Abstract
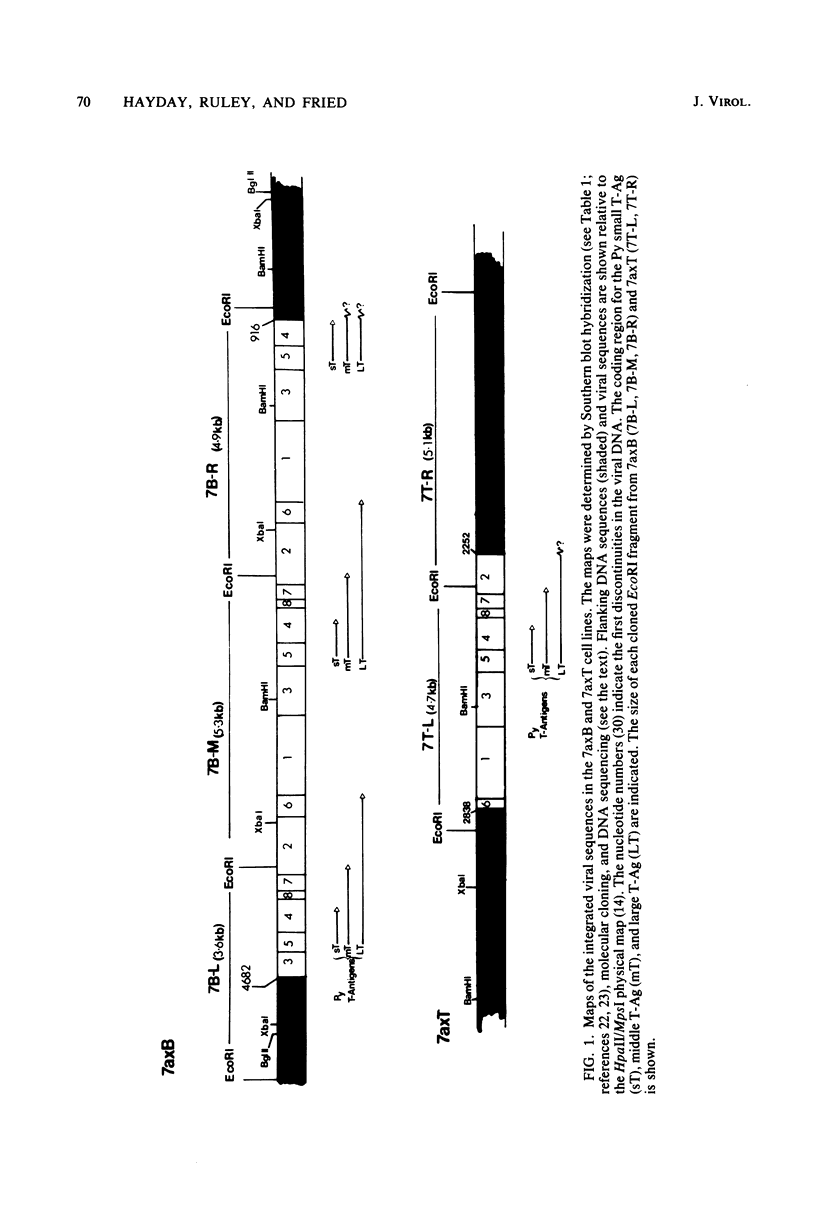
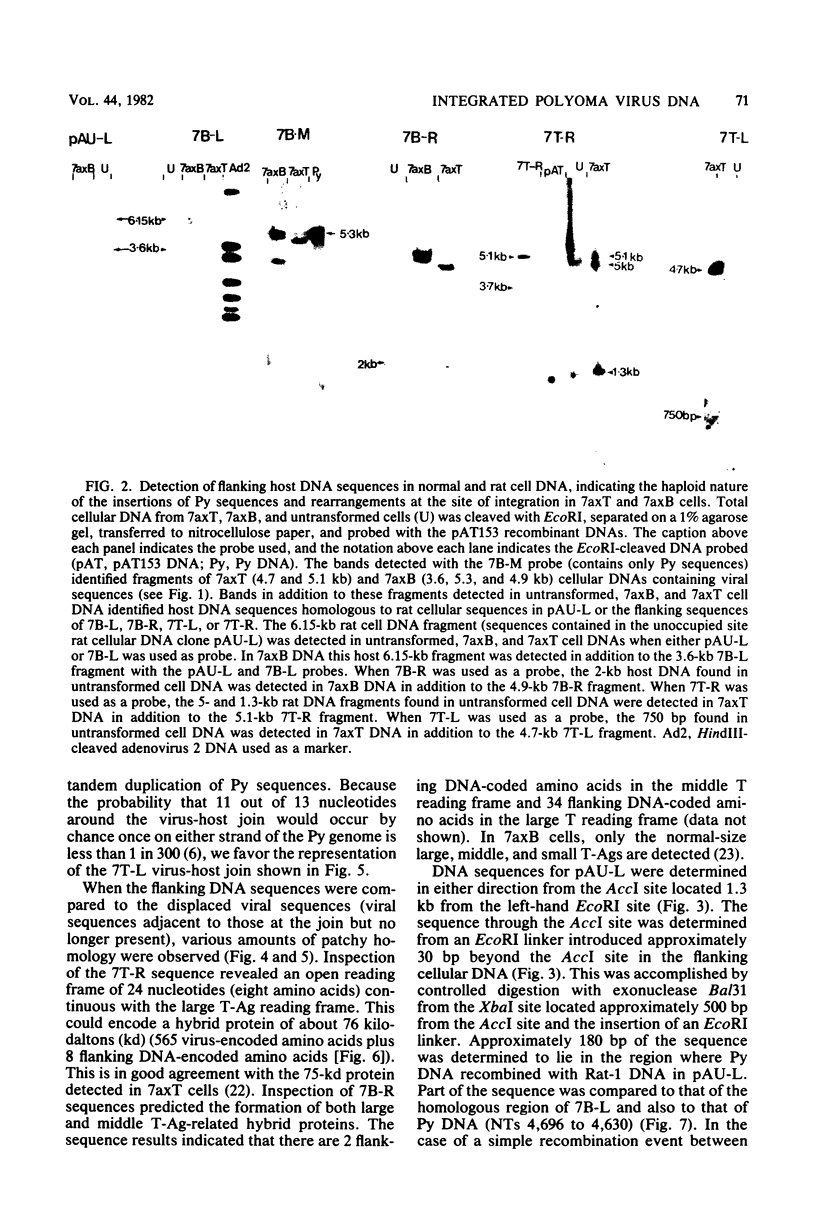
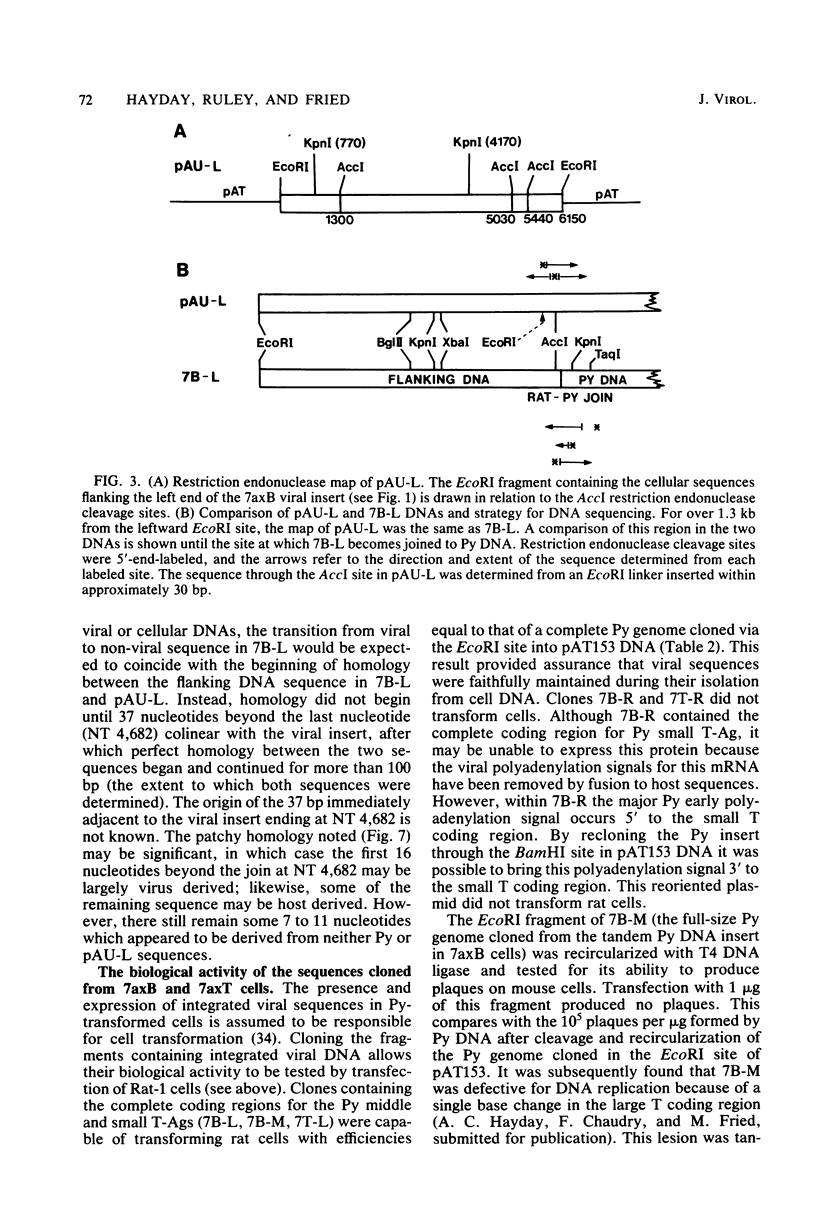
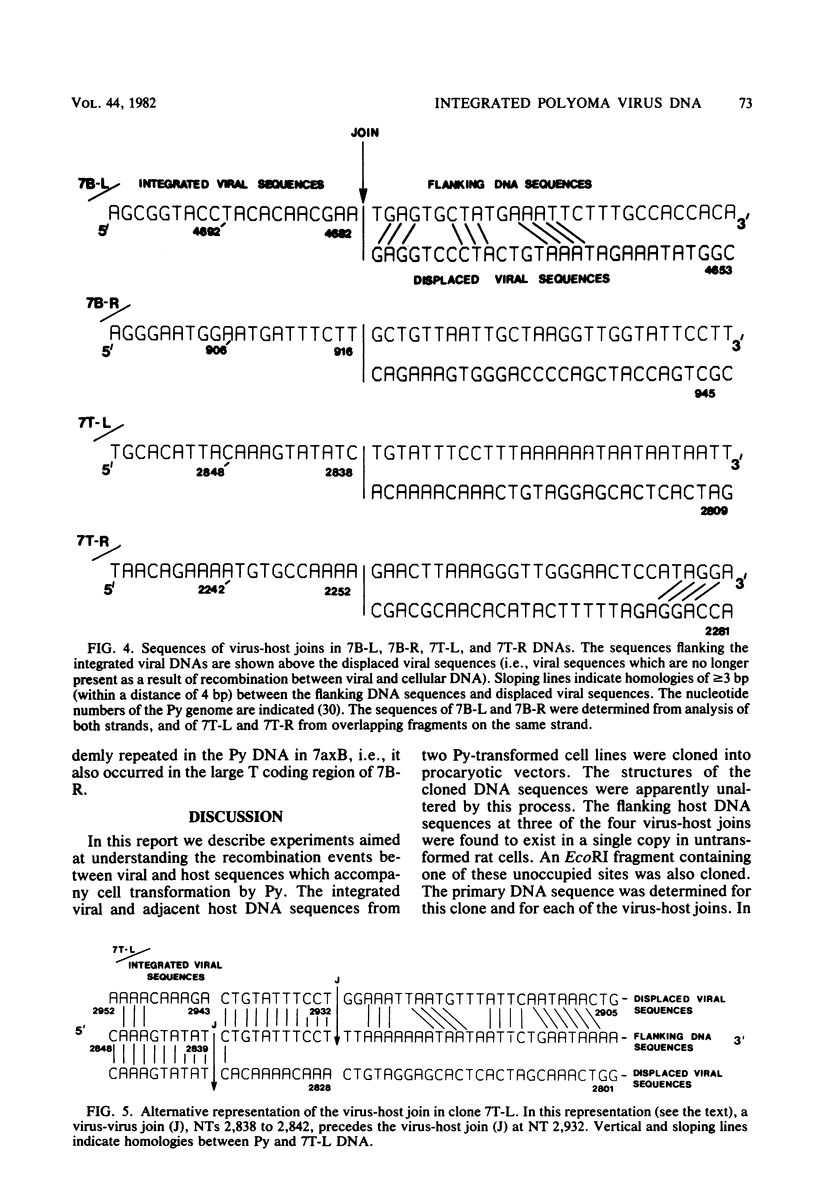
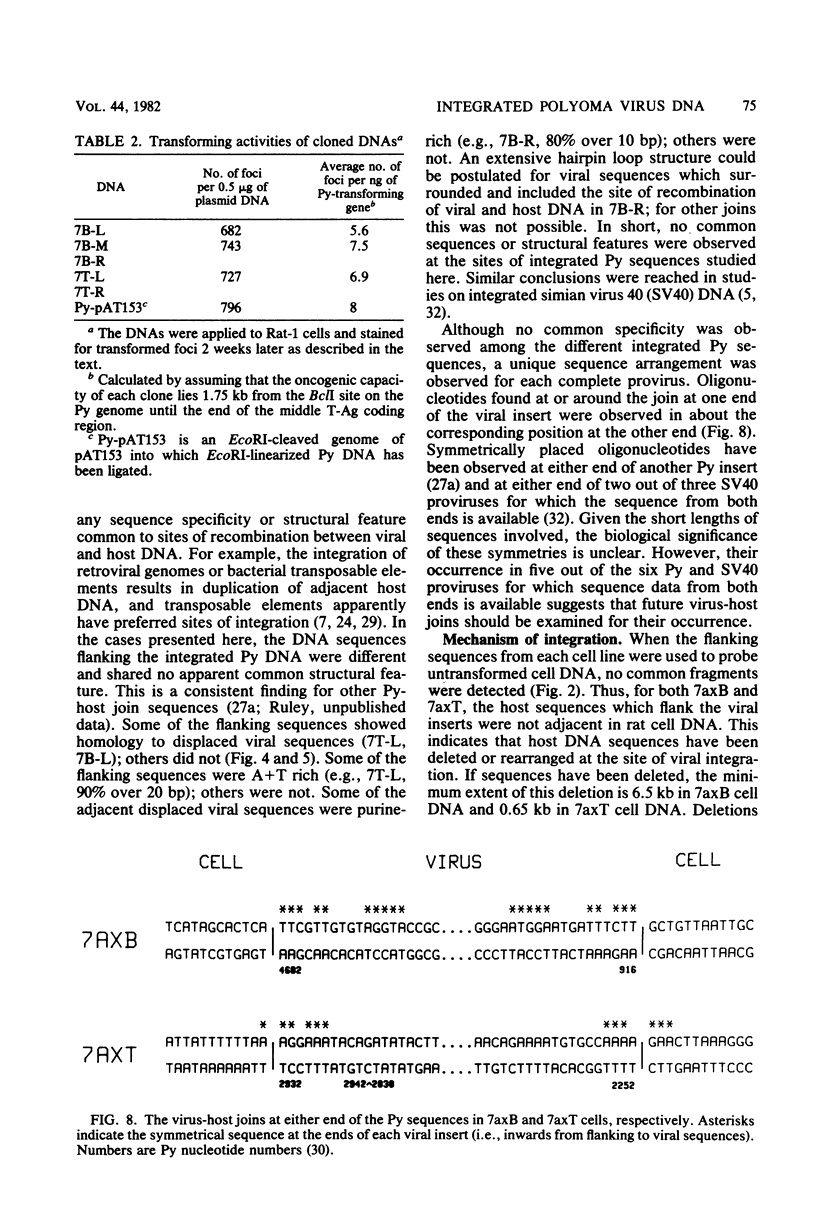
EcoRI fragments containing integrated viral and adjacent host sequences were cloned from two polyoma virus-transformed cell lines (7axT and 7axB) which each contain a single insert of polyoma virus DNA. Cloned DNA fragments which contained a complete coding capacity for the polyoma virus middle and small T-antigens were capable of transforming rat cells in vitro. Analysis of the flanking sequences indicated that rat DNA had been reorganized or deleted at the sites of polyoma virus integration, but none of the hallmarks of retroviral integration, such as the duplication of host DNA, were apparent. There was no obvious similarity of DNA sequences in the four virus-host joins. In one case the virus-host junction sequence predicted the virus-host fusion protein which was detected in the transformed cell line. DNA homologous to the flanking sequences of three out of four of the joins was present in single copy in untransformed cells. One copy of the flanking host sequences existed in an unaltered form in the two transformed cell lines, indicating that a haploid copy of the viral transforming sequences is sufficient to maintain transformation. The flanking sequences from one cell line were further used as a probe to isolate a target site (unoccupied site) for polyoma virus integration from uninfected cellular DNA. The restriction map of this DNA was in agreement with that of the flanking sequences, but the sequence of the unoccupied site indicated that viral integration did not involve a simple recombination event between viral and cellular sequences. Instead, sequence rearrangements or alterations occurred immediately adjacent to the viral insert, possibly as a consequence of the integration of viral DNA.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Backman K., Betlach M., Boyer H. W., Yanofsky S. Genetic and physical studies on the replication of ColE1-type plasmids. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):69–76. doi: 10.1101/sqb.1979.043.01.012. [DOI] [PubMed] [Google Scholar]
- Basilico C., Gattoni S., Zouzias D., Valle G. D. Loss of integrated viral DNA sequences in polyomatransformed cells is associated with an active viral A function. Cell. 1979 Jul;17(3):645–659. doi: 10.1016/0092-8674(79)90272-1. [DOI] [PubMed] [Google Scholar]
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botchan M., Stringer J., Mitchison T., Sambrook J. Integration and excision of SV40 DNA from the chromosome of a transformed cell. Cell. 1980 May;20(1):143–152. doi: 10.1016/0092-8674(80)90242-1. [DOI] [PubMed] [Google Scholar]
- Brutlag D. L., Clayton J., Friedland P., Kedes L. H. SEQ: a nucleotide sequence analysis and recombination system. Nucleic Acids Res. 1982 Jan 11;10(1):279–294. doi: 10.1093/nar/10.1.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calos M. P., Miller J. H. Transposable elements. Cell. 1980 Jul;20(3):579–595. doi: 10.1016/0092-8674(80)90305-0. [DOI] [PubMed] [Google Scholar]
- Dhruva B. R., Shenk T., Subramanian K. N. Integration in vivo into simian virus 40 DNA of a sequence that resembles a certain family of genomic interspersed repeated sequences. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4514–4518. doi: 10.1073/pnas.77.8.4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Mayorca G., Callender J., Marin G., Giordano R. Temperature-sensitive mutants of polyoma virus. Virology. 1969 May;38(1):126–133. doi: 10.1016/0042-6822(69)90134-2. [DOI] [PubMed] [Google Scholar]
- Eckhart W. Complementation and transformation by temperature-sensitive mutants of polyoma virus. Virology. 1969 May;38(1):120–125. doi: 10.1016/0042-6822(69)90133-0. [DOI] [PubMed] [Google Scholar]
- FRIED M. CELL-TRANSFORMING ABILITY OF A TEMPERATURE-SENSITIVE MUTANT OF POLYOMA VIRUS. Proc Natl Acad Sci U S A. 1965 Mar;53:486–491. doi: 10.1073/pnas.53.3.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Griffin B. E., Fried M., Cowie A. Polyoma DNA: a physical map. Proc Natl Acad Sci U S A. 1974 May;71(5):2077–2081. doi: 10.1073/pnas.71.5.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosveld F. G., Dahl H. H., de Boer E., Flavell R. A. Isolation of beta-globin-related genes from a human cosmid library. Gene. 1981 Apr;13(3):227–237. doi: 10.1016/0378-1119(81)90028-7. [DOI] [PubMed] [Google Scholar]
- Gutai M. W., Nathans D. Evolutionary variants of simian virus 40: Cellular DNA sequences and sequences at recombinant joints of substituted variants. J Mol Biol. 1978 Dec 5;126(2):275–288. doi: 10.1016/0022-2836(78)90363-7. [DOI] [PubMed] [Google Scholar]
- Hassell J. A., Topp W. C., Rifkin D. B., Moreau P. E. Transformation of rat embryo fibroblasts by cloned polyoma virus DNA fragments containing only part of the early region. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3978–3982. doi: 10.1073/pnas.77.7.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward G. S., Smith M. G. The chromosome of bacteriophage T5. I. Analysis of the single-stranded DNA fragments by agarose gel electrophoresis. J Mol Biol. 1972 Feb 14;63(3):383–395. doi: 10.1016/0022-2836(72)90435-4. [DOI] [PubMed] [Google Scholar]
- Israel M. A., Vanderryn D. F., Meltzer M. L., Martin M. A. Characterization of polyoma viral DNA sequences in polyoma-induced hamster tumor cell lines. J Biol Chem. 1980 Apr 25;255(8):3798–3805. [PubMed] [Google Scholar]
- Lania L., Gandini-Attardi D., Griffiths M., Cooke B., De Cicco D., Fried M. The polyoma virus 100K large T-antigen is not required for the maintenance of transformation. Virology. 1980 Feb;101(1):217–232. doi: 10.1016/0042-6822(80)90497-3. [DOI] [PubMed] [Google Scholar]
- Lania L., Griffiths M., Cooke B., Ito Y., Fried M. Untransformed rat cells containing free and integrated DNA of a polyoma nontransforming (Hr-t) mutant. Cell. 1979 Nov;18(3):793–802. doi: 10.1016/0092-8674(79)90132-6. [DOI] [PubMed] [Google Scholar]
- Lania L., Hayday A., Bjursell G., Gandini-Attardi D., Fried M. Organization and expression of integrated polyoma virus DNA sequences in transformed rodent cells. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 1):597–603. doi: 10.1101/sqb.1980.044.01.062. [DOI] [PubMed] [Google Scholar]
- Lania L., Hayday A., Fried M. Loss of functional large T-antigen and free viral genomes from cells transformed in vitro by polyoma virus after passage in vivo as tumor cells. J Virol. 1981 Aug;39(2):422–431. doi: 10.1128/jvi.39.2.422-431.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majors J. E., Varmus H. E. Nucleotide sequences at host-proviral junctions for mouse mammary tumour virus. Nature. 1981 Jan 22;289(5795):253–258. doi: 10.1038/289253a0. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Hardison R. C., Lacy E., Lauer J., O'Connell C., Quon D., Sim G. K., Efstratiadis A. The isolation of structural genes from libraries of eucaryotic DNA. Cell. 1978 Oct;15(2):687–701. doi: 10.1016/0092-8674(78)90036-3. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak U., Dilworth S. M., Griffin B. E. Coding capacity of a 35 percent fragment of the polyoma virus genome is sufficient to initiate and maintain cellular transformation. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3278–3282. doi: 10.1073/pnas.77.6.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruley H. E., Lania L., Chaudry F., Fried M. Use of a cellular polyadenylation signal by viral transcripts in polyoma virus transformed cells. Nucleic Acids Res. 1982 Aug 11;10(15):4515–4524. doi: 10.1093/nar/10.15.4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. The use of thin acrylamide gels for DNA sequencing. FEBS Lett. 1978 Mar 1;87(1):107–110. doi: 10.1016/0014-5793(78)80145-8. [DOI] [PubMed] [Google Scholar]
- Shimotohno K., Mizutani S., Temin H. M. Sequence of retrovirus provirus resembles that of bacterial transposable elements. Nature. 1980 Jun 19;285(5766):550–554. doi: 10.1038/285550a0. [DOI] [PubMed] [Google Scholar]
- Soeda E., Arrand J. R., Smolar N., Walsh J. E., Griffin B. E. Coding potential and regulatory signals of the polyoma virus genome. Nature. 1980 Jan 31;283(5746):445–453. doi: 10.1038/283445a0. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stringer J. R. Integrated simian virus 40 DNA: nucleotide sequences at cell-virus recombinant junctions. J Virol. 1981 May;38(2):671–679. doi: 10.1128/jvi.38.2.671-679.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiemeier D. C., Tilghman S. M., Leder P. Purification and cloning of a mouse ribosomal gene fragment in coliphage lambda. Gene. 1977;2(3-4):173–191. doi: 10.1016/0378-1119(77)90016-6. [DOI] [PubMed] [Google Scholar]
- Treisman R., Novak U., Favaloro J., Kamen R. Transformation of rat cells by an altered polyoma virus genome expressing only the middle-T protein. Nature. 1981 Aug 13;292(5824):595–600. doi: 10.1038/292595a0. [DOI] [PubMed] [Google Scholar]
- Twigg A. J., Sherratt D. Trans-complementable copy-number mutants of plasmid ColE1. Nature. 1980 Jan 10;283(5743):216–218. doi: 10.1038/283216a0. [DOI] [PubMed] [Google Scholar]
- Waalwijk C., Flavell R. A. MspI, an isoschizomer of hpaII which cleaves both unmethylated and methylated hpaII sites. Nucleic Acids Res. 1978 Sep;5(9):3231–3236. doi: 10.1093/nar/5.9.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver R. F., Weissmann C. Mapping of RNA by a modification of the Berk-Sharp procedure: the 5' termini of 15 S beta-globin mRNA precursor and mature 10 s beta-globin mRNA have identical map coordinates. Nucleic Acids Res. 1979 Nov 10;7(5):1175–1193. doi: 10.1093/nar/7.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigler M., Sweet R., Sim G. K., Wold B., Pellicer A., Lacy E., Maniatis T., Silverstein S., Axel R. Transformation of mammalian cells with genes from procaryotes and eucaryotes. Cell. 1979 Apr;16(4):777–785. doi: 10.1016/0092-8674(79)90093-x. [DOI] [PubMed] [Google Scholar]
- Zain B. S., Roberts R. J. Characterization and sequence analysis of a recombination site in the hybrid virus Ad2+ND. J Mol Biol. 1978 Mar 25;120(1):13–31. doi: 10.1016/0022-2836(78)90293-0. [DOI] [PubMed] [Google Scholar]




