Abstract
Background
Staphylococcus aureus (S. aureus), is responsible for many infectious diseases, ranging from benign skin infections to life-threatening endocarditis and toxic shock syndrome. Ortho-phenylphenol (OPP) is an antimicrobial agent and an active ingredient of EPA-registered disinfectants with wide human exposure in various agricultural, hospital and veterinary disinfectant products. Despite many uses, an understanding of a cellular response to OPP and it's mechanism of action, targeted genes, and the connectivity between targeted genes and the rest of cell metabolism remains obscure.
Results
Herein, we performed a genome-wide transcriptome analysis of the cellular responses of S. aureus when exposed to 0.82 mM of OPP for 20 and 60 min. Our data indicated that OPP downregulated the biosynthesis of many amino acids, which are required for protein synthesis. In particular, the genes encoding the enzymes of the diaminopimelate (DAP) pathway which results in lysine biosynthesis were significantly downregualted. Intriguingly, we revealed that the transcription of genes encoding ribosomal proteins was upregulated by OPP and at the same time, the genes encoding iron acquisition and transport were downregulated. The genes encoding virulence factors were upregulated and genes encoding phospholipids were downregulated upon 20 min exposure to OPP.
Conclusion
By using microarray analysis that enables us to simultaneously and globally examine the complete transcriptome during cellular responses, we have revealed novel information regarding the mode of action of OPP on Staphylococcus: OPP inhibits anabolism of many amino acids and highly downregulates the genes that encode the enzymes involved in the DAP pathway. Lysine and DAP are essential for building up the peptidoglycan cell wall. It was concluded that the mode of action of OPP is similar to the mechanism of action of some antibiotics. The discovery of this phenomenon provides useful information that will benefit further antimicrobial research on S. aureus.
Background
The U.S. Environmental Protection Agency (EPA) has endeavored to determine the efficacy and the mode of action of antimicrobials. At EPA, 5,000 antimicrobial products are registered, and hospital-level disinfectants are being tested against pathogens such as S. aureus, which is responsible for many infectious diseases, ranging from benign skin infections to life-threatening endocarditis and toxic shock syndrome [1]. One of the reasons EPA has exerted such efforts is that hospital-acquired infections are a serious threat to public health. Therefore, it is important to use appropriate antimicrobial agents with clear understanding of the subsequent effects to prevent infection outbreaks in health care environments [2].
The phenolic compound, ortho-phenylphenol (OPP), is an antimicrobial agent and an active ingredient of EPA-registered disinfectant with wide human exposure in various agricultural, hospital and veterinary disinfectant products. OPP is employed in a variety of applications, including hard surface disinfection, wood preservation, treatment of citrus fruit, vegetables before packaging to prevent microbial decay and textile production due to its bactericidal and fungicidal activity [3-5].
There have been several reports related to the exposure of OPP on humans. It has been reported that OPP increased the incidence of urinary bladder tumors in F344/DuCrj rats when administered in the diet [6]. The results of this study stimulated the initiation of additional testing of OPP for both tumor induction and possible reactivity with DNA. OPP has been found to have estrogenic or anti-androgenic activity, and binds to the androgen or estrogen receptors [7]. In spite of these effects OPP is still used in applications that simultaneously contact both humans and bacteria. It is therefore important to understand the differential effects on each so that its efficacy can be understood and even optimized.
Moreover, a lack of understanding of a cellular response to OPP hinders further development of more innovative methods for combating pathogens. Certainly, better elucidation of the molecular events responsible for establishing and maintaining pathogenicity will help to map affected cell functions and serve to delineate the mechanisms involved in the disinfectant activity.
Microarrays have been effectively employed to simultaneously and globally examine the complete transcriptional response at the genomic level in Pseudomonas aeruginosa and S. aureus upon exposure to antimicrobials [8-15].
In this study, to our knowledge, for the first time, we show that the global transcription response of S. aureus to OPP includes downregulation of genes involved in lysine metabolism, as well as genes involved in amino acid metabolism, by utilizing Affymetrix S. aureus GeneChip arrays. Our findings indicate that: (i) many cellular protective processes were upregulated, (ii) the transcription of genes involved in primary metabolic pathways was downregulated, and (iii) the transcription of genes encoding lysine and histidine biosynthesis was downregulated. Next we performed real-time PCR analysis on selected genes to validate the array results. Based on this result, it was concluded that this study may help to elucidate the mechanism of action by which OPP stops cell wall construction and thereby inhibiting S. aureus growth, and may facilitate the design of more effective antimicrobials.
Results and discussion
Growth inhibition by OPP
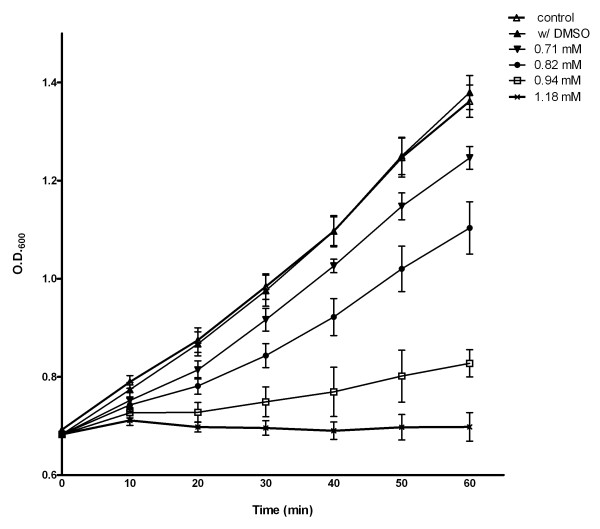
To determine the sublethal inhibitory effect of OPP on S. aureus, we first exposed the exponentially growing cells to different concentrations of OPP dissolved in DMSO (0 up to 1.18 mM). In figure 1, we demonstrate that 0.82 mM concentration of OPP caused a growth inhibition for about 20 min. Note that minimum inhibitory concentration (MIC50) of OPP on S. aureus was reportedly 500 mg/l (3 mM) [16]. In this study, to better understand how S. aureus initially responds to OPP, we chose the rate of cell growth inhibition with 0.82 mM OPP after 20 and 60 min exposure times compared to control (without OPP).
Figure 1.
S. aureus growth (optical density at 600 nm) after treatment with OPP: control (open triangles), with DMSO (closed triangles), 0.71 mM (inverted closed triangles), 0.82 mM (closed circles), 0.94 mM (open squares), and 1.18 mM OPP (X). Growth inhibition was performed during the exponential phase of the cells without DMSO (control) and control with DMSO, 0.71 mM, 0.82 mM, 0.94 mM, and 1.18 mM OPP dissolved in DMSO. The results are the mean of triplicate experiments; the error bars represent standard deviation.
Transcriptional profiles in response to OPP
To investigate early transcriptional changes in response to OPP exposure, we isolated total RNA after 20 min and 60 minutes exposure to 0.82 mM OPP and conducted five independent microarray experiments in the absence (control) and the presence (experimental) of 0.82 mM OPP (see figure 1). To further identify genes with statistically marked changes in expression levels, we applied the following criteria to each of the 20 min, 60 min, and control-experimental microarray data sets: (i) a p-value for a t-test should be equal to or less than 0.05, (ii) an absolute fold change in transcript level should be equal to or greater than 2, and (iii) a gene should have a presence or marginal call [17] from 50% or more replicates on both the experimental and control replicate sets. Of the 7,775 genes represented on the S. aureus GeneChip, 2,348 genes showed statistical significance based on a 1-way ANOVA. We found that mRNA levels 669 genes of S. aureus were significantly altered in response to OPP by two fold or more upregulation or downregulation. The raw data of 7,775 genes control (0 min) and experimental genes after (20 and 60 min exposure to 0.82 mM of OPP) has been deposited in NCBI's Gene Expression Omnibus [18] and is accessible through GEO Series accession number GSE10605 (Additional file 1).
Analysis of gene expression changes in 20 min and 60 min
To examine how genes with transcript level changes are distributed with regard to their functions, we further classified these 669 genes that were either upregulated or downregulated by a fold change of two or more according to the Gene Classification based on COG functional categories in the genome of National Center for Biotechnology information (NCBI) [19] (see also Additional file 2).
In Figure 2, the differences between the numbers of up and downregulated genes in each functional class after 20 and 60 minutes exposure to 0.82 mM of OPP are illustrated. Note that Figure 2 represents a total of 669 genes including the group of "function unknown (36), hypothetical protein (132) and general function predicted only (70)". Some interesting findings are as follows: (i) the genes of amino acid transport and metabolism were highly downregulated at both 20 and 60 min; (ii) the genes of inorganic ion transport and metabolism were downregulated at 20 min and decrease in the number of genes downregulated at 60 min was also observed; (iii) the genes in the class of "translation, ribosomal structure and biogenesis" were significantly upregulated at 20 min; (iv) the number of genes involved in nucleotide transport and metabolism were increased after 20 min compared to after 60 min. In general, figure 2 illustrates that the functional classes contained more downregulated and fewer upregulated genes at 20 min. This result suggests that the functional class profiles were notably different between 20 and 60 min, and this difference might explain why S. aureus underwent the initial growth inhibition followed by partial growth recovery upon exposure to OPP.
Figure 2.
Functional classification of genes with statistically significant upregulated (red) and downregulated (green) when exposed to 0.82 mM OPP at 20 min and 60 min (a total of 669 genes). The number in parenthesis represents the total number of genes affected within the genome in each functional class.
Functional classifications analysis
To further identify genes with similar transcription patterns during the time course, we removed 238 genes (including the group of functional unknown (36), hypothetical protein (132) and general function predicted only (70)). We categorized 431 genes with known functions into 6 groups on the basis of their transcription directions (figure 3). Briefly, group I contained 23 genes upregulated upon both exposure times, while group II had 80 genes with increased expression levels at 20 min and no significant changes upon 60 min exposure. Further, group III possessed 26 genes that were upregulated at 60 min exposure. Group IV contained 128 genes downregulated upon both exposure times, whereas 95 genes of group V exhibited downregulation after 20 min. Finally, group VI had 79 genes that were downregulated upon 60 min exposure. Figure 4 displays the number of genes (431) within groups I through VI in each functional class. As indicated above, additional file 2 contains all 669 genes including the genes classified under the functional group designated as "unknown, hypothetical, and general function prediction only".
Figure 3.

Groups of differentially regulated 431 genes with known functional class, which are categorized by their transcription directions upon 20 and 60 min exposures. Group I contained 23 genes upregulated upon both exposure times, while group II had 80 genes upregulated at 20 min and no significant changes upon 60 min exposure. Further, group III possessed 26 genes that were upregulated in response to 60 min exposure. Group IV contained 128 genes downregulated upon both exposure times, whereas 95 genes of group V exhibited downregulation after 20 min exposure. Finally, group VI had 79 genes that were downregulated upon 60 min exposure.
Figure 4.
Functional classification of genes with statistically significant upregulated (red) and downregulated (green) upon 20 min and 60 min exposures (a total of 431 genes). Note that the functional classes of "hypothetical genes", "general function prediction only" and "function unknown" are not included in this figure.
Since most of the genes discussed in this report are in additional file 2, for further analysis of the data and for the readers convenience, we decided to make table 1 with 138 S. aureus genes that were most strongly upregulated or downregulated in response to OPP after 20 and 60 minutes exposure. These genes were also classified under seven groups based on their transcription directions
Table 1.
List of 138 S. aureus genes that were most strongly affected by OPP and are discussed in this report categorized by their related function. The microarray results are the mean of five replicates of each gene.
| 20 min | 60 min | |||||||
| Affymetrix Probe ID | ORF no. | p-value | Fold change | p-value | Fold change | Description | Gene symbol | Functional class |
| Group I: Upregulation (20 min) – Upregulation (60 min) 18 genes | ||||||||
| sa_c6812s5946_a_at | SA0265 | 8.55E-06 | 2.1 | 8.55E-06 | 3.1 | peptidoglycan hydrolase (surface antigen) | lytM | Cell envelope biogenesis, outer membrane |
| sa_c7382s10191_a_at | SA0423* | 5.16E-07 | 8.1 | 5.16E-07 | 19.4 | hypothetical protein, similar to autolysin (N-acetylmuramoyl-L-alanine amidase) | General function prediction only | |
| sa_c7698s6703_a_at | SA0519 | 0.00293 | 2.8 | 0.00293 | 2.5 | Ser-Asp rich fibrinogen-binding, bone sialoprotein-binding protein | sdrC | Cell envelope biogenesis, outer membrane |
| sa_c8045s7032_at | SA0620 | 1.27E-06 | 7.6 | 1.27E-06 | 5.6 | hypothetical protein, similar to secretory antigen precursor SsaA | General function prediction only | |
| sa_c592s9345_a_at | SA0905 | 4.83E-08 | 5.7 | 4.83E-08 | 7.0 | N-acetylglucosaminidase (major autolysin) | atl | Cell envelope biogenesis, outer membrane |
| sa_c1007s793_a_at | SA1003 | 7.81E-06 | 3.7 | 7.81E-06 | 4.2 | fibrinogen-binding protein precursor | hypothetical protein | |
| sa_c4394s3743_a_at | SA1898 | 7.99E-05 | 6.1 | 7.99E-05 | 5.8 | hypothetical protein, simialr to SceD precursor | hypothetical protein | |
| sa_c4612s9984cs_s_at | SA1972 | 0.00267 | 2.2 | 0.00267 | 2.7 | multidrug resistance protein (efflux transporter) | hypothetical protein | |
| sa_c5066s4362_a_at | SA2093* | 0.00025 | 6.8 | 0.00025 | 9.0 | hypothetical protein, similar to secretory antigen precursor SsaA | ssaA | General function prediction only |
| sa_c5082s4380_a_at | SA2097* | 3.71E-05 | 6.9 | 3.71E-05 | 11.5 | hypothetical protein, similar to secretory antigen precursor SsaA | General function prediction only | |
| sa_c342s182_a_at | SA2142 | 9.36E-07 | 3.6 | 9.36E-07 | 3.6 | multidrug resistance protein B (drug efflux transporter) | hypothetical protein | |
| sa_c5274s4572_a_at | SA2143 | 1.24E-07 | 3.9 | 1.24E-07 | 4.1 | multidrug resistance efflux pump | hypothetical protein | |
| sa_c5652s4904_a_at | SA2206 | 0.00082 | 2.5 | 0.00082 | 2.8 | Immunoglobulin G binding protein A precursor | sbi | Cell envelope biogenesis, outer membrane |
| sa_c6151s5333_a_at | SA2332 | 0.00019 | 7.8 | 0.00019 | 8.6 | hypothetical protein, similar to secretory antigen precursor SsaA | General function prediction only | |
| sa_c6250s5428_a_at | SA2353* | 1.91E-05 | 10.2 | 1.91E-05 | 13.8 | secretory antigen precursor SsaA homolog | General function prediction only | |
| sa_c9402s8223_a_at | SA2355* | 3.23E-06 | 8.5 | 3.23E-06 | 9.4 | transcriptional regulator, MARR family | hypothetical protein | |
| sa_c6259s5439_a_at | SA2356 | 3.43E-06 | 6.5 | 3.43E-06 | 6.9 | immunodominant antigen A | isaA | Cell envelope biogenesis, outer membrane |
| sa_c6506s5675_a_at | SA2423 | 0.00148 | 2.9 | 0.00148 | 3.5 | fibrinogen-binding protein A, clumping factor | clfB | Posttranslational modification, protein turnover, chaperones |
| Group II: Upregulation (20 min) – No change (60 min) 28 genes | ||||||||
| sa_c7511s6531_a_at | SA0459 | 0.000138 | 2.3 | ribosomal protein L25 (general stress protein Ctc) | rplY | Translation, ribosomal structure and biogenesis | ||
| sa_c7621s6634_a_at | SA0497 | 7.95E-05 | 2.2 | 50S ribosomal protein L10; ribosomal protein L10 (BL5) | Translation, ribosomal structure and biogenesis | |||
| sa_c7625s6638_at | SA0498 | 5.07E-05 | 3.5 | 50S ribosomal protein L7:L12; ribosomal protein L7:L12 | Translation, ribosomal structure and biogenesis | |||
| sa_c1147s928_a_at | SA1041 | 0.00142 | 2.8 | uracil phosphoribosyltransferase; Pyrimidine operon regulatory protein pyrR | Nucleotide transport and metabolism | |||
| sa_c1151s932_a_at | SA1042 | 0.000143 | 6.7 | uracil permease (uracil transporter) | pyrP | Nucleotide transport and metabolism | ||
| sa_c9991s8687_a_at | SA1043 | 7.66E-05 | 6.5 | aspartate carbamoyltransferase catalytic chain (Aspartate transcarbamylase) (ATCase) | pyrB | Nucleotide transport and metabolism | ||
| sa_c1155s937_a_at | SA1044 | 0.000101 | 6.0 | dihydroorotase, dihydroorotase-like | pryC | Nucleotide transport and metabolism | ||
| sa_c1159s942_a_at | SA1045 | 0.000161 | 5.1 | carbamoyl-phosphate synthase, arginine-specific, small chain | pyrAA | Amino acid transport and metabolism, Nucleotide transport and metabolism | ||
| sa_c1165s946_a_at | SA1046 | 0.00018 | 3.3 | carbamoyl-phosphate synthase, arginine-specific, large chain | carB | Amino acid transport and metabolism, Nucleotide transport and metabolism | ||
| sa_c1167s950_a_at | SA1047 | 0.000788 | 3.3 | orotidine 5-phosphate decarboxylase | pyrF | Nucleotide transport and metabolism | ||
| sa_c9989s8682_a_at | SA1048 | 0.00107 | 3.0 | orotate phosphoribosyltransferase | pyrE | Nucleotide transport and metabolism | ||
| sa_c1302s1077_a_at | SA1084 | 0.000221 | 2.7 | 50S ribosomal protein L19; ribosomal protein L19 | rplS | Translation, ribosomal structure and biogenesis | ||
| sa_c4792s4098_at | SA2022 | 0.000417 | 2.1 | 50S ribosomal protein L17; ribosomal protein L17 | rplQ | Translation, ribosomal structure and biogenesis | ||
| sa_c4824s4130_a_at | SA2029 | 2.76E-05 | 2.1 | 50S ribosomal protein L15; ribosomal protein L15 | rplO | Translation, ribosomal structure and biogenesis | ||
| sa_c4836s4142_at | SA2032 | 0.00024 | 2.1 | 50S ribosomal protein L18; ribosomal protein L18 | rplR | Translation, ribosomal structure and biogenesis | ||
| sa_c9951s8647_at | SA2033 | 0.000342 | 2.6 | 50S ribosomal protein L6; ribosomal protein L6 (BL8) | rplF | Translation, ribosomal structure and biogenesis | ||
| sa_c4848s4156_at | SA2035 | 0.0021 | 2.1 | 50S ribosomal protein L5; ribosomal protein L5 (BL6) | rpsN | Translation, ribosomal structure and biogenesis | ||
| sa_c4852s4158_at | SA2036 | 0.00267 | 2.1 | 50S ribosomal protein L24; ribosomal protein L24 (BL23) | rplX | Translation, ribosomal structure and biogenesis | ||
| sa_c10191s8871_a_at | SA2038 | 0.0026 | 2.8 | 30S ribosomal protein S17; ribosomal protein S17 (BS16) | rpsQ | Translation, ribosomal structure and biogenesis | ||
| sa_c4860s4166_at | SA2039 | 0.00204 | 2.5 | 50S ribosomal protein L29; ribosomal protein L29 | rpmC | Translation, ribosomal structure and biogenesis | ||
| sa_c4864s4170_at | SA2040 | 0.000749 | 2.7 | 50S ribosomal protein L16; ribosomal protein L16 | rplP | Translation, ribosomal structure and biogenesis | ||
| sa_c4868s4175_a_at | SA2041 | 0.0024 | 2.8 | 30S ribosomal protein S3; ribosomal protein S3 (BS3) | rpsC | Translation, ribosomal structure and biogenesis | ||
| sa_c4876s4184_at | SA2043 | 0.00405 | 2.8 | 30S ribosomal protein S19; ribosomal protein S19 (BS19) | rpsS | Translation, ribosomal structure and biogenesis | ||
| sa_c9959s8654_a_at | SA2044 | 0.000888 | 2.5 | 50S ribosomal protein L2; ribosomal protein L2 (BL2) | rplB | Translation, ribosomal structure and biogenesis | ||
| sa_c10192s8875_a_at | SA2045 | 0.00434 | 2.7 | 50S ribosomal protein L23; ribosomal protein L23 | rplW | Translation, ribosomal structure and biogenesis | ||
| sa_c4880s4187_at | SA2046 | 0.00283 | 2.8 | 50S ribosomal protein L4; ribosomal protein L4 | rplD | Translation, ribosomal structure and biogenesis | ||
| sa_c4888s4195_a_at | SA2047 | 0.00069 | 2.3 | 50S ribosomal protein L3; ribosomal protein L3 (BL3) | rplC | Translation, ribosomal structure and biogenesis | ||
| sa_c9963s8658_a_at | SA2048 | 0.00245 | 2.4 | 30S ribosomal protein S10; ribosomal protein S10 (BS13) | rpsJ | Translation, ribosomal structure and biogenesis | ||
| Group III: No change (20 min) – Upregulation (60 min) 8 genes | ||||||||
| sa_c10571s9056_a_at | SA0845 | 0.000581 | 2.5 | putative oligopeptide ABC transporter integral membrane protein (fragment) | oppB | Amino acid transport and metabolism, Inorganic ion transport and metabolism | ||
| sa_c324s166_a_at | SA0846 | 0.00019 | 2.4 | probable peptide ABC transporter permease ABC transporter protein | oppC | Amino acid transport and metabolism, Inorganic ion transport and metabolism | ||
| sa_c328s170_a_at | SA0847 | 0.00293 | 2.2 | probable peptide ABC transporter ATP-binding ABC transporter protein | oppD | Amino acid transport and metabolism, Inorganic ion transport and metabolism | ||
| sa_c332s172_a_at | SA0848 | 9.13E-05 | 2.2 | probable peptide ABC transporter ATP-binding ABC transporter protein | oppF | Amino acid transport and metabolism | ||
| sa_c5349s4625_a_at | SA0950 | 0.0022 | 2.2 | ABC transporter ATP-binding protein – spermidine:putrescine transport | potA | Amino acid transport and metabolism | ||
| sa_c795s596_a_at | SA0952 | 0.00536 | 2.2 | ABC transporter membrane-spanning permease – spermidine:putrescine transport | potC | Amino acid transport and metabolism | ||
| sa_c803s604_a_at | SA0953 | 0.00987 | 2.2 | spermidine:putrescine ABC transporter, spermidine: putrescine-binding periplasmic protein (potD) homolog | potD | Amino acid transport and metabolism | ||
| sa_c8848s7783_a_at | SA1601 | 0.000923 | 2.3 | CRCB, CrcB-like protein | Cell division and chromosome partitioning | |||
| Group IV: Downregulation (20 min) – Downregulation (60 min) 27 genes | ||||||||
| sa_c5061s4360_a_at | SA0229 | 4.85E-07 | -5.8 | 4.85E-07 | -4.8 | dipeptide ABC transporter, periplasmic dipeptide-binding protein (dppA) | Amino acid transport and metabolism | |
| sa_c736s544_a_at | SA0937 | 0.00365 | -2.9 | 0.00365 | -3.5 | cytochrome D ubiquinol oxidase subunit I | Energy production and conversion | |
| sa_c740s548_a_at | SA0938 | 0.00375 | -2.9 | 0.00375 | -3.4 | cytochrome D ubiquinol oxidase subunit II homolog | Energy production and conversion | |
| sa_c1659s1395_a_at | SA1164* | 0.000431 | -8.9 | 0.000431 | -3.4 | homoserine dehydrogenase (HDH) | dhoM | Amino acid transport and metabolism |
| sa_c1665s1401_a_at | SA1165 | 0.00011 | -8.3 | 0.00011 | -3.2 | threonine synthase (EC 4.2.3.1) homolog thrC | thrC | Amino acid transport and metabolism |
| sa_c1669s1406_a_at | SA1166 | 5.06E-05 | -11.5 | 5.06E-05 | -3.8 | homoserine kinase (thrB) | thrB | Amino acid transport and metabolism |
| sa_c1872s1598_a_at | SA1213 | 0.00127 | -2.8 | 0.00127 | -2.4 | probable peptide ABC transporter permease ABC transporter protein | opp-2C | Amino acid transport and metabolism, Inorganic ion transport and metabolism |
| sa_c1876s1602_a_at | SA1214 | 8.58E-06 | -3.7 | 8.58E-06 | -3.0 | putative oligopeptide ABC transporter integral membrane protein (fragment) | opp-2B | Amino acid transport and metabolism, Inorganic ion transport and metabolism |
| sa_c1912s1635_a_at | SA1225* | 8.90E-06 | -54.6 | 8.90E-06 | -7.7 | aspartokinase II in bifunctional enxyme: aspartokinase II; homoserine dehydrogenase II | lysC | Amino acid transport and metabolism |
| sa_c1918s1640_a_at | SA1226* | 1.78E-05 | -21.5 | 1.78E-05 | -4.3 | aspartate-semialdehyde dehydrogenase (ASA dehydrogenase) (ASA DH) | asd | Amino acid transport and metabolism |
| sa_c1922s1644_a_at | SA1227* | 2.75E-07 | -27.3 | 2.75E-07 | -5.2 | dihydrodipicolinate synthase (DHDPS) | dapA | Amino acid transport and metabolism, Cell envelope biogenesis, outer membrane |
| sa_c1924s1648_a_at | SA1228* | 1.17E-07 | -31.4 | 1.17E-07 | -5.1 | dihydrodipicolinate reductase (DHPR) | dapB | Amino acid transport and metabolism |
| sa_c1928s1652_a_at | SA1229* | 2.55E-07 | -23.5 | 2.55E-07 | -4.4 | 2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N-succinyltransferase | dapD | Amino acid transport and metabolism |
| sa_c1936s1659_a_at | SA1231 | 8.94E-08 | -18.5 | 8.94E-08 | -3.0 | Ala_racemase, Alanine racemase | Cell envelope biogenesis, outer membrane | |
| sa_c3202s2750_a_at | SA1544 | 2.74E-05 | -15.8 | 2.74E-05 | -3.8 | serine-pyruvate aminotransferase; alanine-glyoxylate aminotransferase (Serine-pyruvate aminotransferase) | Amino acid transport and metabolism | |
| sa_c3603s3083_a_at | SA1659 | 7.54E-06 | -2.4 | 7.54E-06 | -3.5 | parvulin-like PPIase precursor (Peptidyl-prolyl cis-trans isomerase plp) (Rotamase plp) | prsA | Posttranslational modification, protein turnover, chaperones |
| sa_c10591s11046_s_at | SA1814 | 1.99E-05 | -3.3 | 1.99E-05 | -2.9 | succinyl-diaminopimelate desuccinylase (dapE) | Amino acid transport and metabolism | |
| sa_c5023s4322_at | SA2082 | 6.38E-07 | -3.5 | 6.38E-07 | -5.5 | urease gamma chain (urea amidohydrolase) | ureA | Amino acid transport and metabolism |
| sa_c5029s4326_a_at | SA2083 | 2.01E-06 | -4.3 | 2.01E-06 | -7.6 | URE2_staxy urease beta subunit | ureB | Amino acid transport and metabolism |
| sa_c5031s4330_a_at | SA2084 | 1.84E-07 | -3.0 | 1.84E-07 | -4.4 | urease alpha chain (urea amidohydrolase) | ureC | Amino acid transport and metabolism |
| sa_c5035s4334_at | SA2085 | 1.71E-05 | -2.9 | 1.71E-05 | -4.2 | urease accessory protein UreE | ureE | Posttranslational modification, protein turnover, chaperones |
| sa_c5039s4340_a_at | SA2086 | 1.40E-05 | -2.3 | 1.40E-05 | -3.8 | urease accessory protein UreF | ureF | Posttranslational modification, protein turnover, chaperones |
| sa_c9293s8136_a_at | SA2088 | 0.000118 | -2.2 | 0.000118 | -2.8 | urease accessory protein UreD | ureD | Posttranslational modification, protein turnover, chaperones |
| sa_c5303s4583_a_at | SA2149 | 1.28E-07 | -35.8 | 1.28E-07 | -64.9 | probable peptide ABC transporter ATP-binding ABC transporter protein | hypothetical protein | |
| sa_c5307s4587_at | SA2150 | 9.33E-07 | -36.5 | 9.33E-07 | -69.0 | ABC-type transporter, permease component | hypothetical protein | |
| sa_c5777s5020_a_at | SA2235 | 2.82E-05 | -5.1 | 2.82E-05 | -3.5 | putative ABC transporter; osmoprotectant-binding protein, | opuCC | Cell envelope biogenesis, outer membrane |
| sa_c6435s5604_a_at | SA2409 | 0.000134 | -4.6 | 0.000134 | -2.2 | anaerobic ribonucleoside-triphosphate reductase activating protein | Posttranslational modification, protein turnover, chaperones | |
| Group V: Downregulation (20 min) – No change (60 min) 35 genes | ||||||||
| sa_c37s34_a_at | SA0010 | 2.44E-06 | -4.7 | branched-chain amino acid permease | Amino acid transport and metabolism | |||
| sa_c4055s3432_a_at | SA0201 | 0.00111 | -2.3 | RGD-containing lipoprotein | rlp | Amino acid transport and metabolism | ||
| sa_c7055s6165_a_at | SA0331 | 0.000236 | -3.0 | probable lipoprotein | Inorganic ion transport and metabolism | |||
| sa_c7100s6210_a_at | SA0344 | 2.80E-06 | -15.2 | methyltetrahydropteroyltriglutamate – homocysteine methyltransferase (vitamin-B12-independent methionine synthase isozyme) | metE | Amino acid transport and metabolism | ||
| sa_c5418s4689_a_at | SA0420 | 7.55E-05 | -2.8 | probable amino acid ABC transporter, ATP-binding protein (abc) | Inorganic ion transport and metabolism | |||
| sa_c7374s6406_a_at | SA0421 | 6.63E-07 | -3.3 | putative amino acid ABC transporter, permease protein, glutamine transport system | Inorganic ion transport and metabolism | |||
| sa_c5431s4700_a_at | SA0769 | 2.32E-06 | -5.4 | probable amino acid ABC transporter, ATP-binding protein | Inorganic ion transport and metabolism | |||
| sa_c8512s7471_a_at | SA0770 | 1.42E-05 | -6.3 | permease protein of ABC transporter system | Inorganic ion transport and metabolism | |||
| sa_c8518s7475_a_at | SA0771 | 5.12E-06 | -7.5 | probable D-methionine-binding lipoprotein metQ precursor (Outer membrane lipoprotein1) | Inorganic ion transport and metabolism | |||
| sa_c350s191_a_at | SA0849 | 8.12E-05 | -2.1 | oligopeptide ABC transporter, periplasmic oligopeptide-binding protein (oppA-2) | Amino acid transport and metabolism | |||
| sa_c352s195_a_at | SA0850 | 0.000757 | -3.4 | periplasmic oligopeptide-binding protein of oligopeptide ABC transporter | Amino acid transport and metabolism | |||
| sa_c1820s1547_a_at | SA1200 | 0.00698 | -2.1 | para-aminobenzoate synthetase glutamine amidotransferase component II | Amino acid transport and metabolism, Coenzyme metabolism | |||
| sa_c1828s1551_a_at | SA1201 | 0.000803 | -2.8 | pir|AE0653 anthranilate synthase component II, | trpD | Amino acid transport and metabolism | ||
| sa_c1832s1558_a_at | SA1202 | 0.00239 | -4.5 | anthranilate synthase; indole-glycerol phosphate synthase; | trpC | Amino acid transport and metabolism | ||
| sa_c1836s1562_at | SA1203 | 0.00279 | -4.3 | indole-3-glycerolphosphate synthetase | trpF | Amino acid transport and metabolism | ||
| sa_c1840s1566_a_at | SA1204 | 0.000663 | -3.6 | tryptophan synthase beta chain; tryptophan synthase (beta subunit) | trpB | Amino acid transport and metabolism | ||
| sa_c1844s1570_a_at | SA1205 | 5.52E-05 | -2.6 | tryptophan synthase alpha chain; tryptophan synthase | trpA | Amino acid transport and metabolism | ||
| sa_c1866s1587_a_at | SA1211 | 0.000158 | -2.3 | ATP-binding ABC transporter protein | opp-2F | Amino acid transport and metabolism | ||
| sa_c4209s3561_a_at | SA1858 | 0.000144 | -6.2 | dihydroxy-acid dehydratase (DAD) | ilvD | Amino acid transport and metabolism, Coenzyme metabolism | ||
| sa_c4213s3565_a_at | SA1859 | 0.000431 | -8.0 | acetolactate synthase isozyme III large subunit (AHAS-III) | ilvB | Amino acid transport and metabolism, Coenzyme metabolism | ||
| sa_c9931s8627_a_at | SA1861 | 0.000255 | -10.3 | ketol-acid reductoisomerase (Acetohydroxy-acid isomeroreductase) | ilvC | Amino acid transport and metabolism, Coenzyme metabolism | ||
| sa_c4223s3575_a_at | SA1862 | 4.06E-05 | -9.8 | 2-isopropylmalate synthase | leuA | Amino acid transport and metabolism | ||
| sa_c4225s3576_a_at | SA1863 | 5.85E-05 | -11.0 | 3-isopropylmalate dehydrogenase | leuB | Energy production and conversion, Amino acid transportand metabolism | ||
| sa_c4229s3580_a_at | SA1864 | 1.39E-05 | -13.2 | 3-isopropylmalate dehydratase large subunit | leuC | Amino acid transport and metabolism | ||
| sa_c4239s3588_a_at | SA1865 | 6.09E-05 | -13.1 | 3-isopropylmalate dehydratase small subunit | leuD | Amino acid transport and metabolism | ||
| sa_c4243s3594_a_at | SA1866 | 1.58E-05 | -7.3 | threonine dehydratase biosynthetic (Threonine deaminase) (TD) | ilvA | Amino acid transport and metabolism | ||
| sa_c9447s10370cs_s_at | SA2464* | 0.00225 | -10.5 | histidine biosynthesis bifunctional protein | hisI | Amino acid transport and metabolism | ||
| sa_c6696s10090cs_s_at | SA2465* | 0.00199 | -10.6 | Imidazole glycerol phosphate synthase subunit hisF | hisF | Amino acid transport and metabolism | ||
| sa_c6706s5846_a_at | SA2466* | 0.00148 | -14.5 | phosphoribosylformimino-5-aminoimidazole carboxamide ribotide isomerase | Amino acid transport and metabolism | |||
| sa_c6708s5850_a_at | SA2467 | 0.00213 | -14.6 | Imidazole glycerol phosphate synthase subunit hisH | hisH | Amino acid transport and metabolism | ||
| sa_c6714s5853_a_at | SA2468* | 0.00639 | -13.6 | Imidazoleglycerol-phosphate dehydratase (IGPD) | hisB | Amino acid transport and metabolism | ||
| sa_c6718s5857_a_at | SA2469 | 0.00106 | -19.8 | histidinol-phosphate aminotransferase | Amino acid transport and metabolism | |||
| sa_c6728s5871_a_at | SA2472 | 0.0059 | -14.5 | ATPphosphoribosyltransferase regulatory subunit | hisZ | Amino acid transport and metabolism | ||
| sa_c6740s5882_a_at | SA2475 | 0.000217 | -6.6 | ABC transporter membrane-spanning permease – unknown substrate | Inorganic ion transport and metabolism | |||
| sa_c5397s4673_a_at | SA2476 | 0.000459 | -7.6 | putative ABC transporter; ATP-binding protein; possible cobalt transport system | Inorganic ion transport and metabolism | |||
| Group VI: No change (20 min) – Downregulation (60 min) 19 genes | ||||||||
| sa_c2346s1974_a_at | SA0144 | 4.02E-05 | -3.1 | capsular polysaccharide synthesis enzyme Cap5A | capA | Cell envelope biogenesis, outer membrane | ||
| sa_c2385s1987_a_at | SA0145 | 4.55E-06 | -2.9 | capsular polysaccharide synthesis enzyme Cap5B | capB | Cell division and chromosome partitioning | ||
| sa_c2399s1991_a_at | SA0146 | 0.00055 | -2.6 | capsular polysaccharide synthesis enzyme Cap8C | capC | Cell envelope biogenesis, outer membrane, Carbohydrate transport and metabolism | ||
| sa_c2413s1997_a_at | SA0147 | 0.0001 | -2.6 | capsular polysaccharide synthesis enzyme Cap5D | capD | Cell envelope biogenesis, outer membrane, Carbohydrate transport and metabolism | ||
| sa_c9546s8318_a_at | SA0148 | 9.77E-05 | -2.8 | putative UDP-glucose 4-epimerase (Galactowaldenase) (UDP-galactose 4-epimerase) | capE | Cell envelope biogenesis, outer membrane, Carbohydrate transport and metabolism | ||
| sa_c2479s2056_a_at | SA0149 | 2.40E-06 | -3.1 | capsular polysaccharide synthesis enzyme Cap5F | capF | Cell envelope biogenesis, outer membrane, Carbohydrate transport and metabolism | ||
| sa_c2516s2092_a_at | SA0150 | 5.92E-05 | -2.4 | UDP-N-acetylglucosamine 2-epimerase (UDP-GlcNAc-2-epimerase) | capG | Cell envelope biogenesis, outer membrane | ||
| sa_c10086s8810_a_at | SA0151 | 0.00025 | -2.8 | chloramphenicol acetyltransferase (Xenobiotic acetyltransferase) (XAT) | capH | General function prediction only | ||
| sa_c10087s8814_a_at | SA0152 | 0.00017 | -2.7 | capsular polysaccharide synthesis enzyme Cap5I | capI | Cell envelope biogenesis, outer membrane | ||
| sa_c10089s8822_a_at | SA0154 | 0.00103 | -2.1 | capsular polysaccharide synthesis enzyme Cap5K | capK | Cell envelope biogenesis, outer membrane | ||
| sa_c898s698_a_at | SA0977 | 0.00044 | -2.7 | 29-kDa cell surface protein | isdA | Cell envelope biogenesis, outer membrane | ||
| sa_c906s704_a_at | SA0978 | 0.00538 | -2.2 | hypothetical protein SirD | isdC | Cell envelope biogenesis, outer membrane | ||
| sa_c3380s9339_a_at | SA1586 | 1.41E-05 | -2.4 | 6,7-dimethyl-8-ribityllumazine synthase (DMRL synthase) | ribH | Coenzyme metabolism | ||
| sa_c3387s2918_a_at | SA1587 | 6.15E-06 | -2.8 | probable riboflavin biosynthesis bifunctional protein | ribA | Coenzyme metabolism | ||
| sa_c3391s2919_a_at | SA1588 | 3.10E-06 | -2.7 | riboflavin synthase alpha chain | ribB | Coenzyme metabolism | ||
| sa_c3395s2925_a_at | SA1589 | 5.10E-07 | -3.0 | riboflavin specific deaminase | ribD | Coenzyme metabolism | ||
| sa_c4369s3721_at | SA1894 | 0.000661 | -2.3 | thiamine-phosphate pyrophosphorylase | thiE | Coenzyme metabolism | ||
| sa_c4373s3725_a_at | SA1895 | 3.05E-05 | -2.5 | hydroxyethylthiazole kinase | thiM | Coenzyme metabolism | ||
| sa_c4379s3726_a_at | SA1896 | 0.000137 | -2.8 | bifunctional enzyme: hydroxy-phosphomethylpyrimidine kinase | thiD | Coenzyme metabolism | ||
| Group VII: Downregulation (20 min) – Upregulation (60 min) 3 genes | ||||||||
| sa_c9442s8255_a_at | SA2459* | 0.0016 | -2.5 | 0.0016 | 2.5 | intercellular adhesion protein IcaA | icaA | Cell envelope biogenesis, outer membrane |
| sa_c6677s5830_a_at | SA2460* | 0.0136 | -2.0 | 0.0136 | 4.7 | IcaD | icaD | Cell envelope biogenesis, outer membrane |
| sa_c6681s9106_a_at | SA2461* | 3.83E-06 | -2.1 | 3.83E-06 | 3.4 | intercellular adhesion protein IcaB | icaB | Cell envelope biogenesis, outer membrane |
* Genes were validated by real-time PCR.
Group I: genes upregulated upon 20 and 60 min exposures
Group I of table 1 contains 18 genes associated with virulence in S. aureus. Interestingly, five of these genes encode the secretory antigen precursor, SsaA. The ssaA gene potentially regulated by the YycG/YycF system encodes proteins involved in cell wall metabolism, membrane-bound transport systems, and pathogenicity, including two major antigenic proteins, SsaA and IsaA. YycF has also been shown to bind specifically to the promoter regions of two genes, encoding the IsaA antigen and the LytM peptidoglycan hydrolase. This is in agreement with the proposed role of this system in controlling virulence and cell wall metabolism [20].
In this study, OPP also upregulated the clfB (clumping factor B) gene expression upon both 20 and 60 min exposure. In our previous results, triclosan upregulated the expression of SA2423 encoding ClfB, which binds fibrinogen [12]. The results of this study show that the production of virulence factors in S. aureus may be a secondary effect of OPP and this may provide new insight into the protective response of S. aureus to OPP.
Group II: genes upregulated upon 20 min exposure
Group II of table 1 indicates that the class of "translation, ribosomal structure and biogenesis" which is responsible for the synthesis of ribosomal proteins was upregulated after 20 minutes. In group II of table 1, for instance four genes encode 30S ribosomal proteins and 15 genes code for 50S ribosomal proteins. In addition, SA0459 (rplY) which encoded general stress protein was upregulated at 20 min. Ribosomal proteins are required for protein translation. Such early response of these ribosomal protein genes may reflect a stress response during exposure. The upregulation of ribosomal protein genes might enhance the translation process or help proper ribosome functioning under stress conditions as exposure to OPP. The suggestion that the expression of ribosomal proteins is activated upon exposure to OPP is surprising as this presumably reflects increased growth rate or virulence.
In group II (table 1), we also observed genes related to primary metabolism that mainly belonged to the functional classes of "purines, pyrimidines, nucleosides, and nucleotides". The gene cluster: SA1041–SA1048 (pyrRPBCAAABFE) which is homologous to the pyrimidine biosynthetic (pyr) operon of Bacillus subtilis [21] was upregulated at 20 min.
Group III: genes upregulated upon 60 min exposure
In group III of table 1, there were some genes from amino acid transport and metabolism, an ATP-binding cassette (ABC) transporters and transcription. The oligopeptide transport system (Opp) of S. aureus is an ABC transporter that transports amino acids, cations- and iron-carrying compounds and peptides with a broad specificity [22]. The peptides are mainly used as nutrients by the multiple amino acid auxotrophic S. aureus. The Opp system consists of four different proteins: OppB and OppC, OppD and OppF. Interestingly, 4 of the 26 genes, including SA0845–SA0848 that code for proteins associated with amino acid transport were upregulated (table 1). Therefore, the suggestion that the expression of these proteins is activated upon exposure to OPP is surprising as this presumably reflects increased growth rate or recovery. We found that a putative operon containing four open reading frames (ORFs) (potABCD) was upregulated (table 1). The potABCD operon encodes a periplasmic binding protein dependent ABC transport systems from Gram-positive bacteria [23]. The SA0950–SA0952–SA0953 shows homology to the genes encoding this potABCD transport system (potA, potC and potD), which are involved in the transport of spermidine and putrescine. Further, we showed the upregulation of ABC transport systems-related genes, which accompanied the growth recovery.
Of further importance was that group III contained genes related to integral membrane protein, which belonged to the functional class of "cell division and chromosome partitioning". SA1601 (crcB) is a putative integral membrane protein possibly involved in chromosome condensation (table 1).
Group IV: genes downregulated upon 20 and 60 min exposures
In group IV in table 1, we noted that genes belonging to the functional classes of "amino acid transport and metabolism", "carbohydrate transport and metabolism", "energy production and conversion", "posttranslational modification protein turnover chaperones", "transcription" classes and putative lipoproteins were downregulated upon both exposure times.
Intriguingly, we observed the high downregulation of SA2149 and SA2150 (hrt A and B), the heme-regulated transport system, which consist of a novel transport system which plays a critical role in staphylococcal heme metabolism (table 1). Among the genes in the class of "amino acid transport and metabolism", SA1225 (lysC)-SA1226 (asd)-SA1227 (dapA)-SA1228 (dapB)-SA1229 (dapD), and SA1814 (dapE) fall within a predicted operon and are all involved in diaminopimelate (DAP) biosynthesis (table 1). The disruption of biosynthetic pathways involved in building up bacterial cell wall components is a common mode of action of antibiotics [24]. Penicillins [25], methicillin [26], cephalosporins [27] and glycopeptide drugs such as vancomycin [28] are all drugs that inhibit major steps in the construction of the peptidoglycan layer of bacterial cell walls. Lysine or its biosynthetic precursor, DAP [29], are essential to most bacteria for the synthesis of the peptidoglycan layer of the cell wall [30-33]. Since mammals neither make nor use DAP and require L-lysine is an essential amino acid that is supplied through dietary intake, inhibitors of the DAP biosynthetic pathway will probably not result in mammalian toxicity. Decisively SA1225 (lysC)-SA1226 (asd)-SA1227 (dapA)-SA1228 (dapB) and SA1229 (dapD) show fold highest decreases as -54.6, -21.5, -27.3, -31.4, and -23.5 folds at 20 min and -7.7, -4.3, -5.2, -5.1, and -4.4 folds at 60 min in this experiment (see also table 2). Our findings suggest that the mode of action of OPP may be related to bacterial biosynthesis of amino acids. Other genes of amino acids, including methionine, threonine, histidine and lysine were also highly down regulated at -15, -7, -19 and -54 folds. Therefore, this outcome in conjunction with the extensive downregulation of the genes encoding DAP biosynthesis suggests that OPP may inhibit construction of the peptidoglycan in cell wall of S. aureus. These genes were less downregulated at 60 min than at 20 min. These results suggest that OPP inhibits the growth of S. aureus at 20 min and that growth recovery occurs at 60 minutes, indicating a possible mechanism of action of OPP in S. aureus. In a similar study carried out using Pseudomonas aeruginosa treated with 0.82 mM OPP, we did not observe extensive downregulation of the genes involved in amino acid biosynthesis and specifically lysine biosynthesis (data not shown). This suggests that the mechanisms of action of 0.82 mM OPP on P. aeruginosa and S. aureus may differ.
Table 2.
Transcript level comparison of S. aureus genes between real-time PCR and microarray analyses.
| mRNA level change with microarray | mRNA level change with real-time PCR | |||||
| Gene | Fold change | Fold change | Sense primer sequence | Antisense primer sequence | ||
| 20 min | 60 min | 20 min | 60 min | |||
| SA0423 | 8.1 | 19.4 | 9.1(± 1.4) | 18.6(± 1.2) | 5'-CGG GTG AAT CAG TGT GGG CAA TTT-3' | 5'-TAT GAT CCG CCA CCT GAG TTC GTT-3' |
| SA1164 | -8.9 | -3.4 | -40.9(± 5.0) | -2.1(± 0.1) | 5'-TAT GAT CCG CCA CCT GAG TTC GTT-3' | 5'-GAG TGT AGC AGG TGG TAT TCC GAT-3' |
| SA1225 | -54.6 | -7.7 | -265.0(± 18.9) | -58.0(± 1.4) | 5'-GAG TGT AGC AGG TGG TAT TCC GAT-3' | 5'-TCA TCA GTT GGA TCC GCT TCA GCA-3' |
| SA1226 | -21.5 | -4.3 | -14.9(± 3.3) | -2.1(± 0.7) | 5'-TCA TCA GTT GGA TCC GCT TCA GCA-3' | 5'-ACT TTA GGC AGA GGC GGT TCT GAT-3' |
| SA1227 | -27.3 | -5.2 | -58.0(± 1.5) | -2.8(± 0.1) | 5'-ACT TTA GGC AGA GGC GGT TCT GAT-3' | 5'-AGT CTT GGG TCA GTG GCA TAC ACA-3' |
| SA1228 | -31.4 | -5.1 | -35.3(± 1.6) | -1.6(± 0.4) | 5'-AGT CTT GGG TCA GTG GCA TAC ACA-3' | 5'-TGG GTG CAA CAG GAT TAG TAG GCA-3' |
| SA1229 | -23.5 | -4.4 | -7.4(± 2.0) | -1.2(± 0.2) | 5'-TGG GTG CAA CAG GAT TAG TAG GCA-3' | 5'-TTC AAC TTC TTG CCC TGC AGA ACG-3' |
| SA2093 | 6.8 | 9.0 | 15.7(± 1.2) | 4.6(± 0.5) | 5'-TTC AAC TTC TTG CCC TGC AGA ACG-3' | 5'-TAT TTG AGG GTG TTG GCG TTG CAC-3' |
| SA2097 | 6.9 | 11.5 | 5.5(± 1.8) | 24.5(± 1.2) | 5'-TAT TTG AGG GTG TTG GCG TTG CAC-3' | 5'-AGG GCT CTC AGC AGT AGT TCC ATT-3' |
| SA2353 | 10.2 | 13.8 | 8.0(± 1.6) | 16.4(± 1.9) | 5'-AGG GCT CTC AGC AGT AGT TCC ATT-3' | 5'-ATT CGT GGA GGT ACG ATT GTC GGT-3' |
| SA2355 | 8.5 | 9.4 | 5.3(± 1.3) | 20.1(± 1.7) | 5'-ATT CGT GGA GGT ACG ATT GTC GGT-3' | 5'-GCT GCT TGT ATA GCA CCA TTC GCA-3' |
| SA2459c | -2.5 | 2.5 | -3.1(± 0.2.) | 2.0(± 0.3) | 5'-TTG TCG ACG TTG GCT ACT GGG ATA-3' | 5'-TGG AAC CAA CAT CCA ACA CAT GGC-3' |
| SA2460c | -2.0 | 4.7 | -1.8(± 0.3) | 2.5(± 0.3) | 5'-ATG GTC AAG CCC AGA CAG AGG GAA TA-3' | 5'-CAC ACG ATA TAG CGA TAA GTG CTG TT-3' |
| SA2461c | -2.1 | 3.4 | -2.6(± 0.5) | 2.1(± 0.5) | 5'-AGC AGT CAC TCC GAA CTC CAA TGA-3' | 5'-TCA TGG AAT CCG TCC CAT CTC T-3' |
| SA2464b | -10.5 | - | -5.5(± 1.4) | - | 5'-GCT GCT TGT ATA GCA CCA TTC GCA-3' | 5'-GAT CGT CGC AAT TCT GCC ATT CCA-3' |
| SA2465b | -10.6 | - | -13.0(± 1.4) | - | 5'-GAT CGT CGC AAT TCT GCC ATT CCA-3' | 5'-TTG TTG CGC CCA TCA TAA CGA CAG-3' |
| SA2466b | -14.5 | - | -5.7(± 1.3) | - | 5'-TTG TTG CGC CCA TCA TAA CGA CAG-3' | 5'-ACC GTA CTG GTG GTT TAG GTG CAA-3' |
| SA2468b | -13.6 | - | -58.1(± 1.3) | - | 5'-ACC GTA CTG GTG GTT TAG GTG CAA-3' | 5'-TGA ACG GCC ATT TGA TGA TGG AGC-3' |
| SA0727a | 1.00 | 1.00 | 1.00 | 1.00 | 5'-GAT GGT GGT TTC CGC GTA AAT GGT-3' | 5'-GCG CCT GCT TCA ATA TGA GCT TGT-3' |
The real time PCR results are the mean of three biological replicates with three technical replicates for each gene. The microarray results are the mean of five replicates of each gene.
aSA0727 was glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and used as the house-keeping gene. bSA2464, SA2465, SA2466, and SA2468 were downregulated at 20 min with no change at 60 min. cSA2459, SA2460, and SA2461 were downregulated after 20 min and upregulated after 60 min exposure.
Additional amino acid biosynthesis genes including: SA1164 (dhoM)-SA1165 (thrC)-SA1166 (thrB) involved in threonine biosynthesis were also in this group (table 1). Further, SA2082–SA2083–SA2084–SA2085–SA2086–SA2088 (ureABCEFD), which make CO2 and NH3 from urea and encode urea amidohydrolase and urease accessory proteins and SA1544 which codes for serine-pyruvate aminotransferase were downregulated at 20 and 60 min (table 1).
Group IV of table 1 also shows that the functional class of "cell envelope biogenesis, outer membrane" was distinctive. In particular, SA1231 which shows an -18-fold decrease after 20 min encodes an alanine racemase that catalyses the conversion of L-alanine into D-alanine, a key component of bacterial peptidoglycan [34]. Additionally, the putative lipoproteins: SA0229 (dppA), SA1213 (opp-2C)-SA1214 (opp-2B), SA1659 (prsA), SA2235 (opuCC), and SA2409 which are cell wall anchoring surface proteins were downregulated in response to OPP (table 1).
In group IV, we also observed genes related to primary metabolism that mainly belonged to the functional classes of "energy metabolism", "lipid metabolism", and "transcription". For example, cytochrome bd complex: SA0937–SA0938 (cydAB) was downregulated upon both 20 min and 60 min exposure (table 1). Cytochrome bd complex is one of two terminal oxidases in the bacterial respiratory chain that reduce molecular oxygen to water, utilizing intermediates shuttled through the electron transport chain [35]. Cytochrome d oxidase catalyses the last step of oxygen respiration and prevails under oxygen-limiting conditions [36]. Interestingly, it was speculated that cytochrome d oxidase is required under conditions of environmental stress and may have crucial roles in cellular physiology other than acting as an oxidase [37]. However, prior studies revealed that the cydAB genes were strongly upregulated upon exposure to hydrogen peroxide strengthens the confidence of the prior assignments about the role of cytochrome d oxidase in oxidative protection processes of both Gram positive and Gram negative bacteria [10,13].
Group V: genes downregulated upon 20 min exposures
In group V in table 1, the most dominant class was "amino acid transport and metabolism", which contained half of the genes in that group. Further, SA1858 (ilvD)-SA1859 (ilvB) and SA1861 (ilvC)-SA1862 (leuA)-SA1863 (leuB)-SA1864 (leuC)-SA1865 (leuD)-SA1866 (ilvA) which were downregulated (table 1) are possibly parts of an operon homologous to the ilv-leu operon encoding enzymes of branched-chain amino acid biosynthesis in Bacillus subtilis [38]. In addition, SA2464 (hisI)-SA2465 (hisF)-SA2466–SA2467 (hisH)-SA2468 (hisB)-SA2469 and SA2472 (hisZ) which are possibly parts of an operon homologous to the histidine biosynthesis were highly downregulated on 20 min (table 1). SA1200–SA1201 (trpD)-SA1202 (trpC)-SA1203 (trpF)-SA1204 (trpB)-AS1205 (trpA) which are also possibly parts of an operon homologous to the tryptophan biosynthesis operon are downregulated on 20 min (table 1). This result along with the downregulation of 46 genes involved in amino acid biosynthesis in group V suggests that amino acid synthesis was repressed upon 20 min exposure to OPP in S. aureus.
Group V shows that 17 genes in the functional class of "inorganic ion transport and metabolism" were downregulated at 20 min. First, SA0771 (metQ) codes for probable D-methionine-binding lipoprotein (outer membrane lipoprotein 1). The proteins encoded by SA0344 (metE)-SA0769 (metN)-SA0770 (metI)-SA0420 (metN)-SA0421 (metI) are involved in D-methionine transporter of S. aureus ABC transporter (table 1). Interestingly, group V contained lipoproteins such as SA0010, SA0331, SA0849, SA0850, SA0201 and SA1211 (table 1). These results, along with downregulation of all the genes of lipoproteins of S. aureus in group IV and V, suggest that OPP exposure may decrease stability of the staphylococcal membrane. Secondly, the proteins encoded by SA2475 (cbiQ)-SA2476 (cbiO) are involved in cobalt and nickel transport (table 1).
Group VI: genes downregulated upon 60 min exposures
Table 1 illustrates that the functional classes of group VI in general contained more downregulated genes at 60 min. In particular, the functional classes of "cell envelope biogenesis outer membrane", "carbohydrate transport and metabolism", "amino acid transport and metabolism", "coenzyme metabolism", "energy production and conversion" and "posttranslational modification protein turnover chaperones" had significantly more downregulated genes at 60 min (see also figure 4). This result suggests that the functional class profiles were notably different between 20 and 60 min.
One of the characteristics of group VI of table 1 was the downregulation of 12 genes belonging to the functional class of "cell wall/membrane/envelope biogenesis". In particular, genes related to envelope biogenesis were distinctive: SA0144 (capA)-SA0145 (capB)-SA0146 (capC)-SA0147 (capD)-SA0148 (capE)-SA0149 (capF)-SA0150 (capG)-SA0151 (capH)-SA0152 (capI)-SA0154 (capK) were downregulated at 60 min. These genes share homology with the capsular polysaccharide synthesis enzyme (cap) operon which in turn is essential for virulence by impeding phagocytosis [39]. This finding is congruent with the previous outcome that triclosan downregulates several virulence factor-related genes (SA0144–SA0153 (capABCDEFGHIJ)) in S. aureus [12]. Moreover, IsdAC encoded by SA0977 and SA0978, the iron-regulated surface determinant (Isd) system, was downregulated at 60 min (table 1). Identification of the Isd system in S. aureus has demonstrated the importance of cell-wall sorted proteins in heme binding and transport [40]. To date, the Isd system comprises the only known heme-iron utilization pathway in S. aureus. Cell-wall sorted proteins of the S. aureus iron-regulated surface determinant system bind human hemoproteins, remove the heme molecule, and transport heme through the cell wall and plasma membrane for accumulation in the bacterial cytoplasm [41].
Particularly important was that many of the genes in the class of "coenzyme metabolism" were also members of group VI (figure 4 and table 1). Intriguingly, the genes were all involved in the riboflavin biosynthesis. SA1586 (ribH)-SA1587 (ribA)-Sa1588 (ribB)-SA1589 (ribD) was downregulated at 60 min exposure (table 1). Riboflavin (vitamin B2) is an essential component of the basic metabolism, being a precursor of coenzymes flavin adenine dinucleotide (FAD) and flavin mononucleotide (FMN). The best studied system of the riboflavin biosynthesis in bacteria is the rib operon of Bacillus subtilis encoding a pyrimidine deaminase/reductase, α-subunit of riboflavin synthase, GTP cyclohydrolase/3,4-dihydroxy 2-butanone 4-phosphate (3,4-DHBP) synthase, and β-subunit of riboflavin synthase [42]. These enzymes form a pathway that creates one riboflavin molecule from one molecule of GTP and two molecules of ribulose 5-phosphate [43]. The proteins encoded by SA1894 (thiE)-SA1895 (thiM)-SA1896 (thiD) were involved in thiamine biosynthesis of coenzyme metabolism at 60 min (table 1). Methicillin-resistant S. aureus small colony variants are frequently auxotrophic for hemin, menadione, thiamine, and CO2 involved in biosynthesis of the electron transport chain element. This phenotype grows slowly, and forms very small, nonhemolytic colonies in routine culture, so it may lead to the misidentification of this organism. As discussed above, group IV also had SA2149 (hrtA)-SA2150 (hrtB), which exhibited expression level decreases upon 20 and 60 min exposures in chorus with the repression of the genes of thiamine biosynthesis. Therefore, this result suggests that growth inhibition was accompanied with the repression of many coenzyme metabolism-related genes.
Group VII: genes downregulated upon 20 min and upregulated upon 60 min exposures
Note that group VII has been included only in table 1 in order to discuss the aberrant behaviour of the ica genes. Group VII is not indicated on figure 3 and 4. Intriguingly, we observed that SA2459, SA2460 and SA2461 (icaADB) which make up the intercellular adhesion (ica) operon and contribute to virulence in S. aureus were downregulated after 20 min and upregulated after 60 min of exposure to OPP (table 1 and table 2). The intercellular adhesion operon (icaRADBC) mediates polysaccharide intercellular adhesion in S. aureus, which leads to cell-cell adhesion and is required for biofilm formation [44] Prior studies have demonstrated that polysaccharide intercellular adhesin/hemagglutinin production is involved in the pathogenesis of S. epidermidis [45,46], and is also upregulated by subinhibitory concentrations of certain antibiotics [47]. Therefore, this finding proposes that biofilm formation may not occur after 20 min of exposure to OPP but is possibly favoured as a protective response as exposure time increases to 60 minutes.
Validation of array data by real-time PCR
As an independent measure of differential gene expression, we examined the relative levels of 18 genes with a range of fold changes (-265.0- to 24.5-fold) by real-time PCR analysis, which were specifically involved in the pathogenesis or metabolism of S. aureus. Table 2 shows that our microarray results were corroborated with real-time PCR analysis, which provides independent verification of transcript level changes of the genes that we discuss in this report.
Conclusion
In this paper, we demonstrated how OPP upregulated and downregulated genes in S. aureus, for the first time, by utilizing whole-genome microarrays. Moreover, we presented how the transcriptome profile of S. aureus was shifted during its cellular response to OPP, which involved the growth inhibition. To our knowledge, this is the first study demonstrating the activation of fermentative metabolism after OPP treatment in S. aureus. In summary, we revealed that amino acid metabolism genes were selectively downregulated between 20 and 60 minutes when exposed to 0.82 mM OPP. We also found that the growth inhibition was accompanied by the downregulation of many membrane function-related genes; however, the majority of these genes returned to normal transcription levels during the growth resumption. Further, we showed that the repression of the iron-regulated surface determinant (Isd) system, hemin and thiamine-related genes accompanied with the growth inhibition. Notably, we discovered the upregulation of virulence genes and ribosomal genes while the cells returned to normal growth. These results suggest that S. aureus might be arrested upon exposure to OPP.
In this study, OPP treatment led to the downregulation of several genes involved in amino acid anabolism. The genes involved in the DAP and lysine biosynthetic pathways were most significantly downregulated. Lysine and DAP are essential for building up the peptidoglycan cell wall. This finding proposes that the mode of action of the antimicrobial, OPP in S. aureus might be attributed to the inhibition of genes of lysine biosynthesis and subsequently peptidoglycan biosynthesis. We can therefore, conclude, that the mode of action of OPP is similar to the mechanism of action of some antibiotics. This study has revealed novel information on the mechanism of action of OPP in S. aureus which will benefit further antimicrobial research on S. aureus.
Methods
Bacterial strains and growth conditions
In this study, we used S. aureus NCTC 8325 obtained from the Network on Antimicrobial Resistance in S. aureus (NARSA). As previously described [10-12], we initiated and maintained S. aureus cultures at 37°C with shaking at 250 rpm using sterilized Lurria-Bertani (LB) broth. For growth inhibition, 0.14 mg/L (0.82 mM) of OPP (Aldrich Chemical Co., St. Louis, MO) was dissolved in DMSO and used for the microarray study and added immediately after OD600 reached 0.8. OD600 was measured by using Lambda 25 spectrophotometer (PerkinElmer, Inc., MA). Note that the pH of S. aureus cultures was around 7.0 at 37°C after the exposure [48].
RNA isolation
Total RNA was isolated after 20 and 60 min incubation with and without (control) OPP using the RiboPure – Bacteria kit (Ambion, Inc., Austin, TX) [11]. The quantity of eluted RNA was determined using the NanoDrop spectrophotometer (NanoDrop Technologies, Inc., Wilmington, DE). RNA 6000 Nano LabChip with an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA).
cDNA synthesis, labeling, hybridization, staining, and scanning
cDNA synthesis, cDNA fragmentation, labeling, hybridization, staining and washing steps were performed according to the manufacturer's protocol for the Affymetrix S. aureus GeneChip arrays (Affymetrix, Inc., Santa Clara, CA).
Affymetrix S. aureus genechip analysis
The arrays were scanned with the Affymetrix GeneChip Scanner 3000. To analyze the array data, GeneChip Operating Software (GCOS) v. 1.2 (Affymetrix, Inc., Santa Clara, CA) and GeneSpring GX v. 7.3 (Agilent Technologies, Inc., Santa Clara, CA) were utilized with the following parameters: alpha 1, 0.04; alpha 2, 0.06; tau, 0.015; target signal, 500. Fold changes were calculated as the ratio between the signal averages of five biological controls (untreated) and five biological experimental (OPP-treated) for 20 and 60 min exposures.
Real-time PCR analysis
To determine the validity of the array data, transcript level changes obtained with the microarray analysis were compared with those from quantitative real-time PCR. Genes and primer sequences employed for the real-time PCR analysis are listed in table 2. The housekeeping gene, glyceraldehyde 3-phosphate dehydrogenase (GAPDH), was used as an endogenous control. The real-time PCR was performed by employing iCycler iQ Real-Time PCR Detection System with iScript cDNA Synthesis Kit and IQ SYBR Green Supermix (BioRad Laboratories, Inc., Hercules, CA). For each gene, three biological replicates with three technical replicates each were employed. Reaction mixtures were initially incubated for 3 min at 95.0°C, followed by 40 cycles of 10 s at 95.0°C, 30 s at 55.0°C, and 20 s at 72.0°C. PCR efficiencies were derived from standard curve slopes in the iCycler software v. 3.1 (BioRad Laboratories, Inc., Hercules, CA). Melt-curve analysis was also performed to evaluate PCR specificity and resulted in single primer-specific melting temperatures. In this report, relative quantification based on the relative expression of a target gene versus GAPDH gene was utilized to determine transcript level changes.
Authors' contributions
HJ performed microarray experiments, and data analysis, and drafted the manuscript. FT initiated and supervised the study, and reviewed the manuscript. CN and WEB reviewed the manuscript.
Supplementary Material
The raw data of 7,775 genes control (0 min) and experimental (after 20 and 60 min. exposure of OPP). It has been also deposited in NCBI's Gene Expression Omnibus http://www.ncbi.nlm.nih.gov/geo and is accessible through GEO Series accession number GSE10605 http://www.ncbi.nlm.nih.gov/projects/geo/query/acc.cgi?acc=GSE10605&targ=self&form=html&view=quick.
Staphylococcus aureus 669 genes that showed statistically significant mRNA level changes upon either 20 or 60 min exposure to OPP. The genes were grouped based on their regulation directions upon 20 and 60 min exposures.
Acknowledgments
Acknowledgements
This research is supported by the United States Environmental Protection Agency Grant number T-83284001-1. Although the research described in this paper has been funded wholly by the United States Environmental Protection Agency, it has not been subjected to the Agency's peer and administrative review and therefore may not necessarily reflect the views of the EPA; nor does the mention of trade names or commercial products constitute endorsement of recommendation of use.
Contributor Information
Hyeung-Jin Jang, Email: jang.hyeungjin@epa.gov.
Chantal Nde, Email: Nde.Chantal@epa.gov.
Freshteh Toghrol, Email: toghrol.freshteh@epa.gov.
William E Bentley, Email: bentley@eng.umd.edu.
References
- Said-Salim B, Dunman PM, McAleese FM, Macapagal D, Murphy E, McNamara PJ, Arvidson S, Foster TJ, Projan SJ, Kreiswirth BN. Global regulation of Staphylococcus aureus genes by Rot. Journal of bacteriology. 2003;185:610–619. doi: 10.1128/JB.185.2.610-619.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dancer SJ. How do we assess hospital cleaning? a proposal for microbiological standards for surface hygiene in hospitals. Journal of Hospital Infection. 2004;56:10–15. doi: 10.1016/j.jhin.2003.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton K, Walker R, Sandt JJ van de, Castell JV, Knapp AG, Kozianowski G, Roberfroid M, Schilter B. The application of in vitro data in the derivation of the acceptable daily intake of food additives. Food Chem Toxicol. 1999;37:1175–1197. doi: 10.1016/s0278-6915(99)00107-6. [DOI] [PubMed] [Google Scholar]
- Bomhard EM, Brendler-Schwaab SY, Freyberger A, Herbold BA, Leser KH, Richter M. O-phenylphenol and its sodium and potassium salts: a toxicological assessment. Critical reviews in toxicology. 2002;32:551–625. doi: 10.1080/20024091064318. [DOI] [PubMed] [Google Scholar]
- Johnson GD, Harsy SG, Geronimo J, Wise JM. Orthophenylphenol and phenylhydroquinone residues in citrus fruit and processed citrus products after postharvest fungicidal treatments with sodium orthophenylphenate in California and Florida. J Agric Food Chem. 2001;49:2497–502. doi: 10.1021/jf0012087. [DOI] [PubMed] [Google Scholar]
- Hiraga K, Fujii T. Induction of tumours of the urinary bladder in F344 rats by dietary administration of o-phenylphenol. Food Chem Toxicol. 1984;22:865–870. doi: 10.1016/0278-6915(84)90166-2. [DOI] [PubMed] [Google Scholar]
- Routledge EJ, Sumpter JP. Structural features of alkylphenolic chemicals associated with estrogenic activity. The Journal of biological chemistry. 1997;272:3280–3288. doi: 10.1074/jbc.272.6.3280. [DOI] [PubMed] [Google Scholar]
- Chang W, Small DA, Toghrol F, Bentley WE. Microarray analysis of Pseudomonas aeruginosa reveals induction of pyocin genes in response to hydrogen peroxide. BMC Genomics. 2005;6:115. doi: 10.1186/1471-2164-6-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W, Small DA, Toghrol F, Bentley WE. Microarray analysis of toxicogenomic effects of peracetic acid on Pseudomonas aeruginosa. Environmental science & technology. 2005;39:5893–5899. doi: 10.1021/es0503534. [DOI] [PubMed] [Google Scholar]
- Chang W, Small DA, Toghrol F, Bentley WE. Global transcriptome analysis of Staphylococcus aureus response to hydrogen reroxide. Journal of bacteriology. 2006;188:1648–1659. doi: 10.1128/JB.188.4.1648-1659.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W, Toghrol F, Bentley WE. Toxicogenomic response of Staphylococcus aureus to peracetic acid. Environmental science & technology. 2006;40:5124–5131. doi: 10.1021/es060354b. [DOI] [PubMed] [Google Scholar]
- Jang HJ, Chang MW, Toghrol F, Bentley WE. Microarray analysis of toxicogenomic effects of triclosan on Staphylococcus aureus. Appl Microbiol Biotechnol. 2008;78:695–707. doi: 10.1007/s00253-008-1349-x. [DOI] [PubMed] [Google Scholar]
- Small DA, Chang W, Toghrol F, Bentley WE. Toxicogenomic analysis of sodium hypochlorite antimicrobial mechanisms in Pseudomonas aeruginosa. Appl Microbiol Biotechnol. 2007;74:176–185. doi: 10.1007/s00253-006-0644-7. [DOI] [PubMed] [Google Scholar]
- Small DA, Chang W, Toghrol F, Bentley WE. Comparative global transcription analysis of sodium hypochlorite, peracetic acid, and hydrogen peroxide on Pseudomonas aeruginosa. Appl Microbiol Biotechnol. 2007;76:1093–105. doi: 10.1007/s00253-007-1072-z. [DOI] [PubMed] [Google Scholar]
- Chang MW, Toghrol F, Bentley WE. Toxicogenomic response to chlorination includes induction of major virulence genes in Staphylococcus aureus. Environmental science & technology. 2007;41:7570–7575. doi: 10.1021/es070929k. [DOI] [PubMed] [Google Scholar]
- Lambert RJ. Comparative analysis of antibiotic and antimicrobial biocide susceptibility data in clinical isolates of methicillin-sensitive Staphylococcus aureus, methicillin-resistant Staphylococcus aureus and Pseudomonas aeruginosa between 1989 and 2000. J Appl Microbiol. 2004;97:699–711. doi: 10.1111/j.1365-2672.2004.02345.x. [DOI] [PubMed] [Google Scholar]
- Affymetrix . Affymetrix GeneChip® expression analysis technical manual. Santa Clara, California: Affymetrix, Inc; 2004. [Google Scholar]
- NCBI's Gene Expression Omnibus http://www.ncbi.nlm.nih.gov/geo
- the Gene Classification based on COG functional categories in the genome of National Center for Biotechnology information (NCBI) http://www.ncbi.nlm.nih.gov/sites/entrez?db=genome&cmd=Retrieve&dopt=Overview&list_uids=19213
- Dubrac S, Msadek T. Identification of genes controlled by the essential YycG/YycF two-component system of Staphylococcus aureus. Journal of bacteriology. 2004;186:1175–1181. doi: 10.1128/JB.186.4.1175-1181.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Switzer RL. Transcriptional pausing in the Bacillus subtilis pyr operon in vitro: a role in transcriptional attenuation? Journal of bacteriology. 2003;185:4764–4771. doi: 10.1128/JB.185.16.4764-4771.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher MP, Pearce SR, Higgins CF. Identification and localization of the membrane-associated, ATP-binding subunit of the oligopeptide permease of Salmonella typhimurium. European journal of biochemistry/FEBS. 1989;180:133–141. doi: 10.1111/j.1432-1033.1989.tb14623.x. [DOI] [PubMed] [Google Scholar]
- Matthysse AG, Yarnall HA, Young N. Requirement for genes with homology to ABC transport systems for attachment and virulence of Agrobacterium tumefaciens. Journal of bacteriology. 1996;178:5302–5308. doi: 10.1128/jb.178.17.5302-5308.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugg TD, Walsh CT. Intracellular steps of bacterial cell wall peptidoglycan biosynthesis: enzymology, antibiotics, and antibiotic resistance. Natural product reports. 1992;9:199–215. doi: 10.1039/np9920900199. [DOI] [PubMed] [Google Scholar]
- Giesbrecht P, Kersten T, Maidhof H, Wecke J. Staphylococcal cell wall: morphogenesis and fatal variations in the presence of penicillin. Microbiol Mol Biol Rev. 1998;62:1371–1414. doi: 10.1128/mmbr.62.4.1371-1414.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seligman SJ. Penicillinase-negative variants of methicillin-resistant Staphylococcus aureus. Nature. 1966;209:994–996. doi: 10.1038/209994a0. [DOI] [PubMed] [Google Scholar]
- Izaki K, Matsuhashi M, Strominger JL. Biosynthesis of the peptidoglycan of bacterial cell walls. 8. Peptidoglycan transpeptidase and D-alanine carboxypeptidase: penicillin-sensitive enzymatic reaction in strains of Escherichia coli. The Journal of biological chemistry. 1968;243:3180–3192. [PubMed] [Google Scholar]
- Neu HC. The crisis in antibiotic resistance. Science. 1992;257:1064–73. doi: 10.1126/science.257.5073.1064. [DOI] [PubMed] [Google Scholar]
- Hoare DS, Work E. The stereoisomers of alpha epsilon-diaminopimelic acid. II. Their distribution in the bacterial order Actinomycetales and in certain Eubacteriales. The Biochemical journal. 1957;65:441–447. doi: 10.1042/bj0650441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox RJ. The DAP pathway to lysine as a target for antimicrobial agents. Natural product reports. 1996;13:29–43. doi: 10.1039/np9961300029. [DOI] [PubMed] [Google Scholar]
- Scapin G, Blanchard JS. Enzymology of bacterial lysine biosynthesis. Advances in enzymology and related areas of molecular biology. 1998;72:279–324. doi: 10.1002/9780470123188.ch8. [DOI] [PubMed] [Google Scholar]
- Born TL, Blanchard JS. Structure/function studies on enzymes in the diaminopimelate pathway of bacterial cell wall biosynthesis. Current opinion in chemical biology. 1999;3:607–613. doi: 10.1016/s1367-5931(99)00016-2. [DOI] [PubMed] [Google Scholar]
- Cox RJ, Sutherland A, Vederas JC. Bacterial diaminopimelate metabolism as a target for antibiotic design. Bioorganic & medicinal chemistry. 2000;8:843–871. doi: 10.1016/s0968-0896(00)00044-4. [DOI] [PubMed] [Google Scholar]
- Hayashi H, Wada H, Yoshimura T, Esaki N, Soda K. Recent topics in pyridoxal 5'-phosphate enzyme studies. Annual review of biochemistry. 1990;59:87–110. doi: 10.1146/annurev.bi.59.070190.000511. [DOI] [PubMed] [Google Scholar]
- Das A, Silaghi-Dumitrescu R, Ljungdahl LG, Kurtz DM., Jr Cytochrome bd oxidase, oxidative stress, and dioxygen tolerance of the strictly anaerobic bacterium Moorella thermoacetica. Journal of bacteriology. 2005;187:2020–2029. doi: 10.1128/JB.187.6.2020-2029.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govantes F, Orjalo AV, Gunsalus RP. Interplay between three global regulatory proteins mediates oxygen regulation of the Escherichia coli cytochrome d oxidase (cydAB) operon. Molecular microbiology. 2000;38:1061–1073. doi: 10.1046/j.1365-2958.2000.02215.x. [DOI] [PubMed] [Google Scholar]
- Cook GM, Loder C, Soballe B, Stafford GP, Membrillo-Hernandez J, Poole RK. A factor produced by Escherichia coli K-12 inhibits the growth of E. coli mutants defective in the cytochrome bd quinol oxidase complex: enterochelin rediscovered. Microbiology. 1998;144:3297–3308. doi: 10.1099/00221287-144-12-3297. [DOI] [PubMed] [Google Scholar]
- Mader U, Hennig S, Hecker M, Homuth G. Transcriptional organization and posttranscriptional regulation of the Bacillus subtilis branched-chain amino acid biosynthesis genes. Journal of bacteriology. 2004;186:2240–2252. doi: 10.1128/JB.186.8.2240-2252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Riordan K, Lee JC. Staphylococcus aureus capsular polysaccharides. Clin Microbiol Rev. 2004;17:218–234. doi: 10.1128/CMR.17.1.218-234.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian SK, Skaar EP, Gaspar AH, Humayun M, Gornicki P, Jelenska J, Joachmiak A, Missiakas DM, Schneewind O. Passage of heme-iron across the envelope of Staphylococcus aureus. Science. 2003;299:906–9. doi: 10.1126/science.1081147. [DOI] [PubMed] [Google Scholar]
- Skaar EP, Schneewind O. Iron-regulated surface determinants (Isd) of Staphylococcus aureus: stealing iron from heme. Microbes and infection/Institut Pasteur. 2004;6:390–397. doi: 10.1016/j.micinf.2003.12.008. [DOI] [PubMed] [Google Scholar]
- Bacher A, Eberhardt S, Eisenreich W, Fischer M, Herz S, Illarionov B, Kis K, Richter G. Biosynthesis of riboflavin. Vitam Horm. 2001;61:1–49. doi: 10.1016/s0083-6729(01)61001-x. [DOI] [PubMed] [Google Scholar]
- Vitreschak AG, Rodionov DA, Mironov AA, Gelfand MS. Regulation of riboflavin biosynthesis and transport genes in bacteria by transcriptional and translational attenuation. Nucleic acids research. 2002;30:3141–3151. doi: 10.1093/nar/gkf433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramton SE, Gerke C, Schnell NF, Nichols WW, Gotz F. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect Immun. 1999;67:5427–5433. doi: 10.1128/iai.67.10.5427-5433.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp ME, Ulphani JS, Fey PD, Bartscht K, Mack D. Characterization of the importance of polysaccharide intercellular adhesin/hemagglutinin of Staphylococcus epidermidis in the pathogenesis of biomaterial-based infection in a mouse foreign body infection model. Infect Immun. 1999;67:2627–2632. doi: 10.1128/iai.67.5.2627-2632.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp ME, Fey PD, Heilmann C, Gotz F. Characterization of the importance of Staphylococcus epidermidis autolysin and polysaccharide intercellular adhesin in the pathogenesis of intravascular catheter-associated infection in a rat model. J Infect Dis. 2001;183:1038–1042. doi: 10.1086/319279. [DOI] [PubMed] [Google Scholar]
- Rachid S, Ohlsen K, Witte W, Hacker J, Ziebuhr W. Effect of subinhibitory antibiotic concentrations on polysaccharide intercellular adhesin expression in biofilm-forming Staphylococcus epidermidis. Antimicrob Agents Chemother. 2000;44:3357–63. doi: 10.1128/aac.44.12.3357-3363.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dukan S, Levi Y, Touati D. Recovery of culturability of an HOCl-stressed population of Escherichia coli after incubation in phosphate buffer: resuscitation or regrowth? Appl Environ Microbiol. 1997;63:4204–9. doi: 10.1128/aem.63.11.4204-4209.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The raw data of 7,775 genes control (0 min) and experimental (after 20 and 60 min. exposure of OPP). It has been also deposited in NCBI's Gene Expression Omnibus http://www.ncbi.nlm.nih.gov/geo and is accessible through GEO Series accession number GSE10605 http://www.ncbi.nlm.nih.gov/projects/geo/query/acc.cgi?acc=GSE10605&targ=self&form=html&view=quick.
Staphylococcus aureus 669 genes that showed statistically significant mRNA level changes upon either 20 or 60 min exposure to OPP. The genes were grouped based on their regulation directions upon 20 and 60 min exposures.