Abstract
Two cytidine analogues, gemcitabine (dFdC) and cytosine arabinoside (AraC), show significant therapeutic effect in a variety of cancers. However, response to these drugs varies widely. Evidence from tumor biopsy samples shows that expression levels for genes involved in the cytidine transport, metabolism and bioactivation pathway contribute to this variation in response. In the present study, we set out to test the hypothesis that variation in gene expression both within and outside of this “pathway” might influence sensitivity to gemcitabine and AraC. Specifically, Affymetrix U133 Plus 2.0 GeneChip and cytotoxicity assays were performed to obtain basal mRNA expression and IC50 values for both drugs in 197 ethnically-defined Human Variation Panel lymphoblastoid cell lines. Genes with a high degree of association with IC50 values were involved mainly in cell death, cancer, cell cycle, and nucleic acid metabolism pathways. We validated selected significant genes by performing real time quantitative RT-PCR and selected two representative candidates, NT5C3 (within the pathway) and FKBP5 (outside of the pathway), for functional validation. Those studies demonstrated that down regulation of NT5C3 and FKBP5 altered tumor cell sensitivity to both drugs. Our results suggest that cell-based model system studies, when combined with complementary functional characterization, may help to identify biomarkers for response to chemotherapy with these cytidine analogues.
Keywords: cytidine analogues, gemcitabine, dFdC, cytosine arabinoside, AraC, lymphoblastoid cell lines, expression array, 5′-nucleotidase, cytosolic III nucleotidase, NT5C3, FK506 binding protein 5, FKBP5 and Ingenuity Pathway Analysis
Introduction
Studies of gene expression may make it possible to identify biomarkers that will help predict clinical response to antineoplastic drug therapy. Many of these drugs have narrow therapeutic indexes. Therefore, it is crucial to identify biomarkers that might help to maximize efficacy and minimize drug-related toxicity. Most previous studies have focused on the relationship of expression signatures in tumor tissue to therapeutic response (1–3). However, individual variation in expression patterns controlled by germline DNA can also play an important role in response. Genes encoding proteins involved in drug transport, metabolism, activation, deactivation, or drug targets and downstream signaling pathways could all potentially influence drug response phenotypes (4). To test the hypothesis that variation in basal gene expression might affect sensitivity or resistance to chemotherapy, we have used a Human Variation Panel lymphoblastoid cell line model system consisting of 197 cell lines for which we obtained gene expression data using Affymetrix U133 Plus 2.0 GeneChips and drug-related cytotoxicity phenotypes. Although their expression profiles are not identical with those of tumor cells, these cell lines provide an opportunity to query the effect on drug response of common variation across the genome, providing data that cannot be obtained with other model systems such as the NCI-60 cell lines. Therefore, these Human Variation Panel cell lines can serve as an initial screen to identify candidate genes for which variation in expression might contribute to variation in drug response phenotypes.
We used two cytidine analogues, gemcitabine (dFdC) and cytosine arabinoside (AraC), to determine whether individual variation in basal gene expression might influence drug sensitivity. These drugs have been used to treat many cancers, and they share similar chemical structures, metabolic pathways, and mechanisms of action (5–8). Both are prodrugs that must be transported into cells, activated by kinases to form active di- and tri-phosphorylated metabolites, and inactivated by dephosphorylation. Seventeen genes are known to be involved in this cytidine analogue “pathway” (Supplementary Table 1). The triphosphates, AraCTP and dFdCTP, can be incorporated into DNA, terminating DNA synthesis (8, 9). dFdCDP can also inhibit ribonucleotide reductases (RRs), enzymes that catalyze the conversion of ribonucleotides to deoxyribonucleotidase (10, 11).
Gemcitabine is used to treat solid tumors (6, 12), while AraC is a major component of the therapy of acute myelogenous leukemia (AML) (6, 7, 13). Clinical response to both drugs varies widely (8, 14, 15). Most previous studies have focused on variation in the expression of genes within the known cytidine analogue metabolism and activation pathway (1, 16, 17). However, very little information is available with regard to genes outside of that pathway. Therefore, we have used these lymphoblastoid cell lines to explore the possible contribution of individual variation in basal gene expression to gemcitabine and AraC sensitivity.
Specifically, drug cytotoxicity and basal expression array data were obtained for 197 lymphoblastoid cell lines. Selected genes significantly associated with cytotoxicity were then validated functionally. A series of functional analyses were performed for two candidate genes, NT5C3, within the cytidine analogue pathway, and FKBP5, outside of that pathway. Specific siRNA “knockdown” confirmed the results of the association study, and the functional effects of both genes on response to gemcitabine and AraC was explored. Therefore, the use of these cell lines to identify pharmacogenomic candidate genes for cytidine analogue cytotoxicity resulted in novel hypotheses that can now be tested in clinical translational studies.
Materials and Methods
Cell lines
Lymphoblastoid cell lines from 60 Caucasian-American (CA), 54 African-American (AA), 60 Han Chinese-American (HCA), and 23 CEPH (CA) unrelated subjects were purchased from the Coriell Institute (Camden, NJ). Human SU86 pancreatic cancer cells were a gift from Dr. Daniel D. Billadeau, Mayo Clinic. Human breast cancer MDA-MB-231 cells were obtained from the American Type Culture Collection (ATCC) (Manassas, VA).
Drugs and cell proliferation assays
AraC was purchased from Sigma-Aldrich (St. Louis, MO) and gemcitabine was provided by Eli Lilly (Indianapolis, IN). Drugs were dissolved in DMSO and were frozen at −20°C. Assays were performed in triplicate at each drug concentration using the CellTiter 96® AQueous Non-Radioactive Cell Proliferation Assay (Promega Corporation, Madison, WI) in 96 well plates (Corning, Corning, NY) at a density of 5×104 cells/well. One hour after plating, 10 μl of gemcitabine (0.1 nM to 1 mM) or AraC (1 nM to 10 mM) was added to the wells, and plates were read in a Safire² microplate reader after 72 h incubations (Tecan AG, Switzerland). Twelve randomly selected lymphoblastoid cell lines were used to repeat the cytotoxicity studies three months later. Human tumor cell line cytotoxicity was determined in a similar fashion except the cells were incubated overnight prior to the addition of drug.
Expression array data
Total RNA was extracted using Qiagen RNeasy Mini kits (QIAGEN Inc. Valencia, CA). RNA quality was tested using an Agilent 2100 Bioanalyzer, followed by hybridization to Affymetrix U133 Plus 2.0 GeneChips. Only 26,653 probe sets that could be verified using the RefSeq RNA database were used in our analyses.
Real-time quantitative reverse transcription-PCR (QRT-PCR)
QRT-PCR was performed with the 1-step, Brilliant SYBR Green QRT-PCR kit (Stratagene, La Jolla, CA) using primers purchased from Qiagen. All experiments were performed in triplicate, with β-actin as an internal control. Reverse transcribed Universal Human reference RNA (Stratagene) was used to generate a standard curve.
Transient transfection and RNA interference
Human breast cancer MDA-MB-231 and SU86 pancreatic cancer cell lines were used to perform siRNA studies. Cells were grown to 30–50% confluence in 6-well plates and lipofectamine RNAMAX reagent (Invitrogen, Carlsbad, CA) was used to perform the transfections.
Western blot analysis
Western blot analysis was performed with lysates from cells transiently expressing siRNA 48 hours after transfection with control, NT5C3, FKBP5, NT5C2 or FKBP1A siRNA. Specifically, 30 μg of protein was subjected to electrophoresis on 12% SDS/PAGE gels. Proteins were transferred to PVDF membranes that were incubated overnight at 4ºC with primary antibodies, followed by the secondary antibody. Bands were detected with ECL (Amersham Biosciences). Antibodies were obtained from GenWay Biotech (San Diego, CA), Abcam (Cambridge, MA) and Novus Biologicals (Littleton, CO).
Intracellular gemcitabine and AraC metabolites
HPLC was used to measure intracellular AraCDP, AraCTP, dFdCDP and dFdCTP concentrations. Nucleotide extracts were prepared with a modification of the method of Van Haperen et al. (18) after treatment for 8 h with the average IC50 concentration for each drug. 5×106 cells were then centrifuged and washed with ice-cold phosphate buffered saline (PBS), followed by resuspension in 135 μL PBS with 15 μL of 100 μM AraCTP or dFdCTP as internal standards. Subsequently, 50 μL of 40% TCA was added, followed by vortexing and centrifugation. The supernatant was neutralized with 400 μL of trioctylamine:trichlorotrifluoroethane (1:4). After centrifugation, the aqueous phase was subjected to HPLC using a ZirChrom SAX HPLC column with photo-diode array detection and a gradient from 100% 10 mM K2HPO4 and 40 mM NaCl, pH 6.8, to 65% 100 mM K2HPO4 and 400 mM K2HPO4, pH 6.8.
Caspase-3/7 activity assay
Caspase-3/7 activity was determined as a measure of apoptosis using the Caspase-Glo®3/7 Assay kit (Promega BioSciences, San Luis Obispo, CA). siRNA-transfected cells were then treated for 72 h with increasing concentrations of gemcitabine or AraC. One hundred μL of Caspase-Glo® 3/7 Reagent was added, and the cells were incubated at room temperature for 1 h, followed by the measurement of luminescence.
Statistical methods
Three different logistic functions were used to fit the cytotoxicity data. The logistic model with the lowest mean square error was used to determine IC50 values. Expression array data were normalized on a log 2 scale, using both GCRMA and Fastlo (19, 20). Normalized expression data were regressed on gender, race, and time since the Coriell Institute acquired the cell line. Residuals were then standardized by subtracting the mean residual for individual probe sets and dividing by the standard deviation to derive a “standardized adjusted expression value”. Analyses were based on adjusted standardized values for both expression array and log transformed IC50 data. Pearson correlation coefficients were calculated, and a Wald test was used to test for a non-zero correlation. Multiple testing using 10,000 permutations was performed for selected probe sets. The contribution of pathway genes to expression was based on the coefficient of determination (R2) using a multiple regression model. Differences in intracellular metabolites between randomly selected resistant and sensitive cell lines were determined with student’s T-test. Correlations between expression array and QRT-PCR or intracellular metabolites were also determined. Agreement between cytotoxicity performed at different times was determined using an intra-class correlation coefficient. Ingenuity pathway analysis was performed by using Fischer’s exact test to calculate p-values to identify genes within given pathways.
Results
Gemcitabine and AraC cytotoxicity
Gemcitabine and AraC cytotoxicity studies were performed to determine the range of variation in IC50 values as an indication of individual cell line variation in drug sensitivity. Average unadjusted IC50 values for gemcitabine and AraC in these 197 cell lines were 25.3 ± 30.7 nM and 8.4 ± 14.3 μM (mean ± SD), respectively. IC50 differed among the groups of subjects studied (p < 0.01 for both drugs), with CEPH samples appearing to be more resistant to both drugs when compared with the three other groups. Gender did not appear to have a significant effect on IC50 values for either gemcitabine (p = 0.39) or AraC (p = 0.88). The time since the Coriell Institute acquired the cell lines had a slight effect on gemcitabine (p = 0.037), but not AraC IC50 values (p = 0.18). We also performed a replication study to exclude the possibility that this phenotype might vary over time. Specifically, 12 ethnically diverse cell lines were selected randomly and cytotoxicity assays were repeated three months after the initial experiments. There was good agreement between results at the two different times. The intra-class correlation coefficient was 0.83 (95% CI: 0.51–0.95) for gemcitabine and 0.71 (95% CI: 0.26–0.91) for AraC.
Association between expression and cytotoxicity
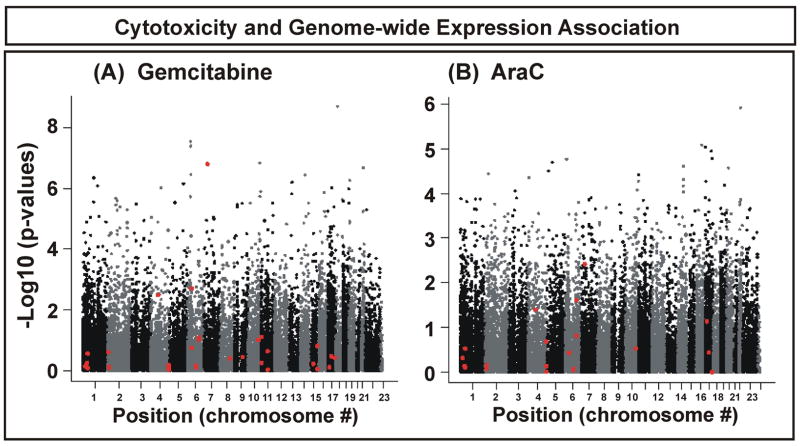
Correlations between basal gene expression and IC50 values for gemcitabine and AraC were determined to identify genes that might contribute to variation in cytotoxicity. The 26,653 RefSeq validated sequences among the 54,000 Affymetrix probe sets were used to perform these correlation studies. The p-values for association for gemcitabine tended to be smaller than those for AraC (Fig. 1A). With the exception of NT5C3, genes encoding proteins in the “cytidine analogue pathway” (Supplementary Table 1), shown as red points in Fig. 1, did not display highly significant p-values. NT5C3 had only one probe set, and it was significantly associated with gemcitabine IC50 values (p = 1.6×10−7) (Fig. 1A). NT5C3 encodes a member of the nucleotidase family that catalyzes the dephosphorylation of monophosphorylated drug metabolites, thus decreasing the concentration of active drug metabolites. NT5C3 expression showed a less significant association with AraC IC50 values although, among all genes within the “pathway” (Supplementary Table 1), it also had the smallest p-value for AraC (p = 0.004). Since most previous studies have focused only on pathway genes, we also estimated the effect of variation in expression for all known pathway genes on variation in IC50 values. Approximately 27% of the variation in gemcitabine IC50 values and approximately 11% of the variation for AraC could be explained by variation in gene expression within this intensively studied metabolic pathway.
Figure 1.
Association between expression array data and IC50 values for gemcitabine and AraC. Each dot on the y-axis represents the –log10 (p-value) for the probe set with the lowest p-value for each gene. Probe sets are plotted on the x-axis with regard to the chromosomal location of their genes. Red dots represent probe sets for genes listed in Supplementary Table 1 that encode proteins within the cytidine analogue metabolism and target pathway.
Among the 26,653 probe sets tested, 55 had p-values ≤ 10−6 for gemcitabine (adjusted multiple testing p-value = 0.0002) and 21 had p-values ≤ 10−5 (adjusted multiple testing p-value = 0.047). Since gemcitabine and AraC function in a similar fashion as anti-neoplastic drugs, we overlapped significant genes for both drugs. To identify top candidate genes for each drug that might be further characterized functionally, we used p values as a way to rank genes with regard to their association with drug cytotoxicity rather than establishing a particular “cutoff” value, since very few of the candidate genes could pass Bonferroni correction. Therefore, we arbitrarily used a p-value cutoff of <10−3 for AraC and <10−4 for gemcitabine to obtain a similar number of genes for each drug, realizing that we might still miss some true candidate genes. Thirty-one probe sets were identified that were common to both gemcitabine and AraC. Among those 31 probe sets, 14 encoding 12 genes were replicated when we used a different method of expression array normalization, Fastlo (Table 1). In addition, three “non-overlapping” genes with highly significant associations for either gemcitabine or AraC are also listed in Table 1.
Table 1.
Significant genes with expression that was associated with gemcitabine or AraC cytotoxicity (IC50 values). R values represent correlation coefficients. Q values represent the false discovery rate.
| Status of Probe Sets | Gene Name | Chromosome | Correlation between QRT-PCR and Microarray
(P-values) |
Gemcitabine | AraC | ||||
|---|---|---|---|---|---|---|---|---|---|
| P-value | R value | Q value | P-value | R value | Q value | ||||
| Specific Probe Sets | FKBP5 gemcitabine) | 6 | 0.045 | 4.12E-08 | −0.38 | 2.04E-04 | — | — | — |
| FKBP5 (gemcitabine) | 6 | 0.028 | 4.15E-08 | −0.38 | 2.04E-04 | — | — | — | |
| NT5C3 (gemcitabine) | 7 | 0.007 | 1.60E-07 | 0.365 | 5.23E-04 | — | — | — | |
| C14orf169 | 14 | 0.067 | 8.45E-06 | −0.313 | 3.25E-03 | 4.84E-05 | −0.285 | 6.96E-02 | |
| ESR2 | 14 | 0.023 | 3.61E-07 | 0.355 | 7.87E-04 | 3.40E-04 | 0.253 | 9.59E-02 | |
| GCAT | 22 | 0.011 | 2.08E-07 | −0.361 | 5.83E-04 | 1.39E-04 | −0.268 | 8.92E-02 | |
| INPP5F | 10 | 0.001 | 1.89E-06 | −0.334 | 1.69E-03 | 8.35E-05 | −0.277 | 8.92E-02 | |
| MYBBP1A | 17 | 0.009 | 5.65E-06 | −0.319 | 2.44E-03 | 9.24E-06 | −0.31 | 5.79E-02 | |
| MYBBP1A | 17 | 0.311 | 3.72E-05 | −0.291 | 7.30E-03 | 2.97E-04 | −0.255 | 8.92E-02 | |
| TLE4 | 9 | 0.003 | 3.11E-06 | −0.327 | 1.85E-03 | 1.77E-04 | −0.264 | 8.92E-02 | |
| ZNF278 | 22 | 0.251 | 5.67E-06 | −0.319 | 2.44E-03 | 1.20E-06 | −0.338 | 2.48E-02 | |
| Non-specific Probe Sets | ARL2BP | 16 | 0.339 | 1.37E-06 | −0.338 | 1.41E-03 | 8.20E-06 | −0.312 | 5.79E-02 |
| CENPB | 20 | 0.677 | 5.52E-07 | −0.349 | 9.85E-04 | 4.37E-04 | −0.248 | 1.01E-01 | |
| MAP4K4 | 2 | 0.118 | 2.57E-06 | −0.329 | 1.85E-03 | 1.74E-04 | −0.264 | 8.92E-02 | |
| MGMT | 10 | 0.232 | 1.30E-06 | 0.338 | 1.41E-03 | 2.21E-04 | 0.26 | 8.92E-02 | |
| TRRAP | 7 | 0.955 | 8.85E-06 | −0.312 | 3.26E-03 | 1.26E-04 | −0.27 | 8.92E-02 | |
| FKBP5 (gemcitabine) | 6 | 0.085 | 3.47E-07 | −0.35 | 7.8E-0.4 | — | — | — | |
| TPMT (AraC) | 6 | 0.387 | — | — | — | 1.72E-05 | 0.301 | 5.93E-02 | |
To verify expression array data for these 15 genes (18 probe sets), 20 lymphoblastoid cell lines were randomly selected to perform real-time QRT-PCR, and the results were compared with the expression array data. However, before performing QRT-PCR, we determined the specificity of these 18 probe sets by aligning the sequences of individual probes with the sequences of their presumed gene targets. We only verified probe specificity for top candidate genes that were considered for further functional validation because it was practically difficult to verify specificity for all 26,653 probe sets. Seven probe sets (Table 1) lacked specificity, defined as at least 5 of 11 probes that were “non-specific”. When we performed real time QRT-PCR and correlated RT-PCR with expression array for these non-specific probe sets, they did not correlate significantly (Table 1). Among the remaining 11 probe sets for 9 genes, 9 showed significant or near significant correlations between QRT-PCR and expression array data (p-value < 0.05), while two (MYBBP1A and ZNF278), failed to show a significant correlation (Table 1).
NT5C3 and FKBP5 candidate gene validation
To confirm results obtained during the association study, we selected two candidate genes for functional validation with specific siRNA, followed by cytotoxicity studies. These genes were selected on the basis of the significance of the observed association and whether the gene was within, or outside of the cytidine analogue metabolism pathway. The first was a pathway gene, NT5C3, a gene encoding an enzyme that dephosphorylates active cytidine analogue metabolites to form the inactive parent drug. Although NT5C3 is a “pathway” gene, no previous reports had suggested that NT5C3 might play a role in sensitivity to cytidine analogues, although other members of the nucleotidase family, e.g., NT5C and NT5C1A, have been associated with clinical response (21, 22). Our association study had shown a positive correlation between the NT5C3 expression levels and gemcitabine IC50 values (p = 1.6×10−7, Bonferroni corrected p-value = 0.0004, r = 0.365), indicating – as anticipated – that high expression of NT5C3 was associated with gemcitabine resistance.
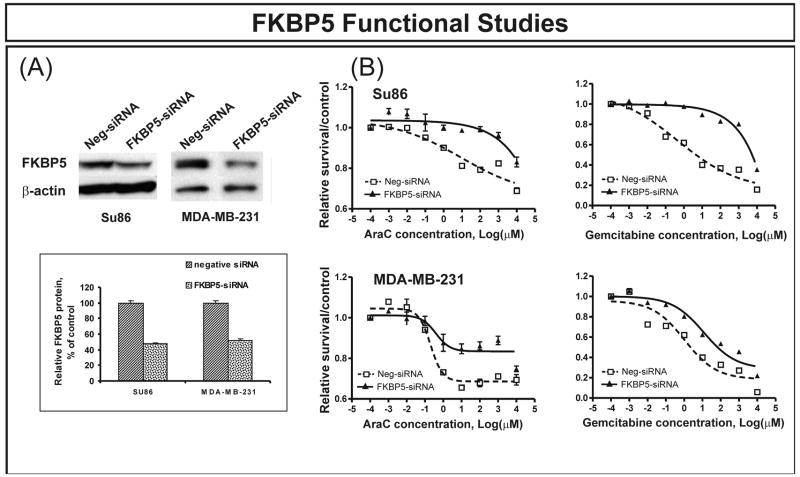
The second gene tested was from outside of the metabolic pathway, FKBP5, a gene encoding a 51 kDa immunophilin. FKBP5 is involved in steroid receptor maturation, and it is a binding partner for rapamycin (23–25). However, there had been no previous indication that FKBP5 might be involved in response to cytidine analogues. In contrast to NT5C3, expression levels for FKBP5 were negatively correlated with gemcitabine IC50 values (r = −0.38), indicating that increased FKBP5 transcription resulted in increased sensitivity to these drugs. Three Affymetrix probe sets targeted FKBP5, although we found that one probe set was not specific (Table 1). Both specific probe sets showed highly significant associations with gemcitabine IC50 values (p = 4.12×10−8 and 4.15×10−8, respectively). After Bonferroni correction, the p-values for these two FKBP5 probe sets remained significant (p = 0.0001). However, neither NT5C3 nor FKBP5 were significantly associated with AraC cytotoxicity (p = 3.47 × 10−3 for NT5C3 and p = 1.26 × 10−2 for FKBP5); although, as described subsequently, both proved to influence AraC response when tested functionally.
To confirm the possible functional significance of these two genes, we performed siRNA knockdown studies, followed by cytotoxicity assays, using two tumor cell lines to confirm the results of the association study and to extend our results beyond the lymphoblastoid cell lines to include cancer cell lines. Although neither gene displayed as significant an association with AraC as with gemcitabine cytotoxicity, functional studies were performed with both drugs. Although AraC is mainly used clinically to treat AML, previous studies have also used solid tumor cell lines for the analysis of both gemcitabine and AraC cytotoxicity (26). Two human cancer cell lines, the MDA-MB-231 breast cancer cell line and the pancreatic cancer SU86 cell line, were used to perform functional studies since gemcitabine is used to treat both types of cancers and since NT5C3 and FKBP5 showed their most significant associations with IC50 values for gemcitabine.
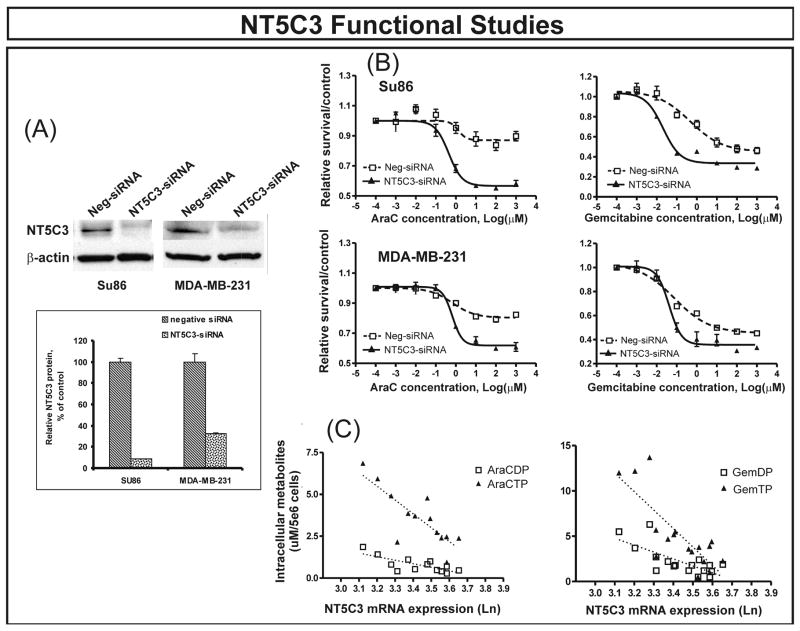
We first performed transient transfections with NT5C3 and FKBP5-specific siRNAs. Western blots verified that both genes were knocked down in both tumor cell lines (Fig. 2A and Fig. 3A). Gemcitabine and AraC cytotoxicity studies were then performed after transient transfection with siRNA. Down regulation of NT5C3 with specific siRNA shifted the dose response curve to the left compared with control siRNA transfection, indicating increased sensitivity to gemcitabine in both cell lines (Fig. 2B), consistent with results obtained during the genome-wide expression association study. In contrast, FKBP5 had demonstrated a negative correlation between level of expression and IC50 values. That relationship was confirmed by knockdown experiments performed with FKBP5-specific siRNA, since down regulation of FKBP5 in the both tumor cell lines desensitized the cells to both gemcitabine and AraC as compared with nonspecific siRNA transfection (Fig. 3B). We also performed siRNA studies with two genes that were not significantly associated with IC50 values for either drug as “negative controls”. One of those genes was FKBP1A, encoding FKBP12, a family member related to FKBP5, and the other was NT5C2, a gene encoding a family member of nucleotidases. Knockdown performed with specific siRNA for FKBP1A and NT5C2 did not significantly alter response to either gemcitabine or AraC (Supplementary Fig. 1).
Figure 2.
NT5C3 functional validation in two tumor cell lines. (A) Western blot analyses showing significantly decreased levels of NT5C3 protein in SU86 pancreatic cancer and MDA-MB-231 breast cancer cells after treatment with specific siRNAs. The “insert” shows average levels of protein as a percentage of control. Error bars represent SEM values for 3 experiments. (B) SU86 and MDA-MB-231 cytotoxicity. Both cell lines were sensitized to gemcitabine and AraC as determined by MTS assay after the down regulation of NT5C3 gene expression. Error bars represent SEM values for 3 independent experiments. (C) Levels of intracellular phosphorylated gemcitabine and AraC metabolites were correlated with NT5C3 gene expression in 14 randomly selected sensitive and resistant lymphoblastoid cell lines. [S2]Rp and p-values for metabolites, with expression adjusted for IC50, were RAraCDP = −0.50, pAraCDP = 0.084; RAraCTP = −0.65, pAraCTP = 0.016; RGemDP = −0.49, pGemDP = 0.045; and RGemTP = −0.52, pGemTP = 0.033. [S3]
Figure 3.
FKBP5 functional validation in two tumor cell lines. (A) Western blot analyses showing significantly decreased levels of FKBP5 protein in SU86 pancreatic cancer and MDA-MB-231 breast cancer cells after treatment with FKBP5-specific siRNA. The “insert” shows average levels of expressed protein as a percentage of control. Error bars represent SEM values for 3 experiments. (B) SU86 and MDA-MB-231 cytotoxicity. Both cell lines became more resistant to gemcitabine and AraC as determined by MTS cytotoxicity assay after the down regulation of FKBP5. Error bars represent SEM values for 3 independent experiments.
Characterization of NT5C3 and FKBP5 cytotoxicity mechanisms
The 5′-nucleotidases catalyze the dephosphorylation of nucleoside monophosphates and, as a result, inactivate active phosphorylated drug metabolites (22). Both clinical and in vitro studies suggest that an increase in nucleotidase activity can reverse nucleoside analogue metabolic activation, resulting in drug resistance (22, 27). Furthermore, NT5C3 hydrolyzes pyrimidine monophosphates like the active metabolites of gemcitabine and AraC (22, 28). As a result, the effect of NT5C3 on gemcitabine and AraC cytotoxicity could result from alterations in levels of active intracellular drug metabolites. Therefore, we randomly selected 7 sensitive and 7 resistant lymphoblastoid cell lines for gemcitabine and another 7 sensitive and 7 resistant lymphoblastoid cell lines for AraC to measure levels of active intracellular di- and the triphosphate metabolites for both drugs. Sensitive and resistant cell lines were defined as cell lines with IC50 values more than 0.85 SD from the mean of the IC50 distribution curve. Using this definition, we identified 32 resistant and 31sensitive cell lines for gemcitabine and 35 resistant and 38 sensitive cell lines for AraC. The sensitive and resistant cell lines used to measure intracellular metabolites were selected randomly from these two groups. HPLC was used to measure levels of intracellular phosphorylated metabolites in these cells after three days of treatment with gemcitabine or AraC. Concentrations of active metabolites were higher in sensitive than in resistant cell lines (p < 0.05) (Table 2). These results imply that genes within the cytidine analogue metabolic pathway, including the 5′-NTs, may contribute to the variation in cytotoxicity that we had observed. Equally important was the fact that intracellular metabolite concentrations were inversely related to levels of NT5C3 mRNA (Fig. 2C). This result was consistent with the conclusion that higher NT5C3 expression in these cells was associated with AraC and gemcitabine dephosphorylation, with decreased concentrations of active drug metabolites, resulting in drug resistance. These findings were also consistent with results of our NT5C3-siRNA functional studies (Fig. 2B).
Table 2.
Intracellular metabolites determined by HPLC in lymphoblastoid cells after incubation with (A) AraC or (B) gemcitabine. Metabolites are μM/5 × 106 cells.
| (A)
Cell Sample |
AraC IC50 (μM) | AraCDP | AraCTP | |||
|---|---|---|---|---|---|---|
| Resistant | Sensitive | Resistant | Sensitive | Resistant | Sensitive | |
| 1 | 7.2 | 0.24 | 0.28 | 1.42 | 0.95 | 5.94 |
| 2 | 13.8 | 0.65 | 0.70 | 1.87 | 2.47 | 6.86 |
| 3 | 20.0 | 0.96 | 0.49 | 0.81 | 2.73 | 4.92 |
| 4 | 20.4 | 1.06 | 0.41 | 1.00 | 2.39 | 3.56 |
| 5 | 26.0 | 1.99 | 1.34 | 0.42 | 2.64 | 2.16 |
| 6 | 51.3 | 2.73 | 0.46 | 1.11 | 2.39 | 3.87 |
| 7 | 76.6 | 2.77 | 0.83 | 0.54 | 4.78 | 3.72 |
| Average ± SEM | 30.8 ± 8.7 | 1.49 ± 0.36 | 0.65 ± 0.13 | 1.02 ± 0.18 | 2.62 ± 0.40 | 4.43 ± 0.56 |
| P-values | 0.020* | 0.133 | 0.032* | |||
|
(B)
Cell Sample |
Gemcitabine IC50 (nM) | GemDP | GemTP | |||
| Resistant | Sensitive | Resistant | Sensitive | Resistant | Sensitive | |
| 1 | 32.2 | 1.24 | 0.5 | 3.7 | 0.5 | 12.2 |
| 2 | 34.3 | 3.88 | 0.5 | 5.5 | 1.0 | 12.0 |
| 3 | 40.7 | 6.43 | 1.1 | 6.3 | 4.4 | 13.7 |
| 4 | 58.6 | 7.32 | 1.2 | 2.7 | 3.6 | 5.7 |
| 5 | 80.7 | 12.0 | 1.9 | 2.2 | 2.3 | 4.7 |
| 6 | 103 | 12.2 | 1.2 | 1.2 | 2.2 | 2.8 |
| 7 | 273.1 | 13.4 | 2.4 | 1.8 | 3.8 | 5.5 |
| Average ± SEM | 80.5 ± 26.8 | 7.61 ± 1.58 | 1.34 ± 0.24 | 3.14 ± 0.66 | 2.71 ± 0.51 | 7.49 ± 1.55 |
| P-values | 0.032* | 0.030* | 0.017* | |||
= indicates p < 0.05 between sensitive and resistant cell lines.
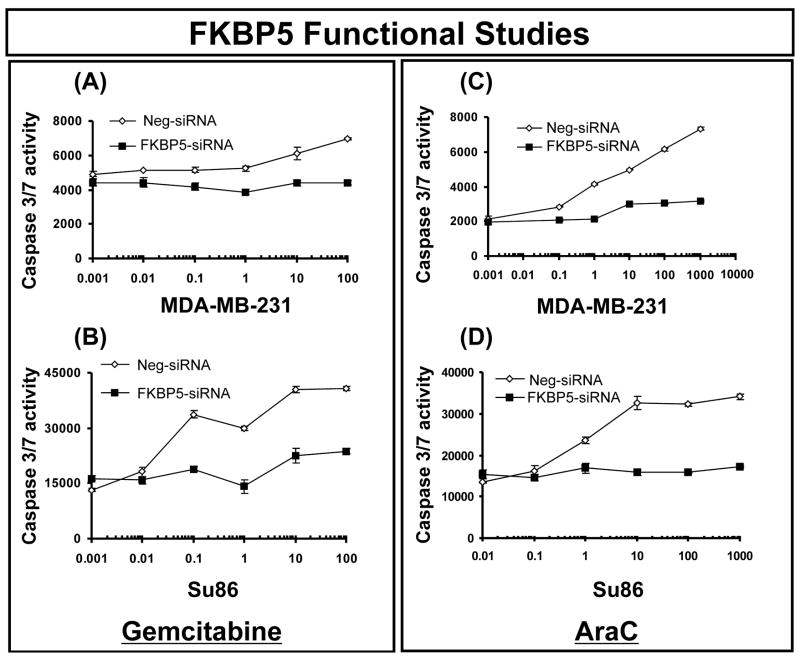
We next attempted to test hypotheses with regard to mechanisms by which FKBP5 might influence sensitivity to gemcitabine and AraC. One possibility would involve the blockade of apoptosis signaling pathways as a result of FKBP5 knockdown. We also performed caspase-3/7 activity assays to determine whether alterations in FKBP5 expression might influence apoptotic signaling, since previous studies had suggested that FKBP5 might be involved in apoptosis (29). Caspase-3/7 activity in FKBP5 siRNA-treated cells was significantly decreased after treatment with increasing concentrations of both gemcitabine and AraC when compared with cells treated with negative siRNA (Fig. 4), suggesting that activation of the apoptotic pathway was affected by the down regulation of FKBP5 expression.
Figure 4.
Caspase activity in SU86 and MDA-MB-231 cells after FKBP5 siRNA treatment. Caspase activity was measured using the Caspase-Glo® 3/7 activity assay in (A, C) for MDA-MB-231 cells and (B, D) for SU86 cells treated with FKBP5 siRNA in the presence of (A, B) gemcitabine or (C, D) AraC. Error bars represent SEM values for 3 independent experiments.
Gene network analysis
In an attempt to help define biological relationships among candidate genes identified during our study, we also applied Ingenuity Pathway Analysis (30). That analysis was focussed on top ranked genes based on p-values, using p-values < 10−5 for gemcitabine and < 10−4 for AraC that were significant with both GCRMA and Fastlo normalization methods. Eighty-four probe sets for gemcitabine and 75 for AraC were used to perform this analysis. A total of 9 and 20 networks were identified for gemcitabine and AraC, respectively. The “top” networks, with p-values < 0.05 based on Fisher’s exact text were associated with cell death, cancer, cell growth and proliferation, cell signaling, DNA replication, DNA recombination, DNA repair, and nucleic acid metabolism (Supplementary Table 2). On the basis of pathways with the highest number of significant candidate genes[S1], cancer was identified as the major disease, and cell signaling and the cell cycle were the major molecular and cellular functions identified. These preliminary results are clearly consistent with the use of these drugs to treat cancer.
Discussion
Drug response predictors have evolved significantly in the post-genomic era. Rather than using single genes, transcripts, proteins or metabolites to predict response, information from across the genome is now available to help predict drug response (4). Variation in response to chemotherapy results from a combination of factors that include gene sequence variation, ultimately resulting in differences in mRNA and protein expression, but also including differences in gender, ethnic group or environmental factors. Variation in mRNA is one important factor that may contribute to variation in response to chemotherapy (31). To screen for possible pharmacogenomic candidate genes that might contribute to variation in drug response, we tested the relationship between variation in basal gene expression and sensitivity to gemcitabine and AraC using Human Variation Panel cell lines, a model system designed to study common human genetic variation.
Gemcitabine and AraC share similar chemical structures and pathways of metabolism, but they are used in very different ways clinically (8, 32). Gemcitabine is mainly used to treat solid tumors, while AraC is first line chemotherapy for AML (7, 12, 13, 33, 34). Clinical response to these two cytidine analogues varies widely (8, 14, 15, 35). We set out to use the Human Variation Panel model system to identify candidate genes that might contribute to this variation. It should be emphasized once again that, although the tumor genome is critical for understanding disease pathophysiology and response to therapy, the germline genome is also critical, especially for response to drug therapy. In addition, variation in gene expression in lymphoblastoid cells is strongly influenced by inheritance (36 ). Therefore, these cell lines represent one model system suitable for the study of the contribution of “pharmacogenomic” variation in expression to individual variation in drug response.
Previous studies have demonstrated that variation in the expression of genes within the cytidine analogue metabolism and bioactivation pathway is associated with variation in response to gemcitabine and AraC (37–39). However, little information is available with regard to genes outside of this pathway. Therefore, rather than focus only on the known pathway, we used 26,635 expression probe sets to perform association analysis with drug response, in this case cytotoxicity. However, this type of approach will also produce false positive results if functional validation experiments, such as those which we applied, are not performed. Therefore, we first identified candidate genes, both within and outside of the known pathway, that might be important for response to these two cytidine analogues, followed by functional validation.
As a first step, our strategy involved obtaining expression data for 197 cell lines, followed by gemcitabine and AraC cytotoxicity assays. By correlating expression and IC50 values for both drugs, we identified genes both within and outside of the currently known cytidine analogue metabolic pathway that were significantly associated with cytotoxicity (Table 1). One pathway gene, NT5C3, a gene not previously identified as a factor in response to gemcitabine or AraC, showed a significant association with cytotoxicity. However, the overall contribution of expression for all pathway genes to variation in IC50 values was only about 27% for gemcitabine and 11% for AraC. These observations emphasize the potential advantage of genome-wide analyses rather than focusing entirely on known biological pathways. Although none of the genes listed in the Table 1 have been previously reported to be associated with gemcitabine or AraC response, our results complement and extend several previous reports of correlations of response to gemcitabine with gene expression profiles in cancer cell lines or tumor tissue samples obtained from patients (3, 40–43).
The lymphoblastoid cell lines used in our study are EBV transformed, so they are neither tumor cell lines nor tumor tissue. Therefore, one potential problem with the use of these cell lines is that EBV transformation could influence drug sensitivity and/or expression profiles, so we might miss some genes of importance, either because they are not expressed in these cell lines or, after transformation, are down-regulated. There is also evidence showing that gemcitabine and doxorubicin can induce the lytic form of EBV transformed cells (44). However, the fact that we were able to functionally validate the two candidate genes that we studied supports the feasibility of our approach. Furthermore, lymphoblastoid cells have been used to identify genetic variation associated with cytotoxicity for other antineoplastic drugs, including daunorubicin, another drug that also potentially induces EBV lytic forms (45, 46). Finally, in any high throughput association study, candidate genes identified require functional validation, as performed for our two candidate genes.
NT5C3 encodes a protein involved in cytidine metabolism, while FKBP5 lies outside of that pathway. Their novelty, and the fact that one of these genes is within, while the other is outside of our current sphere of knowledge, highlights the potential of this model system for hypothesis generation. Although several 5′-NT isoforms have been reported to be associated with AraC and gemcitabine response (22, 47, 48), the role of the “pyrimidine-specific” nucleotidase NT5C3 in response to these drugs is unexplored (28, 49). FKBP5 is a 51 kDa immunophilin (23, 50) that, like NT5C3, has never previously been reported to influence gemcitabine cytotoxicity. Our studies indicated that decreased expression of NT5C3 and FKBP5 after specific siRNA treatment altered response to both gemcitabine and AraC, but through different mechanisms. The functional characterization of both NT5C3 and FKBP5 provided biological evidence in support of their involvement in variation in gemcitabine and AraC cytotoxicity.
Finally, although p-values for AraC were not as significant as those for gemcitabine, functional studies of NT5C3 and FKBP5 demonstrated similar effects for both drugs. That observation reminds us of the limitations of p-values when determining true associations, as well as the fact that genome-wide association studies represent only one step in the identification of biomarkers to help predict clinically relevant variation in response to chemotherapy or new targets that might be used to enhance treatment outcomes. These results can now be applied in translational studies designed to test the hypothesis that variation in the expression of these two genes might be associated with clinically relevant variation in response to therapy with gemcitabine and/or AraC.
Supplementary Material
Acknowledgments
The authors thank Mrs. Luanne Wussow for her assistance with the preparation of this manuscript.
Supported in part by National Institutes of Health (NIH) grants GM61388 (The Pharmacogenetics Research Network), CA102701 (The Pancreatic Cancer SPORE), CA136780, an ASPET-Astellas Award and a PhRMA Foundation “Center of Excellence in Clinical Pharmacology” Award, and R01CA-132780.
References
- 1.Smid K, Bergman AM, Eijk PP, et al. Micro-array analysis of resistance for gemcitabine results in increased expression of ribonucleotide reductase subunits. Nucleosides Nucleotides Nucleic Acids. 2006;25:1001–7. doi: 10.1080/15257770600890269. [DOI] [PubMed] [Google Scholar]
- 2.Gullans SR. Connecting the dots using gene-expression profiles. N Engl J Med. 2006;355:2042–4. doi: 10.1056/NEJMcibr065953. [DOI] [PubMed] [Google Scholar]
- 3.Thuerigen O, Schneeweiss A, Toedt G, et al. Gene expression signature predicting pathologic complete response with gemcitabine, epirubicin, and docetaxel in primary breast cancer. J Clin Oncol. 2006;24:1839–45. doi: 10.1200/JCO.2005.04.7019. [DOI] [PubMed] [Google Scholar]
- 4.Weinshilboum RM, Wang L. Pharmacogenetics and pharmacogenomics: development, science, and translation. Annu Rev Genomics Hum Genet. 2006;7:223–45. doi: 10.1146/annurev.genom.6.080604.162315. [DOI] [PubMed] [Google Scholar]
- 5.Berlin JD, Catalano P, Thomas JP, Kugler JW, Haller DG, Benson AB., 3rd Phase III study of gemcitabine in combination with fluorouracil versus gemcitabine alone in patients with advanced pancreatic carcinoma: Eastern Cooperative Oncology Group Trial E2297. J Clin Oncol. 2002;20:3270–5. doi: 10.1200/JCO.2002.11.149. [DOI] [PubMed] [Google Scholar]
- 6.Kern W, Estey EH. High-dose cytosine arabinoside in the treatment of acute myeloid leukemia: Review of three randomized trials. Cancer. 2006;107:116–24. doi: 10.1002/cncr.21543. [DOI] [PubMed] [Google Scholar]
- 7.Wiley JS, Taupin J, Jamieson GP, Snook M, Sawyer WH, Finch LR. Cytosine arabinoside transport and metabolism in acute leukemias and T cell lymphoblastic lymphoma. J Clin Invest. 1985;75:632–42. doi: 10.1172/JCI111741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mini E, Nobili S, Caciagli B, Landini I, Mazzei T. Cellular pharmacology of gemcitabine. Ann Oncol. 2006;17 Suppl 5:v7–v12. doi: 10.1093/annonc/mdj941. [DOI] [PubMed] [Google Scholar]
- 9.Plunkett W, Huang P, Searcy CE, Gandhi V. Gemcitabine: preclinical pharmacology and mechanisms of action. Semin Oncol. 1996;23:3–15. [PubMed] [Google Scholar]
- 10.Heinemann V, Xu YZ, Chubb S, et al. Inhibition of ribonucleotide reduction in CCRF-CEM cells by 2′,2′-difluorodeoxycytidine. Mol Pharmacol. 1990;38:567–72. [PubMed] [Google Scholar]
- 11.Heinemann V, Xu YZ, Chubb S, et al. Cellular elimination of 2′,2′-difluorodeoxycytidine 5′-triphosphate: a mechanism of self-potentiation. Cancer Res. 1992;52:533–9. [PubMed] [Google Scholar]
- 12.Kindler HL. In focus: advanced pancreatic cancer. Clin Adv Hematol Oncol. 2005;3:420–2. [PubMed] [Google Scholar]
- 13.Castaigne S, Tilly H, Sigaux F, et al. Treatment of malignant hemopathies with aracytine in low doses. Analysis of 159 cases. Nouv Rev Fr Hematol. 1985;27:377–82. [PubMed] [Google Scholar]
- 14.Braess J, Jahns-Streubel G, Schoch C, et al. Proliferative activity of leukaemic blasts and cytosine arabinoside pharmacodynamics are associated with cytogenetically defined prognostic subgroups in acute myeloid leukaemia. Br J Haematol. 2001;113:975–82. doi: 10.1046/j.1365-2141.2001.02866.x. [DOI] [PubMed] [Google Scholar]
- 15.Schoch C, Haferlach T, Haase D, et al. Patients with de novo acute myeloid leukaemia and complex karyotype aberrations show a poor prognosis despite intensive treatment: a study of 90 patients. Br J Haematol. 2001;112:118–26. doi: 10.1046/j.1365-2141.2001.02511.x. [DOI] [PubMed] [Google Scholar]
- 16.Seve P, Mackey JR, Isaac S, et al. cN-II expression predicts survival in patients receiving gemcitabine for advanced non-small cell lung cancer. Lung Cancer. 2005;49:363–70. doi: 10.1016/j.lungcan.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 17.Rosell R, Danenberg KD, Alberola V, et al. Ribonucleotide reductase messenger RNA expression and survival in gemcitabine/cisplatin-treated advanced non-small cell lung cancer patients. Clin Cancer Res. 2004;10:1318–25. doi: 10.1158/1078-0432.ccr-03-0156. [DOI] [PubMed] [Google Scholar]
- 18.van Haperen VW, Veerman G, Vermorken JB, Pinedo HM, Peters G. Regulation of phosphorylation of deoxycytidine and 2′,2′-difluorodeoxycytidine (gemcitabine); effects of cytidine 5′-triphosphate and uridine 5′-triphosphate in relation to chemosensitivity for 2′,2′-difluorodeoxycytidine. Biochem Pharmacol. 1996;51:911–8. doi: 10.1016/0006-2952(95)02402-6. [DOI] [PubMed] [Google Scholar]
- 19.Ballman KV, Grill DE, Oberg AL, Therneau TM. Faster cyclic loess: normalizing RNA arrays via linear models. Bioinformatics. 2004;20:2778–86. doi: 10.1093/bioinformatics/bth327. [DOI] [PubMed] [Google Scholar]
- 20.Zhijin Wu RAI, Rober Gentleman, Francisco Martinez-Murillo. A model-based background adjustment for oligonucleotide. Forrest Spencer Journal of the American Sttatistical Association. 2004;99:909. [Google Scholar]
- 21.Borowiec A, Lechward K, Tkacz-Stachowska K, Skladanowski AC. Adenosine as a metabolic regulator of tissue function: production of adenosine by cytoplasmic 5′-nucleotidases. Acta Biochim Pol. 2006;53:269–78. [PubMed] [Google Scholar]
- 22.Hunsucker SA, Mitchell BS, Spychala J. The 5′-nucleotidases as regulators of nucleotide and drug metabolism. Pharmacol Ther. 2005;107:1–30. doi: 10.1016/j.pharmthera.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 23.Baughman G, Wiederrecht GJ, Campbell NF, Martin MM, Bourgeois S. FKBP51, a novel T-cell-specific immunophilin capable of calcineurin inhibition. Mol Cell Biol. 1995;15:4395–402. doi: 10.1128/mcb.15.8.4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheung J, Smith DF. Molecular chaperone interactions with steroid receptors: an update. Mol Endocrinol. 2000;14:939–46. doi: 10.1210/mend.14.7.0489. [DOI] [PubMed] [Google Scholar]
- 25.Cheung-Flynn J, Roberts PJ, Riggs DL, Smith DF. C-terminal sequences outside the tetratricopeptide repeat domain of FKBP51 and FKBP52 cause differential binding to Hsp90. J Biol Chem. 2003;278:17388–94. doi: 10.1074/jbc.M300955200. [DOI] [PubMed] [Google Scholar]
- 26.Bergman AM, Munch-Petersen B, Jensen PB, et al. Collateral sensitivity to gemcitabine (2′,2′-difluorodeoxycytidine) and cytosine arabinoside of daunorubicin- and VM-26-resistant variants of human small cell lung cancer cell lines. Biochem Pharmacol. 2001;61:1401–8. doi: 10.1016/s0006-2952(01)00627-x. [DOI] [PubMed] [Google Scholar]
- 27.Wallden K, Stenmark P, Nyman T, et al. Crystal structure of human cytosolic 5′-nucleotidase II: insights into allosteric regulation and substrate recognition. J Biol Chem. 2007;282:17828–36. doi: 10.1074/jbc.M700917200. [DOI] [PubMed] [Google Scholar]
- 28.Galmarini CM, Cros E, Thomas X, Jordheim L, Dumontet C. The prognostic value of cN-II and cN-III enzymes in adult acute myeloid leukemia. Haematologica. 2005;90:1699–701. [PubMed] [Google Scholar]
- 29.Giraudier S, Chagraoui H, Komura E, et al. Overexpression of FKBP51 in idiopathic myelofibrosis regulates the growth factor independence of megakaryocyte progenitors. Blood. 2002;100:2932–40. doi: 10.1182/blood-2002-02-0485. [DOI] [PubMed] [Google Scholar]
- 30.Bush CR, Havens JM, Necela BM, et al. Functional genomic analysis reveals cross-talk between peroxisome proliferator-activated receptor gamma and calcium signaling in human colorectal cancer cells. J Biol Chem. 2007;282:23387–401. doi: 10.1074/jbc.M702708200. [DOI] [PubMed] [Google Scholar]
- 31.Potti A, Dressman HK, Bild A, et al. Genomic signatures to guide the use of chemotherapeutics. Nat Med. 2006;12:1294–300. doi: 10.1038/nm1491. [DOI] [PubMed] [Google Scholar]
- 32.Wiley JS, Jones SP, Sawyer WH, Paterson AR. Cytosine arabinoside influx and nucleoside transport sites in acute leukemia. J Clin Invest. 1982;69:479–89. doi: 10.1172/JCI110472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Moorsel CJ, Bergman AM, Veerman G, et al. Differential effects of gemcitabine on ribonucleotide pools of twenty-one solid tumour and leukaemia cell lines. Biochim Biophys Acta. 2000;1474:5–12. doi: 10.1016/s0304-4165(99)00209-3. [DOI] [PubMed] [Google Scholar]
- 34.Li D, Xie K, Wolff R, Abbruzzese JL. Pancreatic cancer. Lancet. 2004;363:1049–57. doi: 10.1016/S0140-6736(04)15841-8. [DOI] [PubMed] [Google Scholar]
- 35.Galmarini CM, Thomas X, Calvo F, et al. In vivo mechanisms of resistance to cytarabine in acute myeloid leukaemia. Br J Haematol. 2002;117:860–8. doi: 10.1046/j.1365-2141.2002.03538.x. [DOI] [PubMed] [Google Scholar]
- 36.Dixon AL, Liang L, Moffatt MF, et al. A genome-wide association study of global gene expression. Nat Genet. 2007 doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 37.Bergman AM, Pinedo HM, Peters GJ. Determinants of resistance to 2′,2′-difluorodeoxycytidine (gemcitabine) Drug Resist Updat. 2002;5:19–33. doi: 10.1016/s1368-7646(02)00002-x. [DOI] [PubMed] [Google Scholar]
- 38.King AE, Ackley MA, Cass CE, Young JD, Baldwin SA. Nucleoside transporters: from scavengers to novel therapeutic targets. Trends Pharmacol Sci. 2006;27:416–25. doi: 10.1016/j.tips.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 39.Achiwa H, Oguri T, Sato S, Maeda H, Niimi T, Ueda R. Determinants of sensitivity and resistance to gemcitabine: the roles of human equilibrative nucleoside transporter 1 and deoxycytidine kinase in non-small cell lung cancer. Cancer Sci. 2004;95:753–7. doi: 10.1111/j.1349-7006.2004.tb03257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Akada M, Crnogorac-Jurcevic T, Lattimore S, et al. Intrinsic chemoresistance to gemcitabine is associated with decreased expression of BNIP3 in pancreatic cancer. Clin Cancer Res. 2005;11:3094–101. doi: 10.1158/1078-0432.CCR-04-1785. [DOI] [PubMed] [Google Scholar]
- 41.Giroux V, Malicet C, Barthet M, et al. p8 is a new target of gemcitabine in pancreatic cancer cells. Clin Cancer Res. 2006;12:235–41. doi: 10.1158/1078-0432.CCR-05-1700. [DOI] [PubMed] [Google Scholar]
- 42.Toshimitsu H, Iizuka N, Yamamoto K, et al. Molecular features linked to the growth-inhibitory effects of gemcitabine on human pancreatic cancer cells. Oncol Rep. 2006;16:1285–91. [PubMed] [Google Scholar]
- 43.Hernandez-Vargas H, Rodriguez-Pinilla SM, Julian-Tendero M, et al. Gene expression profiling of breast cancer cells in response to gemcitabine: NF-kappaB pathway activation as a potential mechanism of resistance. Breast Cancer Res Treat. 2007;102:157–72. doi: 10.1007/s10549-006-9322-9. [DOI] [PubMed] [Google Scholar]
- 44.Feng WH, Hong G, Delecluse HJ, Kenney SC. Lytic induction therapy for Epstein-Barr virus-positive B-cell lymphomas. J Virol. 2004;78:1893–902. doi: 10.1128/JVI.78.4.1893-1902.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Duan S, Bleibel WK, Huang RS, et al. Mapping genes that contribute to daunorubicin-induced cytotoxicity. Cancer Res. 2007;67:5425–33. doi: 10.1158/0008-5472.CAN-06-4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang RS, Duan S, Kistner EO, et al. Genetic variants contributing to daunorubicin-induced cytotoxicity. Cancer Res. 2008;68:3161–8. doi: 10.1158/0008-5472.CAN-07-6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Galmarini CM, Thomas X, Calvo F, et al. Potential mechanisms of resistance to cytarabine in AML patients. Leuk Res. 2002;26:621–9. doi: 10.1016/s0145-2126(01)00184-9. [DOI] [PubMed] [Google Scholar]
- 48.Galmarini CM, Graham K, Thomas X, et al. Expression of high Km 5′-nucleotidase in leukemic blasts is an independent prognostic factor in adults with acute myeloid leukemia. Blood. 2001;98:1922–6. doi: 10.1182/blood.v98.6.1922. [DOI] [PubMed] [Google Scholar]
- 49.Manco L, Ribeiro ML. Gene symbol: NT5C3. Disease: pyrimidine 5′-nucleotidase (P5′N) deficiency. Hum Genet. 2006;119:673–4. [PubMed] [Google Scholar]
- 50.Yeh WC, Li TK, Bierer BE, McKnight SL. Identification and characterization of an immunophilin expressed during the clonal expansion phase of adipocyte differentiation. Proc Natl Acad Sci U S A. 1995;92:11081–5. doi: 10.1073/pnas.92.24.11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.