Abstract
Geographical variation in the mimetic wing patterns of the butterfly Heliconius erato is a textbook example of adaptive polymorphism; however, little is known about how this variation is controlled developmentally. Using microarrays and qPCR, we identified and compared expression of candidate genes potentially involved with a red/yellow forewing band polymorphism in H. erato. We found that transcripts encoding the pigment synthesis enzymes cinnabar and vermilion showed pattern- and polymorphism-related expression patterns, respectively. cinnabar expression was associated with the forewing band regardless of pigment colour, providing the first gene expression pattern known to be correlated with a major Heliconius colour pattern. In contrast, vermilion expression changed spatially over time in red-banded butterflies, but was not expressed at detectable levels in yellow-banded butterflies, suggesting that regulation of this gene may be involved with the red/yellow polymorphism. Furthermore, we found that the yellow pigment, 3-hydroxykynurenine, is incorporated into wing scales from the haemolymph rather than being synthesized in situ. We propose that some aspects of Heliconius colour patterns are determined by spatio-temporal overlap of pigment gene transcription prepatterns and speculate that evolutionary changes in vermilion regulation may in part underlie an adaptive colour pattern polymorphism.
Keywords: heliconius, evo-devo, cinnabar, vermillion, pigmentation, colour patterns
1. Introduction
The butterfly genus Heliconius has been of special interest to biologists for well over a century owing to the extraordinary adaptive variation in their wing patterns. Within any area of the Neotropics, the wing patterns of different heliconiines, ithomiines and even some day-flying moths converge on one of a handful of distinct wing patterns, so-called mimicry rings (Mallet & Gilbert 1995). Most ring participants are unpalatable and the adaptive value of mimetic wing patterns has been demonstrated (Benson 1972; Kapan 2001). The species Heliconius erato is an extreme example of how mimicry can drive colour pattern evolution within a species. There are more than 20 different H. erato colour pattern variants distributed in a largely non-overlapping patchwork across Central and South America (Brown 1979). Crosses between geographical variants of H. erato show that this pattern radiation evolved through allelic changes at a small number of loci (Sheppard et al. 1985; Mallet 1989; Nijhout 1991). It is the goal of the work presented here to achieve a better understanding of the molecular functions of some of these colour pattern genes.
In this study we focus on the activity of genes underlying forewing colour patterns of the species H. erato and Heliconius himera (figure 1). These two species are closely related, and H. himera probably evolved from an isolated H. erato wing-pattern variant within the last 1–2 Myr (Flanagan et al. 2004). Crosses show that the same loci underlying pattern polymorphism in H. erato are also responsible for phenotypic differences between H. himera and H. erato. Therefore, understanding the basis of wing pattern differences between these two species is informative about polymorphism within H. erato itself.
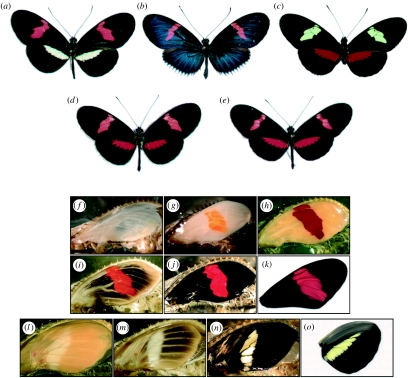
Figure 1.
Wing colour patterns of two races of H. erato: (a) H. erato petiverana and (b) H. erato cyrbia, (c) their sister species H. himera and (d,e) F1 progeny of the interspecific crosses. Pigment development in H. erato petiverana pupal (f–k) forewings and (l–o) hindwings. (f) In early pupae, the wing epithelia are very thin and scales are undeveloped. Scales fated to become (g,l) yellow or red mature first, with red-fated scales progressively darkening from (g) orange to (h) red, and (l–n) yellow-fated scales remaining colourless. (i,j) After the red pigment has matured, melanic scales develop in a ‘wave’ emanating from the centre of the wing. (n) There is a period after the ommochrome and melanin pigments have fully developed that the yellow pigment 3-OHK is undetectable in the wing. (o) Within a few hours before adult emergence 3-OHK appears in yellow-fated scales.
Most forewing pattern differences between H. erato (figure 1a,b) and H. himera (figure 1c) can be explained by allelic differences at two major genes, D and Cr (Sheppard et al. 1985; Jiggins & McMillan 1997; Joron et al. 2006a; Kapan et al. 2006). Many of the effects of D and Cr are beyond the scope of this paper (see Jiggins & McMillan (1997) and Tobler et al. (2005) for more detailed discussions), so here we will focus on how they influence the forewing band phenotype—specifically in terms of the H. himera×Heliconius erato cyrbia cross. The D gene determines if red will be present in the forewing band and at least one Dcyr allele is required for red. Dcyr homozygotes have red forewing bands regardless of the Cr genotype. DcyrDhim heterozygotes can have a mix of red and yellow scales in the forewing band whose ratio will vary depending primarily on which Cr alleles are present—DcyrDhim/CrhimCrhim will be mostly yellow, while DcyrDhim/CrcyrCrhim and DcyrDhim/CrcyrCrcyr will be all or mostly red. Thus, the F1 phenotypes of H. erato cyrbia×H. himera crosses have primarily red forewing bands (figure 1e). Dhim homozygotes will not have red in the forewing band. In these butterflies, the Cr genotype determines a relative blackening of the forewing band so that DhimDhim/CrhimCrhim individuals have a wild-type H. himera yellow band, DhimDhim/CrcyrCrhim have partially blackened forewing bands and DhimDhim/CrcyrCrcyr individuals have most or all of the forewing band blackened out. D and Cr are unlinked (Tobler et al. 2005; Kapan et al. 2006), and Heliconius erato petiverana (figure 1a) and H. e. cyrbia (figure 1b) alleles act similarly on the forewing band (e.g. figure 1d,e). A third unlinked locus, Sd, probably explains differences in the shape of the forewing band (Sheppard et al. 1985; Kapan et al. 2006).
Despite the wealth of information regarding Heliconius wing pattern genetics, little is known about how these patterns develop (Joron et al. 2006b). Most Heliconius wing pattern elements appear to be based on a system of vein-independent proximal–distal boundaries (Gilbert 2003; Reed & Gilbert 2004), and most of the known colour pattern genes affect the shape, position or colour of stripe-like ‘bar’ (hindwing) and ‘band’ (forewing) colour patterns. It is known that the expression of the Notch and Distal-less proteins during imaginal disc development is associated with minor intervenous patterns and scale organization in Heliconius (Reed 2004; Reed & Gilbert 2004; Reed & Serfas 2004); however, no gene expression patterns correlated with the major banding patterns have yet been described.
The ultimate targets of the wing pattern regulatory system probably include genes involved in pigment synthesis. Three pigment types are found in Heliconius: melanin (black), xanthommatin and dihydroxanthommatin (orange and red, respectively) and 3-hydroxykynurenine (3-OHK, yellow; Gilbert et al. 1988). 3-OHK is a precursor of xanthommatin and dihydroxanthommatin, and all three of these pigments share much of the same biosynthetic pathway (Linzen 1974). Although genes involved with the synthesis of these pigments are known to be expressed in the wings of other butterflies (Reed & Nagy 2005), nothing is known about their regulation in Heliconius.
How do a small number of loci regulate pigment pattern diversity in H. erato? One hypothesis, the ‘upstream model’, is that the colour pattern genes act as upstream regulators that activate or repress coordinated sets of effector genes involved with patterning and pigment synthesis. If this model were accurate, for example, genes encoding enzymes and transporters required for pigment synthesis might be expected to be spatially and temporally co-regulated and co-expressed. Alternatively, a ‘downstream model’ would predict that the colour pattern genes would regulate only one or a few of the downstream effector genes, such as those involved in scale maturation or pigment synthesis, to alter the colour pattern.
The major goals of this study were twofold. First, we sought to gain a basic understanding regarding some of the genes and processes that underlie pattern development and polymorphism in H. erato. Second, we sought to use these data to, as much as possible, distinguish between the upstream and downstream models of pigment control in Heliconius. In this paper we cover four sets of experiments and observations. The first concerns the relationship between the colour pattern development and the temporal sequence of scale and pigment development in H. erato pupal wings. The second describes the use of cDNA microarrays to determine transcript levels in band versus non-band tissue in H. erato petiverana. Third, we report on the use of quantitative real time PCR (qPCR) to test for an association of specific transcripts with a red/yellow forewing band colour polymorphism in the H. erato cyrbia and H. himera. Lastly, we sought to determine the source of the yellow pigment in Heliconius wings. Together, our data suggest a model of gene expression that may underlie the characteristics of pattern development and polymorphism in H. erato.
2. Material and methods
(a) Microarrays
cDNA microarrays were constructed representing 99 genes expressed during H. erato wing development (electronic supplementary material). For each microarray experiment, forewings were removed from three late stage H. e. petiverana pupae. Wings were dissected into three regions representing major colour pattern elements, and the tissue samples were grouped into three respective pools from which total RNA was extracted. Labelled cDNA was made by reverse transcription aminoallyl labelling and then used for two-colour microarray hybridizations.
We conducted two comparisons using RNA extracted from pupal wings at a stage approximately 1 day before eclosion. At this time point the scales have matured and ommochrome and melanin pigments have appeared. The first comparison was between the proximal melanic region and the red band, and the second comparison was between the proximal and the distal melanic regions. Data were analysed using GeneSpring (Agilent Technologies), and expression ratios were calculated using median normalization. To determine the significance of transcription differences between tissue types, t-tests were used. Detailed protocols are available in the electronic supplementary material.
(b) Quantitative real-time PCR
Total RNA extracted from identical wing pattern elements from both right and left wings of a single individual was used to synthesize cDNA. Transcription levels were calculated using a standard curve from a cDNA pool standard dilution series. cinnabar and vermilion data were normalized using expression levels of an annexin IX control (see below). Three replicates were executed for each time point. Primer sequences and detailed protocols are available in the electronic supplementary material.
(c) 3-OHK spectrophotometry, radiolabelling and incorporation
Haemolymph 3-OHK concentrations from late-stage H. himera pupae were determined based on 380 nm absorbance. To assay for the presence of 3-OHK in H. himera fat bodies, complete fat bodies were leeched in acidified methanol and spectra were determined from the supernatant. 14C-labelled 3-OHK was generated using a technique similar to that described by Koch (1993). Radiolabelled 3-OHK was injected into late-stage H. himera pupae that were then frozen 1 day after injection. Pupal wings were dissected out of the frozen pupae and exposed to autoradiography film. Detailed protocols are available in the electronic supplementary material.
3. Results
(a) Scale and pigment development in H. erato
Early pupal wing development in H. erato begins with the wing tissue as a thin epidermal bilayer (figure 1f) that thickens rapidly as the scale-forming cells develop during the first few days after pupation (Reed 2004). As in other butterflies (Koch et al. 1998; Reed & Nagy 2005), there is a well-defined progression of scale maturation strongly associated with the colour pattern. In H. erato, the first scales to mature and sclerotize, approximately a week after pupation, are those that will eventually become white, yellow or red in adult wings. These scales appear to mature simultaneously across the surface of the forewing. Red-fated scales begin developing an orange colour, presumably attributable to xanthommatin (figure 1g), and progressively darken until they are dihydroxanthommatin red (figure 1h). Yellow-fated scales mature without visible pigmentation (figure 1l–n) becoming yellow with 3-OHK only a few hours before adult emergence (figure 1o).
After the red, white and (presumptive) yellow scales mature, the melanic scales mature in a ‘wave’ moving outward from the centre of the wing (figure 1i,m). The wing margin and veins are the last regions to display melanin before the final adult phenotype is realized (figure 1j,k,o).
(b) Microarray analysis of transcription in H. erato petiverana wings
Our microarray analysis compared gene expression between the central red band, and the proximal and distal melanic regions of H. erato petiverana pupal forewings at a time point approximately 1 day before adult emergence. This is a developmental stage at which both melanin and ommochrome pigments are evident. This comparison highlighted several transcripts associated with specific regions of the wing (table 1). Of particular interest were two mRNAs, cinnabar and pale, that were found to have significantly higher expression in the red band compared with the proximal and distal melanic regions of the wing. cinnabar encodes an enzyme, kynurenine 3-monooxygenase (kynurenine hydroxylase, EC: 1.14.13.9), implicated in ommochrome synthesis, and has previously been reported to be expressed in butterfly wings (Reed & Nagy 2005). Pale encodes tyrosine 3-monooxygenase (tyrosine hydroxylase, EC:1.14.16.2), an enzyme associated with both melanin and cuticle development in many insects.
Table 1.
Significant differences in gene transcription between colour patterns dissected from late-stage H. erato pativerana pupal wings. (FC, fold change; *p< 0.05; **p< 0.005.)
| tissue type | top blast hita | FC | putative function | accession no. |
|---|---|---|---|---|
| red band versus proximal transcription differences | ||||
| red band | Kynurenine 3-monooxygenase (cinnabar); Vanessa cardui; 2e-63 | 11.4** | ommochrome synthesis enzyme | DQ380224 |
| Tyrosine hydroxylase (pale); Papilio xuthus; 1e-57 | 2.5** | melanin synthesis enzyme | EL598252 | |
| proximal | Larval cuticle protein A1A; Tenebrio molitor; 4e-28 | 4.8** | cuticle protein | EL597152 |
| distal versus proximal transcription differences | ||||
| distal | Gasp; Bombyx mori; 1e-127 | 2.1* | chitin metabolism | CV525685 |
| proximal | Larval cuticle protein A1A; Tenebrio molitor; 4e-28 | 12.7** | cuticle protein | EL597152 |
| Pupal cuticle protein precursor; Bombyx mori; 8e-17 | 3.0** | cuticle protein | CV525741 | |
| Putative cuticle protein; Manduca sexta; 6e-56 | 2.6** | cuticle protein | EL602566 | |
| No Significant Hit | 2.4** | Unknown | CV526418 | |
| Similar to CG1919-PA; Tribolium castaneum; 4e-24 | 2.0** | cuticle protein | EL598743 | |
| Similar to CG1919-PA; Tribolium castaneum; 3e-24 | 2.0** | cuticle protein | EL596529 | |
Additional blast information is available in the electronic supplementary material A.
(c) cinnabar expression associated with the forewing band
In order to identify suitable normalization transcripts for the qPCR experiments, we scanned the microarray data for genes that varied the least between wing regions across comparisons, yet were also transcribed at relatively high levels (table S1 in the electronic supplementary material). Of the candidates tested, the annexin IX transcript produced the most robust and consistent signal in qPCR experiments, so we used this transcript for qPCR normalization.
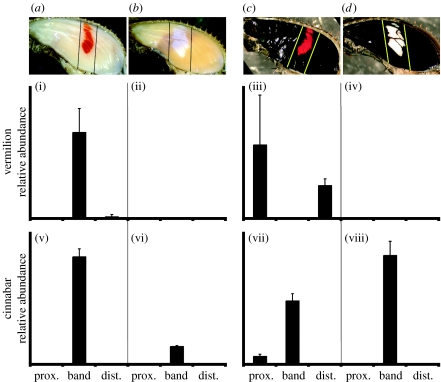
Given the results from the H. erato petiverana microarray experiments, we selected cinnabar as a candidate gene for qPCR comparison between H. erato cyrbia and H. himera forewings. We found that cinnabar transcript abundance was associated with the forewing band in both H. erato cyrbia and H. himera during both early ommochrome and late melanin development (figure 2v–viii).
Figure 2.
(a,b) Early ommochrome development (cyrbia, himera, respectively) and (c,d) late melanin development (cyrbia, himera, respectively). Quantitative RT-PCR comparison of (i–iv) vermilion and (v–viii) cinnabar transcript abundance during pigment development in (i,v,iii,vii) H. e. cyrbia and (ii,vi,iv,viii) H. himera. Transcription levels are presented relative to control gene expression and error bars represent the standard deviation. Synthesis of red pigment occurs when the transcription of both genes spatio-temporally coincides in the forewing band (i,v).
(d) vermilion expression associated with red/yellow polymorphism
Both cinnabar and vermilion encode enzymes implicated in ommochrome pigment synthesis (Reed & Nagy 2005). It was therefore surprising that in the microarray experiments vermilion did not show significantly higher abundance specifically in the red forewing band (table 1). We used qPCR to determine whether cinnabar and vermilion might be co-regulated in some manner not detected by the microarray.
In H. erato cyrbia, vermilion transcript abundance appeared to be generally associated with the timing of scale maturation. During early ommochrome development, when the red band scales were maturing, vermilion had a high relative abundance in the band (figure 2i). Later, when the melanic scales were maturing, vermilion was expressed at high levels primarily in melanic tissue, but not in the red band tissue (figure 2iii). It is this latter time point that was sampled in the H. erato petiverana microarray work, which may explain why the ommochrome-stage red band association was not detected by this method. In H. himera, there was no detectable vermilion expression in the forewings during ommochrome or melanin development (figure 2ii,iv). The fact that vermilion was detectable in red forewing band tissue in H. erato cyrbia but was undetectable in yellow forewing band H. himera demonstrates a polymorphism-related difference in transcription of this gene.
(e) 3-OHK circulation and incorporation into yellow scales
Activity of the vermilion enzyme is required for 3-OHK synthesis. This leads to the question: if vermilion is not expressed at detectable levels in H. himera wings, then what is the source of the yellow pigment in the wings?
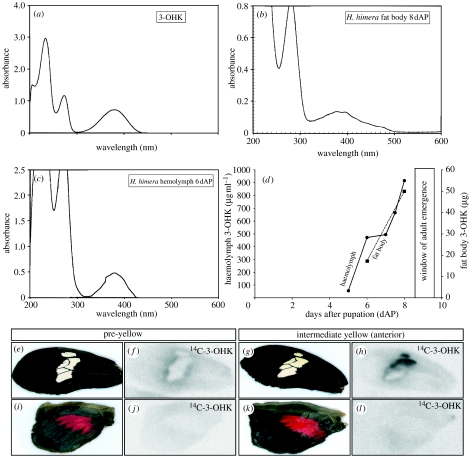
We noted that haemolymph sampled from mid- and late-stage Heliconius pupae was bright yellow, resembling the colour of 3-OHK in solution. Pure 3-OHK (figure 3a) and late-pupal H. himera haemolymph (figure 3c) show similar absorbance spectra, with nearly identical absorbance peaks at 380 nm, suggesting that 3-OHK is a major component of the Heliconius haemolymph late in the pupal development. Analysis of 380 nm absorbance peaks from a time series of H. himera haemolymph samples (figure 3d) shows that 3-OHK titres are relatively low (approx. 50 μg ml−1) before pigment development (day 5), moderately high (approx. 700 μg ml−1) during ommochrome development (day 7) and very high (approx. 900 μg ml−1) shortly before adult emergence (day 8). Furthermore, the amount of 3-OHK extracted from fat bodies (figure 3b) increases in concert with the haemolymph concentration (figure 3d).
Figure 3.
The yellow pigment 3-OHK is stored in the fat bodies, circulated in the haemolymph and taken up from the haemolymph into yellow scales. The absorbance spectrum of (a) pure 3-OHK is similar to (b) fat body extract and (c) haemolymph, suggesting that 3-OHK is a major component of both. (d) There is a rapid increase of 3-OHK in the haemolymph (solid line) and the fat bodies (broken line) of late-stage pupae, as inferred from spectrophotometric analysis of the 380 nm absorbance peak. 14C-3-OHK injected into pupal haemolymph is incorporated specifically into yellow scales. (e) At a stage shortly preceding the appearance of yellow in the H. himera forewing band, (f) only low levels of 14C-3-OHK are incorporated around the edges of the band. At an intermediate stage of yellow development, when 3-OHK is observed only in the anterior portion of the forewing band, high levels of (h) 14C-3-OHK incorporation are perfectly correlated with (g) yellow pigmentation. (i–l) No incorporation of 14C-3-OHK was detected in red hindwing scales.
To test whether 3-OHK circulating in the haemolymph is incorporated into yellow scales, we injected 14C-labelled 3-OHK into the haemolymph of H. himera pupae 1 day before emergence, during late melanin development. Pupal wings at a stage shortly preceding the appearance of yellow in the forewing band show only low levels of 14C-3-OHK incorporation around the margin of the band (figure 3e,f). Very high levels of 14C-3-OHK incorporation, perfectly associated with yellow pigmentation, were observed at a later stage of yellow development when 3-OHK is observed in the anterior portion of the band (figure 3g,h). No significant incorporation was detected in red or black scales on the forewing or hindwing (figure 3e–l). In sum, these results support the idea that 3-OHK circulating in the haemolymph is incorporated into yellow scales in a pattern-specific manner.
4. Discussion
(a) cinnabar and vermilion regulation during Heliconius wing pattern development
The forewing band colour patterns are among the largest and the most common pattern elements in Heliconius and are important for both mimicry and mate recognition (Jiggins et al. 2001; Kapan 2001; McMillan et al. 2002). We report here that transcription of the cinnabar gene is associated with the Heliconius forewing band. This represents the first report of gene expression associated with a major Heliconius wing pattern. Additionally, the apparent association of vermilion expression with the yellow/red polymorphism provides a candidate gene for an element of the developmental pathway controlled by the D and Cr colour pattern genes.
How might the regulation of cinnabar and vermilion relate to the activity of D and Cr? cinnabar expression was not qualitatively different between the genotypes we compared, so very little can be surmised about how this gene might be regulated by D or Cr. vermilion, on the other hand, showed a dramatic difference between the butterflies surveyed, and we speculate that D could be involved in the regulation of this gene. Genetically, D is critical for red pigmentation in the forewing band, where at least one Dcyr allele is required for ommochrome synthesis, regardless of the Cr genotype. If the red/yellow polymorphism operates in part by preventing ommochrome synthesis through the repression of vermilion expression, it would make sense that Dcyr could be a positive regulator of vermilion. Further work is required to test this hypothesis.
(b) Yellow wing pigment is taken up from the haemolymph
One novel finding from this study concerns the timing of 3-OHK deposition in yellow pattern elements. Because 3-OHK is a precursor of dihydroxanthommatin (figure 4a), our initial hypothesis was that 3-OHK would be visible in scales before, or at the same time as, xanthommatin during wing development. The finding that 3-OHK appeared long after dihydroxanthommatin was surprising and suggests that there is a fundamental difference in the production of 3-OHK versus ommochromes in Heliconius scales. Indeed, vermilion transcripts were undetectable in H. himera forewings (figure 2ii,iv), even very late in pigment development, suggesting that in situ activity of the ommochrome biosynthesis pathway is not required for the occurrence of 3-OHK in scales.
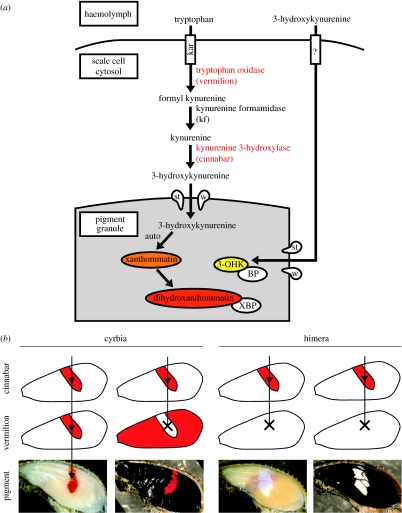
Figure 4.
(a) A hypothetical model of ommochrome synthesis in butterfly wing scales based on the work from the Drosophila eye. Tryptophan, the ommochrome precursor, is thought to be taken up into cells by the karmoisin transporter. Tryptophan is then processed by several enzymes, including those encoded by vermilion and cinnabar into 3-OHK, which is then transported into granules by the scarlet/white heterodimer. 3-OHK is processed into xanthommatin in a granule, where a binding protein (XBP) is thought to modulate the redox state of xanthommatin, thereby determining whether the pigment will appear more orange or red in hue. 3-OHK can be taken up directly into scales, and we have previously speculated that a binding protein (BP) may be required to stabilize this molecule in a pigment granule. See Reed & Nagy (2005) for a detailed discussion. (b) A working model of pigment regulation through the overlap of vermilion and cinnabar prepatterns. Red pigment is synthesized at the time and place transcription of vermilion and cinnabar (shown in red) overlap.
Where does the yellow pigment come from in Heliconius wings? We observed that 3-OHK concentration in the haemolymph increases rapidly in late pupal development (figure 3d), and that radiolabelled 3-OHK is taken up from the haemolymph into yellow scale cells (figure 3h). These findings support the model that yellow pigment is produced in the pupa somewhere other than the wing, circulates in the haemolymph and is transported into scales just prior to adult emergence. There is a precedent for this model in that wings of some nymphalid butterflies incorporate radiolabelled 3-OHK into ommochrome-bearing scales (Koch 1991, 1993). We speculate that the fat bodies may be the major source of 3-OHK in Heliconius owing to their orange ommochrome-like coloration (not shown), and because the 3-OHK in the fat bodies increases in concert with the haemolymph 3-OHK concentration (figure 3d).
(c) Pigmentation associated with prepattern overlap and not coordinated gene regulation: evolutionary implications
The synthesis of ommochrome pigments requires the concerted activity of multiple transporters and enzymes (see Reed & Nagy 2005 for discussion), and it would be reasonable to hypothesize that the genes encoding some of these molecules would be genetically co-regulated, either directly or indirectly, by a common upstream regulatory mechanism. Under this upstream model, one might predict that cinnabar and vermilion would be co-expressed, as they both contribute towards a shared biosynthetic function (figure 4a). Our data provide evidence to the contrary: both transcripts showed different spatio-temporal patterns of expression, implying that they are not co-regulated in a modular fashion and that some aspects of colour pattern regulation may occur through changes in the regulation of individual downstream effector genes. Nevertheless, both vermilion and cinnabar were transcribed in the red band of the wing when the red pigment was being synthesized. Both genes are also expressed in the absence of the other in regions uncorrelated with the red pigmentation. Together, these data support the hypothesis that dihydroxanthommatin synthesis occurs in areas of overlapping cinnabar and vermilion expression (figure 4b).
Interestingly, the putative ommochrome precursor transporters, scarlet, white and atet-like, were not detected by the microarray as being significantly upregulated in specific association with the red band during late melanin development (table 1). These genes are expressed during wing development; however, more work is required to better understand their expression patterns over time. Overall, we visualize the expression of ommochrome-related genes as wing-wide system of overlapping prepatterns, where the area and time point of expression overlap, in the context of the timing of scale maturation (see Koch et al. 2000), contributes to ommochrome synthesis (figure 4b).
This overlapping prepattern model of pigment development has some evolutionary implications. First of all, it suggests that polymorphism can be achieved through the modulation of a ‘limiting factor’ downstream effector gene, possibly vermilion in this case. By downregulating a single enzyme, a biosynthetic process can be prevented from running to completion. This might be an easier feat from an evolutionary perspective than changing the regulation or function of highly pleiotropic upstream regulatory genes or turning an entire enzymatic cascade off and on. A similar effect is found in Drosophila, where pale expression acts as a limiting factor over a prepattern of DDC expression in the production of melanic wing patterns (True et al. 1999).
Another implication of this model is that natural selection on regulation of the prepatterns could be relatively weak. Prepatterns may have little or no effect on the phenotype when they occur outside the regions of transcriptional overlap. For instance, it is hard to imagine a function for vermilion expression in the melanic regions of the Heliconius pupal wing during late development, and there would not necessarily be strong selection against this presumably non-functional expression. It may be possible that some gene expression has no significant phenotypic effect or adaptive value. If this model of ‘sloppy’ gene expression were applicable, we might predict observing cryptic variation in pigment gene prepatterning within and between races and species of Heliconius. Cryptic variation in prepatterning has been noted in butterfly eyespot development (Reed et al. 2007), and future work will hopefully address this issue in Heliconius.
Acknowledgements
We are grateful to the Ecuador Ministerio del Ambiente for collecting permits and to Ana Maria Quiles, Karla Maldonado and Lournet Martinez for their help rearing Heliconius at UPR. We thank Brian Coullahan and the University of Arizona Genomic Analysis and Technology Core for microarray assistance, The Sequencing and Genotyping Facility at UPR for EST sequencing and Fred Nijhout for helpful discussions. This work was funded by United States National Science Foundation grants DEB 0209441, DEB 9806792, IBN 0344705, DEB 0640788, and a University of Arizona IGERT Genomics Fellowship to R.D.R.
Supplementary Material
Candidate genes for qPCR normalization
Detailed materials and methods: (a) Microarrays; (b) Quantitative real-time PCR; (c) 3‐OHK spectrophotometry, radiolabeling, and incorporation
Features on microarray
References
- Benson W.W. Natural selection for Müllerian mimicry in Heliconius erato in Costa Rica. Science. 1972;176:936–939. doi: 10.1126/science.176.4037.936. doi:10.1126/science.176.4037.936 [DOI] [PubMed] [Google Scholar]
- Brown, K. S. J. 1979 Ecologia Geográfica e Evolução nas Florestas Neotropicais PhD dissertation. Universidade Estadual de Campinas, Campinas.
- Flanagan N.S, Tobler A, Davison A, Pybus O.G, Kapan D.D, Planas S, Linares M, Heckel D, McMillan W.O. Historical demography of Mullerian mimicry in the neotropical Heliconius butterflies. Proc. Natl Acad. Sci. USA. 2004;101:9704–9709. doi: 10.1073/pnas.0306243101. doi:10.1073/pnas.0306243101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert L.E. Adaptive novelty through introgression in Heliconius wing patterns: evidence for a shared genetic “toolbox” from synthetic hybrid zones and a theory of diversification. In: Boogs C.L, Watt W.B, Ehrlich P.R, editors. Butterflies: ecology and evolution taking flight. The University of Chicago Press; Chicago, IL: 2003. [Google Scholar]
- Gilbert L.E, Forrest H.S, Schultz T.D, Harvey D.J. Correlations of ultrastructure and pigmentation suggest how genes control development of wing scale of Heliconius butterflies. J. Res. Lepidopt. 1988;26:141–160. [Google Scholar]
- Jiggins C.D, McMillan W.O. The genetic basis of an adaptive radiation: warning colour in two Heliconius species. Proc. R. Soc. B. 1997;264:1167–1175. doi:10.1098/rspb.1997.0161 [Google Scholar]
- Jiggins C.D, Naisbit R.E, Coe R.L, Mallet J. Reproductive isolation caused by colour pattern mimicry. Nature. 2001;411:302–305. doi: 10.1038/35077075. doi:10.1038/35077075 [DOI] [PubMed] [Google Scholar]
- Joron M, et al. A conserved supergene locus controls wing pattern diversity in Heliconius butterflies. PLoS Biol. 2006a;4:1831–1840. doi: 10.1371/journal.pbio.0040303. doi:10.1371/journal.pbio.0040303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joron M, Jiggins C.D, Papanicolaou A, McMillan W.O. Heliconius wing patterns: an evo-devo model for understanding phenotypic diversity. Heredity. 2006b;97:157–167. doi: 10.1038/sj.hdy.6800873. doi:10.1038/sj.hdy.6800873 [DOI] [PubMed] [Google Scholar]
- Kapan D.D. Three-butterfly system provides a field test of mullerian mimicry. Nature. 2001;409:338–340. doi: 10.1038/35053066. doi:10.1038/35053066 [DOI] [PubMed] [Google Scholar]
- Kapan D.D, et al. Localization of Müllerian mimicry genes on a dense linkage map of Heliconius erato. Genetics. 2006;173:735–757. doi: 10.1534/genetics.106.057166. doi:10.1534/genetics.106.057166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch P.B. Precursors of pattern specific ommatin in red wing scales of the polyphenic butterfly Araschnia levana L.: haemolymph tryptophan and 3-hydroxykynurenine. Insect Biochem. 1991;21:785–794. doi:10.1016/0020-1790(91)90120-4 [Google Scholar]
- Koch P.B. Production of [C-14] labeled 3-hydroxy-l-kynurenine in a butterfly, Heliconius charitonia L. (Heliconidae), and precursor studies in butterfly wing ommatins. Pigment Cell Res. 1993;6:85–90. doi: 10.1111/j.1600-0749.1993.tb00586.x. doi:10.1111/j.1600-0749.1993.tb00586.x [DOI] [PubMed] [Google Scholar]
- Koch P.B, Keys D.N, Rocheleau T, Aronstein K, Blackburn M, Carroll S.B, ffrench-Constant R.H. Regulation of dopa decarboxylase expression during colour pattern formation in wild-type and melanic tiger swallowtail butterflies. Development. 1998;125:2303–2313. doi: 10.1242/dev.125.12.2303. [DOI] [PubMed] [Google Scholar]
- Koch P.B, Lorenz U, Brakefield P.M, ffrench-Constant R.H. Butterfly wing pattern mutants: developmental heterochrony and co-ordinately regulated phenotypes. Dev. Genes Evol. 2000;210:536–544. doi: 10.1007/s004270000101. doi:10.1007/s004270000101 [DOI] [PubMed] [Google Scholar]
- Linzen B. The tryptophan–ommochrome pathway in insects. Adv. Insect Physiol. 1974;10:117–246. [Google Scholar]
- Mallet J. The genetics of warning colour in Peruvian hybrid zones of Heliconius erato and H. melpomene. Proc. R. Soc. B. 1989;236:163–185. doi:10.1098/rspb.1989.0019 [Google Scholar]
- Mallet J, Gilbert L.E. Why are there so many mimicry rings—correlations between habitat, behavior and mimicry in Heliconius butterflies. Biol. J. Linn. Soc. 1995;55:159–180. doi:10.1006/bijl.1995.0034 [Google Scholar]
- McMillan W.O, Monteiro A, Kapan D.D. Development and evolution on the wing. Trends Ecol. Evol. 2002;17:125–133. doi:10.1016/S0169-5347(01)02427-2 [Google Scholar]
- Nijhout H.F. Smithsonian Institution Press; Washington, DC: 1991. The development and evolution of butterfly wing patterns. [Google Scholar]
- Reed R.D. Evidence for Notch-mediated lateral inhibition in organizing butterfly wing scales. Dev. Genes Evol. 2004;214:43–46. doi: 10.1007/s00427-003-0366-0. doi:10.1007/s00427-003-0366-0 [DOI] [PubMed] [Google Scholar]
- Reed R.D, Gilbert L.E. Wing venation and Distal-less expression in Heliconius butterfly wing pattern development. Dev. Genes Evol. 2004;214:628–634. doi: 10.1007/s00427-004-0439-8. doi:10.1007/s00427-004-0439-8 [DOI] [PubMed] [Google Scholar]
- Reed R.D, Nagy L.M. Evolutionary redeployment of a biosynthetic module: expression of eye pigment gene vermilion cinnabar, and white during butterfly wing development. Evol. Dev. 2005;7:301–311. doi: 10.1111/j.1525-142X.2005.05036.x. doi:10.1111/j.1525-142X.2005.05036.x [DOI] [PubMed] [Google Scholar]
- Reed R.D, Serfas M.S. Butterfly wing pattern evolution is associated with changes in a Notch/Distal-less temporal pattern formation process. Curr. Biol. 2004;14:1159–1166. doi: 10.1016/j.cub.2004.06.046. doi:10.1016/j.cub.2004.06.046 [DOI] [PubMed] [Google Scholar]
- Reed R.D, Chen P.-H, Nijhout H.F. Cryptic variation in butterfly eyespot development: the importance of sample size in gene expression studies. Evol. Dev. 2007;9:2–9. doi: 10.1111/j.1525-142X.2006.00133.x. [DOI] [PubMed] [Google Scholar]
- Sheppard P.M, Turner J.R.G, Brown K.S, Benson W.W, Singer M.C. Genetics and evolution of muellerian mimcry in Heliconius butterflies. Phil. Trans. R. Soc. B. 1985;308:433–613. doi:10.1098/rstb.1985.0066 [Google Scholar]
- Tobler A, Kapan D, Flanagan N.S, Gonzalez C, Peterson E, Jiggins C.D, Johntson J.S, Heckel D.G, McMillan W.O. First-generation linkage map of the warningly colored butterfly Heliconius erato. Heredity. 2005;94:408–417. doi: 10.1038/sj.hdy.6800619. doi:10.1038/sj.hdy.6800619 [DOI] [PubMed] [Google Scholar]
- True J.R, Edwards K.A, Yamamoto D, Carroll S.B. Drosophila wing melanin patterns form by vein-dependent elaboration of enzymatic prepatterns. Curr. Biol. 1999;9:1382–1391. doi: 10.1016/s0960-9822(00)80083-4. doi:10.1016/S0960-9822(00)80083-4 [DOI] [PubMed] [Google Scholar]
Notice of correction
The gene names of cinnabar and vermilion are now presented in the correct form. 31 October 2007
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Candidate genes for qPCR normalization
Detailed materials and methods: (a) Microarrays; (b) Quantitative real-time PCR; (c) 3‐OHK spectrophotometry, radiolabeling, and incorporation
Features on microarray