Abstract
In the course of liquid culture, serial passage experiments with Escherichia coli K-12 bearing a mutator gene deletion (ΔmutS) we observed the evolution of strains that appeared to kill or inhibit the growth of the bacteria from where they were derived, their ancestors. We demonstrate that this inhibition occurs after the cells stop growing and requires physical contact between the evolved and ancestral bacteria. Thereby, it is referred to as stationary phase contact-dependent inhibition (SCDI). The evolution of this antagonistic relationship is not anticipated from existing theory and experiments of competition in mass (liquid) culture. Nevertheless, it occurred in the same way (parallel evolution) in the eight independent serial transfer cultures, through different single base substitutions in a gene in the glycogen synthesis pathway, glgC. We demonstrate that the observed mutations in glgC, which codes for ADP-glucose pyrophosphorylase, are responsible for both the ability of the evolved bacteria to inhibit or kill their ancestors and their immunity to that inhibition or killing. We present evidence that without additional evolution, mutator genes, or known mutations in glgC, other strains of E. coli K-12 are also capable of SCDI or sensitive to this inhibition. We interpret this, in part, as support for the generality of SCDI and also as suggesting that the glgC mutations responsible for the SCDI, which evolved in our experiments, may suppress the action of one or more genes responsible for the sensitivity of E. coli to SCDI. Using numerical solutions to a mathematical model and in vitro experiments, we explore the population dynamics of SCDI and postulate the conditions responsible for its evolution in mass culture. We conclude with a brief discussion of the potential ecological significance of SCDI and its possible utility for the development of antimicrobial agents, which unlike existing antibiotics, can kill or inhibit the growth of bacteria that are not growing.
Keywords: Escherichia coli, allelopathy, parallel evolution, glycogen, population biology, mathematical models
1. Introduction
When bacteria compete in liquid culture, the evolution of mutants that replicate faster on existing or by-product resources made available by the metabolism or death of other members of the community is anticipated and has been observed (Helling et al. 1987; Lenski et al. 1991; Zambrano & Kolter 1996; Farrell & Finkel 2003). Not expected in these liquid (mass) culture conditions is the evolution of antagonistic relationships, mechanisms that enable bacteria to kill or inhibit the growth of competitors, like the production of bacteriocins or antibiotics. In the absence of spatial heterogeneity, all members of the community gain equally from the resources made available by the death or growth inhibition of other members. As a result, individual bacteria producing these allelopathic agents and engendering the cost of their production would not gain advantage that would enable them to increase in frequency when they are rare (Chao & Levin 1981; Levin 1988; Kerr et al. 2002). Nevertheless, in broth cultures of Escherichia coli we observed the rapid evolution of mutants that appeared to inhibit the growth or kill the bacteria from where they were derived—their ancestors. This inhibition does not occur until these bacteria are at stationary phase, a time when they are refractory to other toxic agents like antibiotics (Bigger 1944; McDermott 1958; Levin & Rozen 2006).
In this report, we examine the properties of this evolved antagonistic interaction, determine its genetic basis, explore its generality in other strains of E. coli and with the aid of a mathematical model explore the population dynamics of this process and the conditions responsible for its evolution. We conclude with a brief discussion of the potential ecological implications and practical applications of this phenomenon.
2. Material and methods
In the following we describe some of the materials and methods used in this investigation. More information about these methods can be found in the electronic supplementary material.
(a) Culture medium
Unless noted, bacterial strains were grown at 37°C in Luria–Bertani (LB) media (Miller 1972) or Kornberg (KB) media (1.1% K2HPO4, 0.85% KH2PO4, 0.6% yeast extract and 1% glucose).
(b) Sampling methods
Estimates of the densities of the bacterial cultures and the relative frequencies of the different populations were obtained by serial dilution and plating on LB agar, LB agar with streptomycin (100 μg ml−1), ampicillin (100 μg ml−1), chloramphenicol (25 μg ml−1) or tetracycline (10 μg ml−1). Tetrazolium arabinose (TA; Levin et al. 1977) agar or McConkey agar (DIFCO 1984) containing 1% arabinose was also used to estimate the relative frequencies of strains that differed by an arabinose marker. Unless otherwise noted, liquid cultures were in 50 ml Erlenmeyer flask containing LB (10 ml) and were maintained at 37°C with shaking at between 150 and 250g.
(c) Bacteria
AB1157. thr-1 leu-6 thi-1 lacY1 galK2 ara-14 xyl-5 mtl-1 proA2 his-4 argE3 str-31 tsx-33 supE44 rec+F-.
AB1157ΔmutS::spc gyrA (a derivative of AB1157 provided by Ivan Matic). For convenience, we designate this most prominently used strain as ABM. We also used spontaneous Ara+ revertants of this strain for mixed culture experiments.
MC1061. F− araD139 Δ(ara-leu)7696 galE15 galK16 Δ(lac)X74 rpsL (Strr) hsdR2 (rK− mK+) mcrA mcrB1.
MG1655. ilvG rfb-50 rph− λ−-F-.
C600. F-, thi-1, thr-1, leuB6, lacY1, tonA21, supE44, λ−.
Escherichia coli B. No mutations known CGSC#: 5713.
(d) Mixed culture experiments
Unless otherwise noted in the electronic supplementary material, strains in the mixed culture experiments were distinguished by an arabinose marker, pink Ara+ and deep red Ara− colonies on the TA agar. For this, we used spontaneous Ara+ mutants of the ancestral and evolved ABM. Reciprocal Ara+ evolved and Ara− ancestral and Ara− evolved and Ara+ ancestral mixed culture experiments were performed to control for the effects of the Ara− marker. None were observed.
(e) Fluorescence-activated cell sorting analysis
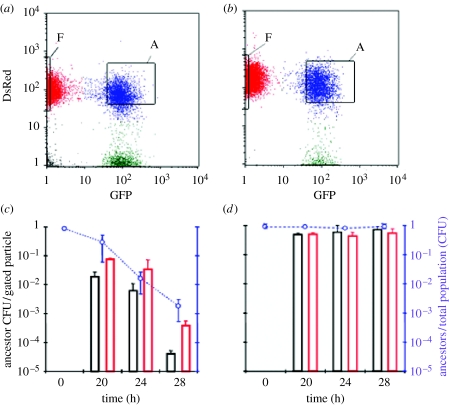
Discosoma red (DsRed)-labelled E. coli were obtained by transformation of ABM cells with plasmid pMW211; a colE1 derivative that expresses a DsRed variant protein and confers resistance to ampicillin (a kind gift from Dr Arthur Altenhoefer, University of Wuerzburg). Green fluorescent protein (GFP)-labelled E. coli were obtained following transformation of ABM cells with the multicopy plasmid pGFP (AmpR), (a kind gift from S. Méresse, centre d'Immunologie de Marseille-Luminy). Flow cytometry was carried out using a Becton Dickinson fluorescence-activated cell sorting (FACS) Vintage SE instrument with a 70 μm sorting nozzle at low pressure (12 psi). GFP and DsRed were excited using a 488 nm blue laser and detected using 530/30 and 585/42 nm filters, respectively. DsRed-labelled ancestral ABM and GFP-labelled evolved ABM E. coli were prepared by overnight growth in LB broth supplemented with ampicillin (75 μg ml−1). Mixed cultures (10 ml) initiated at a 10 : 1 ancestor-to-evolved ratio were incubated in 500 ml culture flasks at 37°C with shaking at 250g for the times indicated (figure 3). Evolved-bound ancestor cells and free ancestor cells were isolated by sorting approximately 30 000 particles of each cell population into separate tubes containing phosphate-buffered saline (0.3 ml). Serial dilutions of the samples were plated on McConkey agar plates containing arabinose to discriminate between Ara− (evolved) and Ara+ (ancestors) bacteria. For the control experiment shown in figure 3b,d, the GFP-labelled ancestors were Ara−, to distinguish them from the DsRed-labelled Ara+ ancestors. The viability of DsRed-ancestral cells was determined by counting colony-forming units per particle sorted from each gated population.
Figure 3.
FACS and colony formation analyses of mixed cultures of evolved and ancestral ABM cells. (a) Ancestral cells constitutively expressing DsRed were mixed with evolved cells constitutively expressing GFP at a 10 : 1 ancestor-to-evolved ratio and were analysed during 28 hours of mixed growth using flow cytometry. The relative GFP and DsRed fluorescence is shown on the x- and y-axes. The ‘F’ and ‘A’ windows enclose the free ancestors and aggregated cell populations, respectively, that were gated during the subsequent FACS sorting (see below). The ‘A’ population contained at least one evolved cell and one or more ancestor cells per particle. (b) As in (a), except that ancestor cells constitutively expressing GFP were used in place of evolved cells. (c) ‘A’ and ‘F’ cell populations were isolated from mixed cultures of GFP-labelled evolved and DsRed-labelled ancestor cells at the indicated times of incubation using FACS sorting. The plating efficiency was scored as the number of CFUs per sorted particle for a given population. Free ancestor cells are shown as red bars and evolved-bound ancestor cells are shown as black bars. The right axis shows the frequency of ancestral cells in the mixed cultures during the sampling period. (d) As in (c), except that GFP-labelled ancestral cells were used in place of GFP-labelled evolved cells. Data are means±s.d., n≥2 (time 28 hours, n=3).
3. Results
(a) The phenomenon
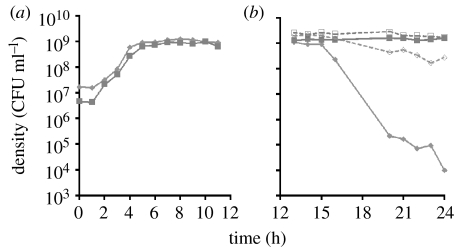
The evolved antagonistic interaction reported here was an unanticipated (serendipitous) outcome of in vitro evolution experiments. Eight independent cultures of a mutator strain of E. coli K-12 AB1157ΔmutS::spc (Bregeon et al. 1999; here referred to as ABM) were maintained in 50 ml flasks by daily transfer (100 μl–10 ml) in LB broth. After 62 passages (approx. 412 generations), 50 μl of overnight cultures derived from single colonies from ‘evolved’ cultures were mixed in 10 ml LB with 50μl of ABM and grown together for 24 hours. For all eight cultures, the density (CFUs) of the ancestral cells after 24 hours of growth was less than 10−3 that of the evolved. This rapid decline in the density of the ancestral cells did not occur when these bacteria were in single clone culture, and in mixed culture it only occurred after the populations stopped growing (figure 1a,b). The details of the methods for this and the other experiments reported here and more information about the phenomenon can be found in the §2 and in the electronic supplementary material.
Figure 1.
Changes in the density of ancestral (Ara+) and evolved (Ara−) ABM in mixed and single clone cultures. (a) Mixed culture during exponential growth and early stationary phase. Solid diamonds, density of ancestral (Ara+); solid square, evolved (Ara−). (b) Mixed and single clone cultures of ancestral and evolved cells at stationary phase. Solid diamond and solid squares with solid lines are, respectively, the density of ancestral and evolved cells in mixed cultures. Open diamonds and open squares with broken line are, respectively, the density of ancestral and evolved cells in single clone culture. In this figure and other data presented, the evolved strain used was that isolated as a single colony from the 62nd transfer (approx. 412 generations) of one of the evolution experiments.
This precipitous decline in the density of the ancestral bacteria also occurs when ancestral cells are mixed at stationary phase with evolved bacteria from any of the eight independent serial transfer cultures or after stationary phase when they are grown together. This inhibition or killing does not occur when evolved cells from independent cultures are mixed with each other (data not shown). Moreover, this apparent killing of the ancestral cells by evolved is frequency and density dependent; it does not occur when the frequency of the evolved cells in the mixture is low or when the total density of bacteria in sterile filtrates of stationary phase cells is less than approximately 5×107 (figure 2).
Figure 2.
Changes in the ratio of evolved and ancestral ABM mixed at different initial frequencies or average densities. (a) Frequency dependence: ancestral (Ara+) and evolved (Ara−) cells from 24 hour stationary phase cultures were mixed at different ratios of ancestral and evolved cells and put into fresh LB (1/100). (b) Density dependence: mixtures of ancestral (Ara−) and evolved (Ara+) cells from 14 hour stationary phase were diluted at different initial densities in the sterile filtrate of a 14 hour mixture of ancestral and evolved cells. The average density is that calculated over the entire sampling period.
(b) Killing or inhibition?
Because the preceding results are based on colony counts, it is possible that in the liquid cultures being sampled the ancestral cells were viable, but their ability to form colonies was inhibited by the evolved cells upon entry into the stationary phase of growth. To ascertain whether this is the case or if the ancestral cells are in fact killed, we tested for the selective inclusion of propidium iodide (PI), which does not occur in viable cells (Boulos et al. 1999). The ancestral ABM cells do not selectively include PI in the mixed stationary phase cultures with evolved (see figure S9 and more detailed information in the electronic supplementary material). Thus, we cannot conclude that the ancestral bacteria are killed by the evolved, but may well be converted to a viable but not culturable (VBNC) state (see §4). For this reason, we shall refer to this phenomenon as inhibition rather than killing.
(c) Stationary phase contact-dependent inhibition
In theory (the results of mathematical modelling studies), the observed frequency- and density-dependent inhibition of ancestral bacteria by evolved can be attributed to either the evolved bacteria releasing extracellular toxins, allelopathic agents that inhibit the ancestral bacteria, or inhibition through physical contact with the evolved bacteria (Levin 1988; electronic supplementary material, figure S13). Two lines of evidence support the hypothesis that the observed inhibition cannot be attributed to extracellular toxins: (i) the rate of decline in the density of ancestral cells in filtrates of stationary phase cultures of evolved cells or mixtures of evolved and ancestral cells is no different than that in filtrates of their own cells (electronic supplementary material, figure S1) and (ii) the density of ancestral cells does not decline at this accelerated rate when they are separated from the evolved bacteria by 0.45 μm filters in ‘U-tube’ experiments (electronic supplementary material, figure S2).
The above experiments, however, do not formally rule out inhibition by a highly labile, secreted allelopathic agent and are therefore only suggestive of a contact-dependent mechanism. Direct evidence that this inhibition involves contact of ancestral bacteria with the evolved comes from FACS experiments (Aoki et al. 2005). Evolved ABM were labelled with the GFP and the ancestral ABM with the DsRed fluorescent protein and grown together in LB. Three different bacterial populations were observed: free green evolved cells; free red ancestral cells; and aggregates of evolved and ancestral cells (figure 3a). This aggregation also occurred in control cultures with mixtures of DsRed- and GFP-labelled ancestral cells (figure 3b). The viability of the free ancestral cells was compared (from CFU data) with that of the clustered, evolved-bound ancestors following FACS of mixed cultures of evolved and ancestral bacteria. The results indicated that the evolved-bound ancestors declined at a faster rate than their unbound counterparts following the onset of stationary phase contact-dependent inhibition (SCDI; figure 3c). A lesser but significant decline in the density of recovered free ancestral cells was also observed in these experiments. We attribute this to their release (dissociation) from the aggregates with evolved as a result of agitation during sampling. Consistent with this interpretation is the observation that there was no loss of viability in control cultures with mixtures of GFP- and DsRed-labelled ancestral cells (figure 3d).
(d) SCDI in other E. coli strains
To begin to explore the generality of the SCDI phenomenon, we grew mixtures of overnight LB cultures of the evolved and the ancestral ABM with five different stocks of E. coli K-12 and a strain of E. coli B. (i) The evolved ABM inhibited the AB1157 mutS+ from where they were derived and two of its derivatives (AB1157-D and JC5129) and an E. coli K-12 strain, MC1061, which has a different genetic ancestry than AB1167. (ii) The ancestral ABM as well as AB1157 mutS+ are inhibited at stationary phase when they are mixed with the E. coli K-12 strains MG1655 and C600 and a wild-type E. coli B (electronic supplementary material, figures S13 and S14). In all of these cases, inhibition does not occur until after the bacteria are at stationary phase nor does it occur when the inhibited strain is in sterile supernatants of the challenging E. coli. Finally, the replication of the evolved ABM is not inhibited when it is mixed with either MG1655, C600 or E. coli B, despite the ability of these bacteria to inhibit its unevolved ancestor.
(e) The genetic basis of SCDI
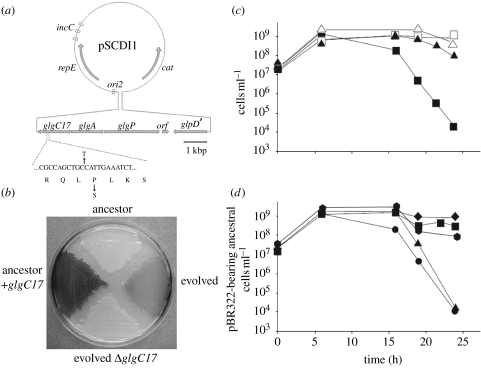
As a first attempt to identify the gene(s) responsible for the observed SCDI, we took a genetic approach that was based on the assumption that the alleles which evolved in the serial passage, evolution experiments with ABM would be dominant (details in the electronic supplementary material). We found that in this evolved ABM, SCDI can be attributed to single base substitution mutations in glgC, which encodes ADP-glucose pyrophosphorylase, a regulatory enzyme that catalyses the first reaction of bacterial glycogen synthesis (Ballicora et al. 2003). The details of the procedures used to identify this gene and demonstrate that it is responsible for both the immunity of the evolved strains to killing and their ability to kill the ancestral strain are presented in the electronic supplementary material. In summary: (i) a shotgun library of genomic DNA from an evolved 62nd transfer clone was constructed with a mini-F plasmid which was used to transform the ancestral ABM. (ii) Independent transformants were pooled, mixed with evolved cells and passaged twice to enrich for bacteria carrying potential genes for immunity to SCDI. (iii) The mini-F plasmid pCDI1 of the pSCDI-refractory clone obtained contained the glgCAP operon for glycogen synthesis which bore a glgC allele that differed from the ancestral glgC by a C to T transition in the 17th codon (proline to serine; figure 4a). (iv) This mutation, glgC17, was cloned in a multicopy pBR322 plasmid, pMLM141. (v) Both the pMLM141 transformants of the ancestral AB1157 ΔmutS, and the original evolved strain produced high concentrations of glycogen as measured by iodine staining, while the ancestral cell line and the evolved strains deleted for the chromosomal glgC17 produced little or no glycogen (figure 4b). (vi) Ancestral cell transformants bearing pMLM141 were not inhibited by the evolved strain while those bearing pBR322 without the glgC17 were (figure 4c). (vii) Ancestral cells carrying pMLM141 inhibited those bearing pBR322 without glgC17 as well as evolved cells that were deleted for glgC17 (figure 4d).
Figure 4.
Genetic basis of SCDI in ABM E. coli. (a) Map of the pSCDI1 plasmid. The genes glgC, glgA and glgP encode the enzymes ADP-glucose pyrophosphorylase, glycogen synthase and glycogen phosphorylase, respectively. orf denotes an open reading frame that encodes a putative membrane protein with no significant homology to any known protein (data not shown). glpD′ is the truncated 3′ portion of the gene for the glycerol-3-phosphate dehydrogenase. ori2, repE and incC are the cis-elements that are necessary for the mini-F type plasmid replication. cat, chloramphenicol acetyl transferase. The partial sequence below the glgC gene shows the base pair substitution leading to the Pro to Ser change in GlgC17. (b) Iodine staining of the pBR322-carrying ABM derivatives ancestor, evolved and evolved ΔglgC17, and of the ancestor carrying the glgC17-bearing multicopy plasmid pMLM141 (ancestor+glgC17). (c) glgC17 protects from SCDI. Plating efficiency of naive cells containing pMLM141 (triangles) or the pBR322 vector (squares) in mixed cultures with evolved cells (solid symbols) or in single cultures (empty symbols). (d) glgC17 is responsible for SCDI. Plating efficiency of pBR322-containing ancestral Ara+ cells was monitored in single cultures (diamonds) or in mixed cultures with Ara− ancestral cells containing pBR322 (squares); evolved, evolved ΔglgC17 cells containing pBR322 (hexagons); Ara− ancestral cells containing pMLM141 (triangles); evolved cells containing pBR322 (circles).
Our results indicate that evolution of SCDI in the eight serial passage cultures occurred independently but was convergent phenotypically as well as genetically. Although the extent varied among the eight independently evolved SCDI strains, all overproduced glycogen relative to the ancestral strain as well as to an E. coli K-12 MG1655 control (data not shown). Furthermore, all eight evolved SCDI strains bore single non-synonymous base substitutions in glgC; two in the 17th codon and one each in the 14th, 66th, 125th, 318th, 330th and 336th codons (electronic supplementary material, table 1).
A biochemical clue to the reason for the high glycogen phenotype of these mutants is provided by the finding that the G336D glgC mutation increases enzymatic activity and alters the allosteric behaviour of ADP-glucose pyrophosphorylase (Ballicora et al. 2003). This overproduction of glycogen appears to be necessary for the inhibition of ancestral cells. AB1157 glgC17 mutants deleted for glgA (glycogen synthase), that no longer synthesize glycogen, do not cause SCDI when mixed with ancestral ABM (electronic supplementary material, figure S8). On the other hand, inactivation of glgP had no impact on SCDI. This gene encodes glycogen phosphorylase, the major exolytic enzyme of glycogen catabolism Alonso-Casaju´s et al. 2006, which removes glucosyl units from the non-reducing ends of the polymer (electronic supplementary material, figure S8). This suggests that glycogen catabolism may not be required for SCDI.
While it is clear that glgC mutations are responsible for evolved SCDI in ABM, our results suggest that these mutations are most probably suppressors that compensate for defects in ABM. AB1157 and MC1061 and possibly other strains may have similar or identical defects that make them susceptible to SCDI. The DNA sequences of glgC in MG1655 and the ancestral ABM as well as AB1157 are identical (data not shown). Thus, mutations in glgC are not necessary for SCDI by MG1655. The genetic basis for these defects awaits further elucidation before any plausible mechanistic model for SCDI can be proposed.
(f) Population and evolutionary dynamics of SCDI
As noted in §1, the evolution of an antagonistic interaction such as SCDI in liquid (mass) culture is inconsistent with the proposition that mechanisms to kill or inhibit the growth of competitors will not evolve under these conditions (Chao & Levin 1981; Levin 1988; Frank 1994; Kerr et al. 2002). In accordance with this theory and these experiments, if there is a cost in the fitness of the evolved SCDI strain associated with its ability to inhibit the growth or kill the ancestral strain, when rare SCDI mutants should not be able to increase to frequencies where they would be detected. To illustrate this and other properties of the population dynamics of SCDI, we used numerical solutions to the differential equations of a simple mathematical model, a computer simulation. In this model we consider a serial transfer, liquid culture populations of the sort used in our evolution experiments with 1/100 dilutions into fresh medium occurring every 24 hours. We assume a mass action process so that interaction between the ancestral and evolved bacteria occurs at a rate proportional to the product of their densities. In this model, ancestral cells are killed instantly upon contact with evolved and this contact-dependent killing does not start until 16 hours after the populations starts to grow, when the bacteria when the culture would be at stationary phase. The simulated populations are initiated with only ancestral cells. The evolved SCDI cells are produced at random by recurrent mutation. For more details about this simulation and the values of the parameters used in these illustrations, see the electronic supplementary material.
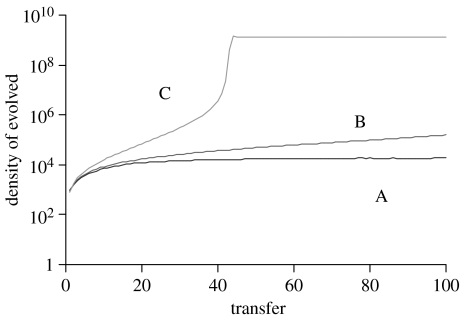
If SCDI engenders a 1% cost in the exponential growth rate, evolved cells continue to be present, due to recurrent mutation, but their population does not increase in frequency, despite the SCDI advantage (figure 5A). If the ability to inhibit ancestral cells engenders no cost or benefit to the density- and frequency-independent fitness of the evolved bacteria, then as a consequence of SCDI the density of the evolved population increases at a rate which itself increases with the density of the evolved cells (figure 5B). However, if SCDI is the only advantage possessed by the evolved bacteria, their rate of ascent will be low and they would probably not have been detected, much less dominated the population by the 62nd transfer (as we observed). On the other hand, if in addition to SCDI the ancestral bacteria had a higher rate of mortality than the evolved, the frequency of the evolved would increase due to the combination of this density-independent fitness advantage and the density/frequency-dependent advantage resulting from SCDI (figure 5C). As can be seen in the electronic supplementary material, in single clone culture the evolved bacteria in fact die at a lower rate than the ancestral strain (electronic supplementary material; figure 4).
Figure 5.
Evolution of SCDI simulation results: change in the density of the evolved strain. (A) Evolved strain has a 1% lower growth rate than the ancestral strain. (B) Evolved and ancestral strains have the same fitness. (C) The ancestral strain has a 25% higher rate of mortality than the evolved. For more details about this model and the values of the parameters, see the electronic supplementary material.
4. Discussion
The observation that initiated this investigation was serendipitous. Based on existing theory and observations (Chao & Levin 1981; Levin 1988; Frank 1994; Kerr et al. 2002), the evolution in mass (liquid) culture of bacteria that inhibit the growth or kill the cells from where they are derived, their ancestors, would not have been anticipated or sought. However, in retrospect, the observation that contact-dependent inhibition (CDI), SCDI, evolved independently by different mutations in the same gene in eight cultures of a mutator strain of E. coli is an excellent and novel, but not at all surprising, example of parallel evolution. For other examples of parallel changes occurring in independent experimental populations of bacteria derived from the same ancestor lineage, see Cunningham et al. (1997), Wichman et al. (1999, 2000, 2005), Cooper et al. (2003), Crozat et al. (2005), Sachs & Bull (2005), Woods et al. (2006) and Bantinaki et al. (2007).
Owing to a yet unknown mutation or a pleiotropic effect of a known mutation in the course of its long existence in laboratory culture, the mutS+ AB1157 strain of E. coli K-12 may have become sensitive to CDI at stationary phase by at least some other strains of E. coli including other K-12 derivatives (MG1655, C600 as well as the glgC mutants that evolved in these experiments). This sensitivity was maintained in the mutator construct of AB1157, ΔmutS::spc (Bregeon et al. 1999) used in the evolution experiments where this phenomenon was first observed. In the course of these experiments, missense mutations were generated in glgC that both suppressed the sensitivity of these bacteria to CDI during the stationary phase these serial transfer experiments and permitted these bacteria to inhibit the ancestral strain. Although we have not formally demonstrated it, by moving the glgC mutant genes responsible for SCDI to separate AB1157 backgrounds, our results and parsimony suggest that these glgC mutations also provided ABM with a density- and frequency-independent fitness advantage by reducing their rate of mortality. As a consequence of the latter, these glgC mutations were able to increase in frequency when they were rare and achieve densities where they had the additional advantage of inhibiting/killing the dominant population with wild-type glgC loci. The primary, if not exclusive, role of the mutS gene in this process was to increase the rate at which the suppressing glgC mutations occurred, which in addition to providing the variation needed for this evolution, reduced the likelihood of their loss in the bottlenecks associated with serial passage (Levin et al. 2000). Presumably SCDI would eventually evolve if mutS+ strains of AB1157 or other strains with this sensitivity were maintained long enough in serial transfer cultures of this type.
In some ways, the SCDI that evolved in these experiments is similar to the growth advantage in stationary-phase (GASP) phenomenon studied by Kolter and colleagues (Zambrano & Kolter 1996; Farrell & Finkel 2003); it too is manifest at stationary phase and requires high pH (greater than 8.5; electronic supplementary material, figure S5). SCDI is, however, clearly different from GASP both functionally and genetically. (i) The decline in plating efficiency of ancestral cells when mixed with evolved (figure 1b) occurs at a rate nearly ten times as great as the highest reported for GASP (Zambrano & Kolter 1996). (ii) Contrary to what would be anticipated by the scavenging mechanism (differential use of resources made available from dead cells) postulated for GASP, there was little or no growth of evolved cells when they were inoculated at low densities into filtrates of stationary phase ancestral cells or filtrates of sonicated ancestral cells (data not shown). (iii) The lrp and rpoS mutations that account for GASP are not present in the evolved ABM, nor in their ancestors ABM and AB1157 (data not shown).
The underlying mechanisms by which cells are inhibited in the SCDI reported here and the CDI reported by Aoki et al. (2005) have not yet been elucidated. However, it is clear that these two inhibitory processes are functionally and genetically distinct. CDI of E. coli K-12 (MG1655) by a naturally occurring strain of E. coli from a urinary tract infection occurred when the bacteria were growing, rather than at stationary phase. Moreover, the cdiA and cdiB genes that were found to be responsible for CDI by the wild E. coli are not present in E. coli K-12, wherein SCDI evolved in the present study.
In this report we use the term ‘inhibition’ rather than ‘killing’ in our description of the observed SCDI because the results of the PI test suggested that the ancestral bacteria are VBNC, when mixed with the evolved cells at stationary phase. It should be noted, however, that the concept of VBNC bacteria is controversial; whether these cells are dormant or are progressively undergoing cell death is unclear (Nystrom 2003; Oliver 2005). What is clear from our studies is that the decline in the density of the inhibited strain in these cultures cannot be attributed to their failure to form colonies on agar. Their densities continue to decline in successive serial passages (electronic supplementary material, figure S10).
The mechanism by which the glgC mutations that evolved in these experiments convert the ancestral strain into an inhibitor requires further research. Excessive glycogen synthesis, the apparent phenotype derived from the glgC mutants, could trigger the inhibitory function. Glycogen excess mutations of E. coli have been mapped to the glgC gene encoding ADP-glucose pyrophosphorylase, a regulatory enzyme for bacterial glycogen synthesis (Ballicora et al. 2003 and references therein). In particular, certain amino acid substitutions of GlgC are known to alter the allosteric behaviour of the protein and increase glycogen synthesis (Meyer et al. 1993; Wu & Preiss 1998). However, to the best of our knowledge there is no evidence that glycogen synthesis is correlated with any known bactericidal activity. In solution, glycogen is devoid of antimicrobial activity (data not shown). High glycogen accumulation might alter the cell structure, leading to effects involved in killing. Indeed, evolved hyperglycogenic strains are more susceptible than the ancestors to SDS 0.2% (data not shown), suggesting possible alterations of the bacterial cell envelope. We cannot discount at the present time that the excess glycogen synthesis may be related to oxidative or other envelope stress pathways. Hence, our report here of the SCDI phenomenon in E. coli populations sets the basis for future research towards the precise mechanism of SCDI in this and eventually other bacteria.
How important the CDI reported by Aoki et al. (2005) and that observed in this study are to the ecology of bacteria is unclear at this time but is certainly intriguing and worthy of further investigation. If in natural populations CDI and SCDI have effects of the magnitude observed in these laboratory studies, they could have a profound effect on the structure of bacterial communities. CDI could facilitate the invasion of strains into habitats occupied by other members of their species and lead to the elimination of these competitors. From a medical perspective CDI may well point to as yet unexploited chinks in the bacterial armour. By elucidating the mechanisms responsible for the sensitivity of E. coli K-12 to SCDI, new targets for antimicrobial agents could be identified. Most if not all classic and contemporary antibiotics have little effect on bacteria that are not replicating (Bigger 1944; McDermott 1958; Levin & Rozen 2006). In practice (Eagle 1952; McDermott 1958; Tuomanen 1986) as well as in theory (Levin 2004; Wiuff et al. 2005; Levin & Rozen 2006) tolerant, persistent and latent populations of non-replicating, antibiotic-refractory bacteria can extend the term of treatment as well as lead to treatment failure. The mechanism responsible for the SCDI reported here is particularly appealing because the inhibition or killing occurs when the bacteria are not replicating.
Acknowledgments
We are grateful to Roberto Kolter and Maria Zambrano not only for their useful comments, suggestions during the early phase of this project, but also for their inspiration. We also thank Luba Beylina, Nina Walker and Mariastella Tucker for their superb technical support, Julie Cazareth at the cell sorting facility from CNRS UMR-6097 and Grégoire Lauvau at INSERM E-344 (IPMC, Valbonne, France) for invaluable help and suggestions during the FACS experiments, Anne Doye (INSERM, U627) for excellent assistance with the confocal microscopy analysis and for preparing high-quality micrographs, Xin Wang for advice and assistance in constructing glgA and glgP deletions, Lauren Ancel, Mark Jensen, Mary Reynolds, Jeff Smith, Mark Tanaka and Renata Zappala for their comments and suggestions. We are also grateful to Ivan Matic, Mary Berlyn and her colleagues at the Coli Genetic Stock Center for providing the strains of bacteria used in this investigation, and to Arthur Altenhoefer and Stéphane Méresse for constructs needed for the fluorescent proteins. We express a particular depth of gratitude for the intellectual and practical (space and facilities) generosity of Ramón Díaz Orejas (CIB, Madrid) in whose laboratory most of the molecular genetic experiments were performed. This research was supported by grants from the US Nation Institutes of Health, AI40662, GM33782 (B.R.L.), GM59969 (T.R.), CO3/14 of the Spanish Fondo de Investigaciones Sanitarias and SAF-2002-04649, BFU2005-03911/BMC (R. D. O.), BFU2004-00879 (J.B.) of the Spanish Ministerio de Educación y Ciencia, and a grant from the European Union (QLK2-CT-2001-873, and BIO2005-04278 and LSHM-CT-2005-518152 (F.B.)). M.L. was supported by a postdoctoral fellowship from La Ligue Nationale Contre le Cancer. M.R.B. was in part supported by the Alfonso X el Sabio University in Madrid.
Supplementary Material
1. Additional evidence for contact dependence; 2. The SCDI phenomenon and bacteria; 3. The genetic basis of SCDI and immunity to SCDI in E. coli ABM; 4. Glycogen production and SCDI; 5. Cell viability analysis during SCDI using confocal microscopy/SCDI is not due to plating efficiency differences; 6. Evidence for SCDI with other strains of E. coli; 7. Population and Evolutionary Dynamics of SCDI
References
- Alonso-Casaju´s N, Dauville´e D, Viale A.M, Muñoz F.J, Baroja-Ferna´ndez E, Mora´n-Zorzano M.T, Endallin G, Ball S, Pozueta-Romero J. Glycogen phosphorylase, the product of the glgp gene catalyses glycogen breakdown by removing glucose units from the nonreducing ends in Escherichia coli. J. Bacteriol. 2006;188:5266–5272. doi: 10.1128/JB.01566-05. doi:10.1128/JB.01566-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki S.K, Pamma R, Hernday A.D, Bickham J.E, Braaten B.A, Low D.A. Contact-dependent inhibition of growth in Escherichia coli. Science. 2005;309:1245–1248. doi: 10.1126/science.1115109. doi:10.1126/science.1115109 [DOI] [PubMed] [Google Scholar]
- Ballicora M.A, Iglesias A.A, Preiss J. ADP-glucose pyrophosphorylase, a regulatory enzyme for bacterial glycogen synthesis. Microbiol. Mol. Biol. Rev. 2003;67:213–225. doi: 10.1128/MMBR.67.2.213-225.2003. doi:10.1128/MMBR.67.2.213-225.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantinaki E, Kassen R, Knight C.G, Robinson Z, Spiers A.J, Rainey P.B. Adaptive divergence in experimental populations of Pseudomonas fluorescens. III. Mutational origins of wrinkly spreader diversity. Genetics. 2007;176:441–453. doi: 10.1534/genetics.106.069906. doi:10.1534/genetics.106.069906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigger J.W. Treatment of staphylococcal infections with penicillin. Lancet. 1944;2:497–500. doi:10.1016/S0140-6736(00)74210-3 [Google Scholar]
- Boulos L, Prevost M, Barbeau B, Coallier J, Desjardins R. LIVE/DEAD BacLight: application of a new rapid staining method for direct enumeration of viable and total bacteria in drinking water. J. Microbiol. Methods. 1999;37:77–86. doi: 10.1016/s0167-7012(99)00048-2. doi:10.1016/S0167-7012(99)00048-2 [DOI] [PubMed] [Google Scholar]
- Bregeon D, Matic I, Radman M, Taddei F. Insufficient mismatch repair: genetic defects and down regulation. J. Genet. 1999;78:21–28. [Google Scholar]
- Chao L, Levin B.R. Structured habitats and the evolution of anticompetitor toxins in bacteria. Proc. Natl Acad. Sci. USA. 1981;78:6324–6328. doi: 10.1073/pnas.78.10.6324. doi:10.1073/pnas.78.10.6324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper T.F, Rozen D.E, Lenski R.E. Parallel changes in gene expression after 20,000 generations of evolution in Escherichia coli. Proc. Natl Acad. Sci. USA. 2003;100:1072–1077. doi: 10.1073/pnas.0334340100. doi:10.1073/pnas.0334340100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crozat E, Philippe N, Lenski R.E, Geiselmann J, Schneider D. Long-term experimental evolution in Escherichia coli. XII. DNA topology as a key target of selection. Genetics. 2005;169:523–532. doi: 10.1534/genetics.104.035717. doi:10.1534/genetics.104.035717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham C.W, Jeng K, Husti J, Badgett M, Molineux I.J, Hillis D.M, Bull J.J. Parallel molecular evolution of deletions and nonsense mutations in bacteriophage T7. Mol. Biol. Evol. 1997;14:113–116. doi: 10.1093/oxfordjournals.molbev.a025697. [DOI] [PubMed] [Google Scholar]
- DIFCO. Difco Laboratories; Detroit, MI: 1984. Difco manual: dehydrated culture media and reagents for microbiology. [Google Scholar]
- Eagle H. Experimental approach to the problem of treatment failure with penicillin. Am. J. Med. 1952;13:389–399. doi: 10.1016/0002-9343(52)90293-3. doi:10.1016/0002-9343(52)90293-3 [DOI] [PubMed] [Google Scholar]
- Farrell M.J, Finkel S.E. The growth advantage in stationary-phase phenotype conferred by rpoS mutations is dependent on the pH and nutrient environment. J. Bacteriol. 2003;185:7044–7052. doi: 10.1128/JB.185.24.7044-7052.2003. doi:10.1128/JB.185.24.7044-7052.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank S.A. Spatial polymorphism of bacteriocin and other allelopathic traits. Evol. Ecol. 1994;8:369–386. doi:10.1007/BF01238189 [Google Scholar]
- Helling R.B, Vargas C.N, Adams J. Evolution of Escherichia coli during growth in a constant environment. Genetics. 1987;116:349–358. doi: 10.1093/genetics/116.3.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr B, Riley M.A, Feldman M.W, Bohannan B.J. Local dispersal promotes biodiversity in a real-life game of rock–paper–scissors. Nature. 2002;418:171–174. doi: 10.1038/nature00823. doi:10.1038/nature00823 [DOI] [PubMed] [Google Scholar]
- Lenski R.E, Rose M.R, Simpson S.C, Tadler S.C. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2000 generations. American Naturalist. 1991;91:1315–1341. [Google Scholar]
- Levin B.R. Frequency-dependent selection in bacterial populations. Phil. Trans. R. Soc. B. 1988;319:459–472. doi: 10.1098/rstb.1988.0059. doi:10.1098/rstb.1988.0059 [DOI] [PubMed] [Google Scholar]
- Levin B.R. Microbiology. Noninherited resistance to antibiotics. Science. 2004;305:1578–1579. doi: 10.1126/science.1103077. doi:10.1126/science.1103077 [DOI] [PubMed] [Google Scholar]
- Levin B.R, Rozen D.E. Non-inherited antibiotic resistance. Nat. Rev. Microbiol. 2006;4:556–562. doi: 10.1038/nrmicro1445. doi:10.1038/nrmicro1445 [DOI] [PubMed] [Google Scholar]
- Levin B.R, Stewart F.M, Chao L. Resource—limited growth, competition, and predation: a model and experimental studies with bacteria and bacteriophage. Am. Nat. 1977;977:3–24. doi:10.1086/283134 [Google Scholar]
- Levin B.R, Perrot V, Walker N. Compensatory mutations, antibiotic resistance and the population genetics of adaptive evolution in bacteria. Genetics. 2000;154:985–997. doi: 10.1093/genetics/154.3.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott W. Microbial persistence. Yale J. Biol. Med. 1958;30:257–291. [PMC free article] [PubMed] [Google Scholar]
- Meyer C.R, Ghosh P, Nadler S, Preiss J. Cloning, expression, and sequence of an allosteric mutant ADPglucose pyrophosphorylase from Escherichia coli B. Arch. Biochem. Biophys. 1993;302:64–71. doi: 10.1006/abbi.1993.1181. doi:10.1006/abbi.1993.1181 [DOI] [PubMed] [Google Scholar]
- Miller J.H. Cold Spring Harbor Press; Cold Spring Harbor, NY: 1972. Experiments in molecular genetics. [Google Scholar]
- Nystrom T. Conditional senescence in bacteria: death of the immortals. Mol. Microbiol. 2003;48:17–23. doi: 10.1046/j.1365-2958.2003.03385.x. doi:10.1046/j.1365-2958.2003.03385.x [DOI] [PubMed] [Google Scholar]
- Oliver J.D. The viable but nonculturable state in bacteria. J. Microbiol. 2005;43:93–100. [PubMed] [Google Scholar]
- Sachs J.L, Bull J.J. Experimental evolution of conflict mediation between genomes. Proc. Natl Acad. Sci. USA. 2005;102:390–395. doi: 10.1073/pnas.0405738102. doi:10.1073/pnas.0405738102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuomanen E. Phenotypic tolerance: the search for beta-lactam antibiotics that kill nongrowing bacteria. Rev. Infect. Dis. 1986;8(Suppl. 3):S279–S291. doi: 10.1093/clinids/8.supplement_3.s279. [DOI] [PubMed] [Google Scholar]
- Wichman H.A, Badgett M.R, Scott L.A, Boulianne C.M, Bull J.J. Different trajectories of parallel evolution during viral adaptation. Science. 1999;285:422–424. doi: 10.1126/science.285.5426.422. doi:10.1126/science.285.5426.422 [DOI] [PubMed] [Google Scholar]
- Wichman H.A, Scott L.A, Yarber C.D, Bull J.J. Experimental evolution recapitulates natural evolution. Phil. Trans. R. Soc. B. 2000;355:1677–1684. doi: 10.1098/rstb.2000.0731. doi:10.1098/rstb.2000.0731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichman H.A, Millstein J, Bull J.J. Adaptive molecular evolution for 13,000 phage generations: a possible arms race. Genetics. 2005;170:19–31. doi: 10.1534/genetics.104.034488. doi:10.1534/genetics.104.034488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiuff C, Zappala R.M, Regoes R.R, Garner K.N, Baquero F, Levin B.R. Phenotypic tolerance: antibiotic enrichment of noninherited resistance in bacterial populations. Antimicrob. Agents Chemother. 2005;49:1483–1494. doi: 10.1128/AAC.49.4.1483-1494.2005. doi:10.1128/AAC.49.4.1483-1494.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods R, Schneider D, Winkworth C.L, Riley M.A, Lenski R.E. Tests of parallel molecular evolution in a long-term experiment with Escherichia coli. Proc. Natl Acad. Sci. USA. 2006;103:9107–9112. doi: 10.1073/pnas.0602917103. doi:10.1073/pnas.0602917103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M.X, Preiss J. The N-terminal region is important for the allosteric activation and inhibition of the Escherichia coli ADP-glucose pyrophosphorylase. Arch. Biochem. Biophys. 1998;358:182–188. doi: 10.1006/abbi.1998.0846. doi:10.1006/abbi.1998.0846 [DOI] [PubMed] [Google Scholar]
- Zambrano M.M, Kolter R. GASPing for life in stationary phase. Cell. 1996;86:181–184. doi: 10.1016/s0092-8674(00)80089-6. doi:10.1016/S0092-8674(00)80089-6 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1. Additional evidence for contact dependence; 2. The SCDI phenomenon and bacteria; 3. The genetic basis of SCDI and immunity to SCDI in E. coli ABM; 4. Glycogen production and SCDI; 5. Cell viability analysis during SCDI using confocal microscopy/SCDI is not due to plating efficiency differences; 6. Evidence for SCDI with other strains of E. coli; 7. Population and Evolutionary Dynamics of SCDI