskey RP, Helmke E, Laatsch H. J. Antibiot. 2003;56:942–949. doi: 10.7164/antibiotics.56.942. [DOI] [PubMed] [Google Scholar]
- 26.Kunstmann MP, Mitscher LA. J. Org. Chem. 1966;31:2920–2925. doi: 10.1021/jo01347a043. [DOI] [PubMed] [Google Scholar]
- 27.Rix U, Fischer C, Remsing LL, Rohr J. Nat. Prod. Rep. 2002;19:542–580. doi: 10.1039/b103920m. [DOI] [PubMed] [Google Scholar]
- 28.Campbell JA, Davies GJ, Bulone V, Henrissat B. Biochemical J. 1997;326:929–939. doi: 10.1042/bj3260929u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Campbell JA, Davies GJ, Bulone V, Henrissat B. Biochem. J. 1998;329:719. doi: 10.1042/bj3290719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henkel T, Zeeck A. J. Antibiot. 1990;43:830–837. doi: 10.7164/antibiotics.43.830. [DOI] [PubMed] [Google Scholar]
- 31.Oki T, Matsuzawa Y, Yoshimoto A, Numata K, Kitamura I, Hori S, Takamatsu A, Umezawa H, Ishizuka M, Naganawa H, Suda H, Hamada M, Takeuchi T. J. Antibiot. 1975;28:830–834. doi: 10.7164/antibiotics.28.830. [DOI] [PubMed] [Google Scholar]
- 32.Künzel E, Faust B, Oelkers C, Weissbach U, Bearden DW, Weitnauer G, Westrich L, Bechthold A, Rohr J. J. Am. Chem. Soc. 1999;121:11058–11062. [Google Scholar]
- 33.Hoffmeister D, Dräer G, Ichinose K, Rohr J, Bechthold A. J. Am. Chem. Soc. 2003;125:4678–4679. doi: 10.1021/ja029645k. [DOI] [PubMed] [Google Scholar]
- 34.Baig I, Kharel M, Kobylyanskyy A, Zhu LL, Rebets Y, Ostash B, Luzhetskyy A, Bechthold A, Fedorenko VA, Rohr J. Angew. Chem. Int. Ed. 2006;45:7842–7846. doi: 10.1002/anie.200603176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mittler M, Bechthold A, Schulz GE. J. Mol. Biol. 2007;372:67–76. doi: 10.1016/j.jmb.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 36.Nonomura H. J. Fermentation Technol. 1974;52:78–92. [Google Scholar]
- 37.Shirling EB, Gottlieb D. Int. J. Syst. Bacteriol. 1966;16:313–340. [Google Scholar]
- 38.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. Norwich, UK: The John Innes Foundation; 2000. [Google Scholar]
- 39.Skehan P, Storeng R, Scudiero D, Monks A, McMahon J, Vistica D, Warren JT, Bokesch H, Kenney S, Boyd MR. J. Natl. Cancer Inst. 1990;82:1107–1112. doi: 10.1093/jnci/82.13.1107. [DOI] [PubMed] [Google Scholar]
- 40.Rubinstein LV, Shoemaker RH, Paull KD, Simon RM, Tosini S, Skehan P, Scudiero DA, Monks A, Boyd MR. J. Natl. Cancer Inst. 1990;82:1113–1118. doi: 10.1093/jnci/82.13.1113. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Taxonomy of strain Streptomyces sp. KY 002 (cultural characteristics, physiological properties, electron-microscopic photograph). This material is available free of charge via Internet at http://pubs.acs.org.