Abstract
We describe here the first characterization of CLEC9A, a group V C-type lectin-like receptor located in the “Dectin-1 cluster” of related receptors, which are encoded within the natural killer (NK)-gene complex. Expression of human CLEC9A is highly restricted in peripheral blood, being detected only on BDCA3+ dendritic cells and on a small subset of CD14+CD16- monocytes. CLEC9A is expressed at the cell surface as a glycosylated dimer and can mediate endocytosis, but not phagocytosis. CLEC9A possesses a cytoplasmic immunoreceptor tyrosine-based activation-like motif that can recruit Syk kinase, and we demonstrate, using receptor chimeras, that this receptor can induce proinflammatory cytokine production. These data indicate that CLEC9A functions as an activation receptor.
The C-type lectins are a superfamily of proteins containing at least one C-type lectin domain (CTLD),2 which have been classified into groups depending on the arrangement of their CTLDs (1). The group V proteins, in particular, are single extracellular CTLD-containing type II transmembrane receptors, which are found only in higher vertebrates and are encoded primarily within a single gene cluster, the NK-gene complex (NKC) on chromosome 12 in human and chromosome 6 in mouse (2). Most of the group V C-type lectin receptors (CTLR) appear to be expressed exclusively by NK cells and certain subsets of T-cells and play a role in tumor and antiviral immunity (3). These receptors detect endogenous ligands, such as MHC class I molecules, and are involved in the detection of “altered-self” or “missing self” (3–5). Immune responses mediated by these CTLRs are largely determined by the balance of signals from “pairs” of inhibitory and activation receptors, which often recognize the same ligand (6). Inhibitory receptors within this group possess immunoreceptor tyrosine-based inhibitory motifs in their cytoplasmic tails, whereas the activation receptors lack cytoplasmic signaling motifs, but associate through charged residues in their transmembrane domains with signaling partners, such as DAP12 (3).
More recently, CTLRs have also been identified on other cell types, including a related subgroup of receptors, the “Dectin-1 cluster” (7). This cluster, which includes Dectin-1, LOX-1, CLEC-1, CLEC-2, MICL, CLEC12B, and CLEC9A, appear to have more diverse ligands and cellular functions (8), and the study of these receptors has provided a number of surprising new insights into innate immunity and homeostasis (9). For example, Dectin-1 has been shown to be involved in the detection of “non-self” and function as a signaling pattern recognition receptor in anti-fungal immunity, whereas LOX-1 acts as a scavenger receptor, involved in the recognition of a variety of ligands including modified lipoproteins, apoptotic cells, activated platelets, hsp70, and bacteria (10). Furthermore, Dectin-1 and CLEC-2 have been shown to be capable of directly triggering cellular activation through cytoplasmic immunoreceptor tyrosine-based activation (ITAM)-like motifs, involving novel interactions with Syk kinase (11–13). Here we characterize CLEC9A, a novel group V CTLR located within the “Dectin-1 cluster” of receptors and show that the molecule functions as an activation receptor on a small subset of myeloid cells.
EXPERIMENTAL PROCEDURES
Primary Cells, Cell Lines, and Growth Conditions—Peripheral blood mononuclear cells (PBMC) were isolated from buffy coats (Western Province Blood Transfusion Service, Cape Town, South Africa) using Ficoll-Paque™ Plus (Amersham Biosciences), as previously described (14). Monocyte-derived macrophages and dendritic cells were generated from PBMCs as described (14). NIH3T3 and HEK293T fibroblasts, RAW264.7 macrophages, HEK293T-based Phoenix ecotropic retroviral packaging cell lines (a gift from Dr. Gary Nolan, Stanford University), Syk-deficient (C35) and Syk-reconstituted (WT8) B-cell lines (15) were maintained in Dulbecco's modified Eagle's medium or RPMI1640 medium (Cambrex) supplemented with 10% heat-inactivated fetal calf serum (Invitrogen), 20 mm HEPES, 2 mm l-glutamine, 100 units/ml penicillin, and 0.1 mg/ml streptomycin (Cambrex). All cells were grown at 37 °C in 5% CO2.
Generation of Constructs and Transduced Cell Lines—The complete CLEC9A open reading frame (ORF) was isolated from human PBMC cDNA by PCR and cloned into the pFBneo (Stratagene) retroviral vector containing an HA tag (16) using the following primers; AAAGAATTCCCACCATGCACGAGGAAGAAATATAC and AAACTCGAGGACAGAGGATCTCAACGC. C-terminally HA-tagged murine CLEC9A (mCLEC9A) was similarly cloned from mouse splenic cDNA using the following primers; AAAGTCGACCACCATGCATGCGGAAGAAATA and GTACTCGACGATGCAGGATCCAAATGC.
The CLEC9A/Dectin-1 chimera in pFBneo was generated using overlap extension PCR with primers CTGCTAAACTTTACAGAAAACCACAAGCCCACA and TCTGGGCTTGTGGTTTTCTGTAAAGTTTAGCAG such that the sequence translated as CLEC9A-79LLNFTECLEC9A-84/Dectin-91NHKPTDectin-95. The Fc-CLEC9A expression construct was generated by PCR using the following primers; TTTGGTACCAGCAGCAAGAAAAACTC and GCGGAATTCGACAGAGGATCTCAACGC, and cloned into the pSecTag2 vector (Invitrogen) upstream from the human IgG1 Fc region, generated as described (17). The fidelity of all constructs was verified by sequencing. Human and murine CLEC9A and isoforms have been deposited at GenBank™ under the following accession numbers: hCLEC9A, EU339276; mCLEC9A, EU339277; mCLEC9Aβ, EU339278; mCLEC9Aγ, EU339279; mCLEC9Aδ, EU339280; mCLEC9Aε, EU339281.
To generate stable cell lines, constructs were packaged into virons, using HEK293T-based Phoenix ecotropic cells, and the various cell lines were transduced as previously described (16). All cell lines were used as nonclonal populations to reduce founder effects and were generated and tested at least twice to confirm phenotype. Where required, cell lines were selected and maintained in 0.6 mg/ml Geneticin (G418) (Merck) or 4 μg/ml puromycin (Invitrogen).
Human CLEC9A expression was analyzed by PCR of human tissue cDNA panels (Clontech) using the following primers encoding the entire ORF, ATGCACGAGGAAGAAATATACACC and GACAGAGGATCTCAACGCATA. mCLEC9A expression was analyzed by PCR of mouse tissue cDNA panels (Clontech) using the same murine primers described above. Expression of G3PDH was used as a positive control.
Generation of a Monoclonal Antibody against CLEC9A—The monoclonal antibody (mAb), 9A11, specific for CLEC9A, was generated by immunization of 129Sv mice with a soluble Fc-CLEC9A fusion protein. NS1 hybridomas were generated, and supernatants from clonally diluted cells were screened by ELISA. The mAb 9A11 (IgG1) was subsequently selected based on its ability to function in ELISA, flow cytometry, and Western blotting, under nonreduced conditions.
Western Blotting and Deglycosylation—To prepare cellular extracts, cells were washed with PBS and then lysed in Nonidet P-40 buffer (1% Nonidet P-40, 0.15 m NaCl, 10 mm EDTA, 10 mm NaN3, 10 mm Tris-HCl, pH 8), supplemented with protease inhibitors (Roche Applied Science), for 30 min at 4 °C. Cellular debris/nuclei were removed by centrifugation, and supernatants were collected and stored at -20 °C. Protein concentrations of cell extracts were determined by BCA assay (Pierce), and equal quantities of protein were run on SDS-PAGE gels, according to standard protocols. After transfer to HybondC+ nitrocellulose membranes (Amersham Biosciences), CLEC9A was detected by immunostaining with 9A11 or anti-HA (clone 16B12, Covance) followed by goat anti-mouse horseradish peroxidase (Jackson). Blots were developed with the ECL-plus kit (Amersham Biosciences). Cell lysates were deglycosylated with with PNGase F and/or O-glycosidase (Roche Applied Science), as described by the manufacturer.
Antibodies, Flow Cytometry, and Cell Sorting—Cells were examined by multicolor flow cytometry on a FASCCaliber (BD Biosciences), performed according to conventional protocols at 4 °C in the presence of 2 mm NaN3. Cells were blocked in PBS containing 5 mm EDTA, 0.5% BSA, and 5% heat-inactivated rabbit serum (murine cells) or murine serum (peripheral blood), and 50 μg/ml human IgG (Sigma; in vitro cultured human cells and PBMCs) prior to the addition of primary antibodies. Cells were fixed with 1% formaldehyde, 0.25% BSA in PBS prior to analysis.
The following antibodies were used in these experiments: 9A11-biotin, D1.3-biotin (a gift from L. Martinez-Pomares, Oxford University), CD1a, CD2, CD3-fluorescein isothiocyanate (FITC) (Serotec), CD4-phycoerythrin (PE) (Serotec), CD8-PE (Serotec), CD11b-FITC (Serotec), CD11c-FITC (Serotec), CD14-FITC (BD Pharmingen), CD16-PE (BD Pharmingen), CD19-PE (BD-Pharmingen), CD56-PE (BD-Pharmingen), CD83-PE (Serotec), CD86-PE (Serotec), BDCA-1-FITC (BD Pharmingen), BDCA-2-PE (BD Pharmingen), DC-SIGN-FITC (BD Pharmingen), HLA-DR-FITC (BD Pharmingen), BDCA3-PE (Miltenyi) and irrelevant biotin-, PE- or FITC-labeled mouse IgG1 (D1.3) IgG2a (BD Pharmingen) and IgG2b (BD Pharmingen) control antibodies. Biotinylated antibodies were detected using streptavidin-allophycocyanin (APC; BD Pharmingen). Data were analyzed using Cellquest (BD Bioscience).
To isolate 9A11+ cells for analysis, 30 million PBMCs, isolated as described above, were blocked with 5% mouse serum or 50 μg/ml human IgG for 15 min at 4 °C. Cells were then stained with biotinylated 9A11 mAb for 30 min at 4 °C, followed by streptavidin-APC (BD Pharmingen), and APC+ cells were sorted using FACSVantage S.E. (Beckton Dickinson). The sorted cells were kept at 4 °C throughout the sorting procedure and then stained for various surface markers, as described above. The sorted cells represented 0.21% ± 0.07% of the starting population of PBMCs and were 72.89 ± 6.21% pure. Only 9A11+ cells within the sorted population were analyzed further (see Fig. 3B).
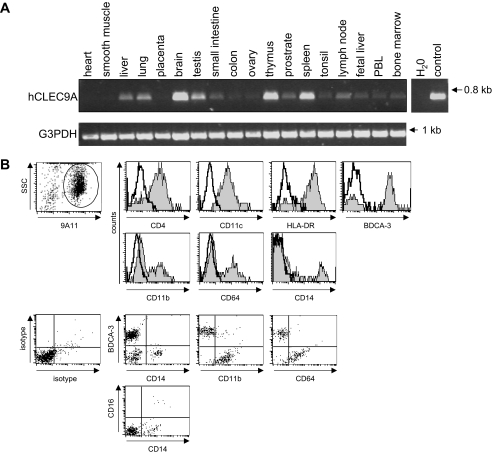
FIGURE 3.
Expression of hCLEC9A on BDCA3+ DC and a subset of CD14+CD16- monocytes. A, RT-PCR analysis showing expression of CLEC9A in most tissues, but predominantly in the brain, spleen, and thymus. B, using a novel monoclonal antibody (9A11), CLEC9A was found to be expressed on a minor population of PBMC (see supplemental Fig. S4C), and cells expressing this receptor were therefore sorted prior to analysis by multicolor flow cytometry. Using various markers, as indicated, all CLEC9A+ cells (gated) were found to be CD4+HLA-DR+CD11c+, and further analysis indicated that the receptor was predominantly expressed on BDCA3+ dendritic cells (59 ± 10%), but also on a subset of CD64+CD11b+CD14+CD16- monocytes (14 ± 6%) and a third population of CD64+CD11b+ CD14- CD16- cells (23 ± 3%), which were not identified further. The data shown are representative of data obtained from at least six different donors.
Endocytosis Assays—For the endocytosis assays, transduced NIH3T3 or RAW264.7 cells were plated at 4 × 105 cells/well and 5 × 105 cells/well, respectively, in 6-well plates the day before the experiment. At the start of the assay, cells were washed and then blocked with PBS containing 5 mm EDTA, 10 mm NaN3, 0.5% BSA, and 5% heat-inactivated rabbit serum for 30 min at 4 °C. Cells were stained with biotinylated 9A11 or anti-mDectin-1 (2A11; 10 μg/ml) and incubated for 50 min at 4 °C and subsequently washed with FACS wash (PBS, 05% BSA, 10 mm NaN3, and 5 mm EDTA), to remove unbound antibody. A cross-linking sheep anti-mouse antibody (Jackson; 5 μg/ml) was added and the cells incubated for another 30 min at 4 °C. After washing in culture medium to remove unbound secondary antibody and azide, the cells were incubated in culture medium for the times indicated at either 37 or 4 °C. Cells were then stained with streptavidin-PE (BD Pharmingen), and examined for remaining receptor surface expression by flow cytometry, as described above.
For microscopy, cells were plated at 2 × 105/well on glass coverslips in 6-well plates the day before the experiment. Cells were treated the same as for the FACS-based assay, only stained with 9A11 for 1 h at 4 °C and subsequently washed with FACS wash to remove unbound antibody. Cells were incubated for 30 min at either 37 °C or 4 °C in culture medium. After incubation, the cells were fixed in 1 ml of Fixative (4% paraformaldehyde; 250 mm HEPES in dH2O) for 20 min. The aldehyde groups were quenched with PBS-glycine (25 mm glycine powder in PBS) for 10 min and then permeabilized with 0.5% Triton X-100 for 20 min. Cells were washed with 0.1% PBS-Tween and blocked thereafter with 0.5% Saponin for 20 min. Cells were stained with LAMP-1 (clone ID4B; Developmental Studies Hybridoma Bank, University of Iowa, Iowa City) for 1 h at room temperature and washed with 0.1% PBS-Tween. Cells were then stained with donkey anti-mouse Cy3 and donkey anti-rat Alexa488 (1:100 dilution; both from Jackson) for 50 min at room temperature and subsequently washed with 0.1% PBS-Tween. Coverslips were mounted with Vectashield (Vector Laboratories) and examined by conventional fluorescence microscopy on a Zeiss Axiovert 40. Images were processed using Adobe Photoshop version 6.0.
Zymosan Binding and Cytokine Assays—RAW264.7 and NIH3T3 transfectants were plated at 5 × 104 cells/well in 24-well plates the day before the experiment. Cells were washed, soluble β-glucans (glucan phosphate; a kind gift from David Williams, ETSU; 5 μg/ml) added where appropriate, and the cells incubated for 20 min at 4 °C to allow inhibition of the Dectin-1 CRD (18). Following the addition of FITC-labeled zymosan (Invitrogen; 25 particles/cell), cells were incubated at 4 °C for 60 min to allow binding, and then washed extensively to remove unbound particles. For the determination of TNF, RAW264.7 cells were incubated for another 3 h, and the amount of TNF released into the supernatants was determined by ELISA (BD Biosciences). For measuring zymosan binding, cells were lysed in 3% Triton X-100 (Sigma), and the amount of bound zymosan was quantified by fluorometry using a Titertek Fluoroskan II (Labsystems Group Ltd.).
For the analysis of the Syk-sufficient and Syk-deficient B-cells, 2 × 106 transduced cells were stimulated with unlabeled zymosan (Sigma; 1 particle per cell) in 24-well suspension plates for 24 h at 37 °C. IL-2 secreted into the supernatants was quantified by ELISA (BD Biosciences). Where indicated, piceatennol (Sigma) was included at 50 μm.
Phagocytosis Assays—To quantify phagocytosis by flow cytometry, transduced NIH3T3 fibroblasts or RAW264.7 macrophages were seeded at 2 × 105cells/well in 6-well plates the day before the experiment. When required, cells were pretreated with 5 μm cytochalasin D (Calbiochem) for 40 min at 37 °C, to inhibit phagocytosis and was maintained throughout the assay. Cells were then washed and FITC-labeled zymosan was added (1 particle/cell) and allowed to bind for 1 h at 4 °C. Following this incubation, unbound zymosan was removed by washing, and the cells incubated at 37°C for 2 h (NIH3T3 cells) or 30 min (RAW264.7 cells) to allow particle uptake. The amount of internalized zymosan was then determined by flow cytometry, as previously described (19). Briefly, external zymosan was stained with polyclonal rabbit anti-zymosan (Invitrogen), which was subsequently detected with APC-labeled goat anti-rabbit antibodies (Invitrogen). Flow cytometric analysis was performed by gating on the FITC-positive cell populations, which had bound zymosan, and the percentage of internalization was determined by comparing the APC-negative (internalized particles) versus the APC-positive (non-internalized particles) cell populations.
The examination of phagocytosis by immunofluoresence microscopy was performed similarly, except that the transduced cells were seeded at 3 × 104 cells/well on glass coverslips, 10 particles per cell of FITC-labeled zymosan were added, and the cells allowed to internalize the particles for 45 min at 37 °C. Following this incubation, the cells were washed, fixed with 4% paraformaldehyde, and permeabilized with 0.25% Saponin in FACS block for 30 min at room temperature. Actin was stained with 1 μm tetramethylrhodamine isothiocyanate (TRITC)-labeled phalloidin (Sigma). Coverslips were mounted with Vectashield (Vector Laboratories) and examined by conventional fluorescence microscopy on a Zeiss Axiovert 40. Images were processed using Adobe Photoshop version 6.0.
Immunoprecipitations—1 × 107 RAW264.7 cells were stimulated with pervanadate for 1 min at 37 °C, and the cells lysed in ice-cold lysis buffer (25 mm Tris, pH 8.0, 140 mm NaCl, 4 mm EDTA, 1.1% Nonidet P-40, 10 mm NaF, 1 mm Na3VO4) with protease inhibitors (Roche Applied Science). Nuclei and cell debris were removed by centrifugation, and the recovered supernatants were added to streptavidin beads (Sigma), precoupled with biotinylated phosphorylated or unphosphorylated peptides (25 μm). The CLEC9A peptides were generated commercially (Sigma) and corresponded to the following region of the cytoplasmic tail of the receptor, 1MHEEEIYRSLQWD13. The Dectin-1 peptides have been described previously (11). The beads were rotated for 2 h at 4 °C and then washed, prior to analysis by Western blotting. Proteins in the immunoprecipitates were detected with anti-phosphotyrosine (clone 4G10), and anti-Syk or anti-Lyn (Santa Cruz Biotechnology), followed by appropriate horseradish peroxidase-linked secondary antibodies (Jackson).
RESULTS
Identification and Characterization of CLEC9A—We had previously identified clec9A as an uncharacterized gene consisting of 6 exons spanning ∼13 kb within the “Dectin-1 cluster” of C-type lectin-like receptors, in the NK complex on chromosome 12 (7, 16) (Fig. 1). Sequence analysis indicated that clec9A encoded a type II transmembrane protein of 241 amino acids with a predicted molecular mass of ∼30 kDa. Human CLEC9A (hCLEC9A) possesses a structure typical of group V C-type lectin-like receptors (2), consisting of a single extracellular CTLD connected to the transmembrane domain by a stalk region, and an intracellular cytoplasmic tail with potential signaling motifs. The CTLD of CLEC9A possesses the characteristic six cysteine residues, which are thought to be involved in disulfide bond formation and stabilization of the CTLD fold, but lacks the conserved residues involved in calcium ion coordination and carbohydrate binding found in the classical C-type lectins (1). The stalk region of CLEC9A possesses two cysteines, potentially involved in receptor dimerization, and a single predicated N-linked glycosylation site. The cytoplasmic tail contains a single tyrosine residue within a sequence showing similarity to the ITAM-like sequence of Dectin-1 (18).
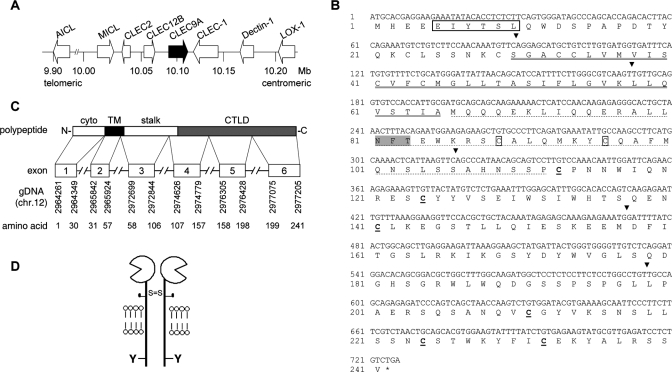
FIGURE 1.
Human CLEC9A genomic localization, gene structure, and polypeptide sequence. A, schematic view of the Dectin-1 cluster on human chromosome 12p13.31, indicating the position of CLEC9A (black arrow). B, nucleotide and translated protein sequence of CLEC9A. The putative ITAM-like motif is boxed; the transmembrane region is underlined by a double solid line; and the stalk region is underlined by a dashed line. The two cysteine residues in the stalk region are boxed, and the six conserved cysteine residues in the CTLD are in bold and underlined. A putative N-glycosylation site is highlighted in gray. The black arrows indicate the exon boundaries of CLEC9A. C, schematic representation of the genomic structure of CLEC9A aligned with the encoded polypeptide, showing the positions of the exons in the genomic DNA (gDNA) and corresponding translated amino acid sequence. D, cartoon structure of predicted human CLEC9A (S, cysteine residue; Y, tyrosine residue).
To characterize the structural features of hCLEC9A, we cloned and expressed an HA-tagged version of the receptor in NIH3T3 fibroblasts. Murine Dectin-1 (18) was included as a control. By flow cytometry, we could demonstrate that hCLEC9A was expressed at the cell surface (Fig. 2A), suggesting that similar to the other receptors in the Dectin-1 cluster, CLEC9A did not require an adaptor protein for expression (8). Western blot analysis of lysates from the transfected NIH3T3 cells, under nonreducing conditions, indicated that hCLEC9A was expressed as protein of ∼100 kDa, more than twice its predicted mass (Fig. 2B). However, under reducing conditions, hCLEC9A resolved to two bands of mass of ∼40 and ∼45 kDa. Treatment of the transduced cell lysates with PNGase, which removes all N-linked glycosylation, further reduced the mass of both bands to ∼30 kDa (the predicted molecular mass of this protein) and ∼35 kDa, respectively. The presence of a second band in these cell lysates, which was also observed with Dectin-1, may be due to other post-translational modifications. However, this is unlikely to be O-glycosylation, as treatment with O-deglycosidase did not affect the mass of this protein. Thus these data indicate that hCLEC9A is expressed as a glycosylated dimer at the cell surface.
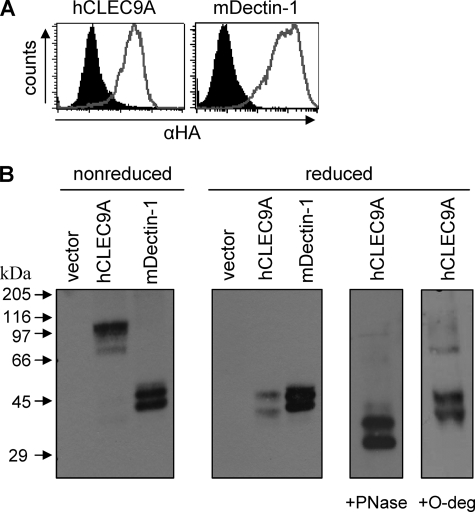
FIGURE 2.
Expression of hCLEC9A as a glycosylated dimer. A, flow cytometry of live NIH3T3 cells stably expressing HA-tagged hCLEC9A or mDectin-1, as indicated (gray lines), demonstrating expression of these receptors at the cell surface. The filled histograms indicate vector-only transduced controls. B, anti-HA Western blotting of lysates from these cells demonstrating that CLEC9A is expressed as a dimer of ∼100 kDa under nonreducing conditions, which can be resolved to two bands of ∼45 and 40 kDa under reducing conditions. Treatment of the cell lysates with PNGase, which removes N-linked glycosylation, reduced the mass of these bands to close to that expected from the predicted amino acid sequence, indicating that CLEC9A is N-glycosylated. The post-translation modification giving rise to the second band seen in these lysates was not due to O-glycosylation, but was also seen with mDectin-1, which is expressed as a monomer (32) and was included as a control for these experiments.
The Dectin-1 cluster of receptors is conserved between species (20), and we identified the murine orthologue based on homology and genomic position. The protein encoded by murine clec9A is 53% identical to human CLEC9A, and retains most of the structural features described above, including the intracellular signaling motifs (supplemental Fig. S1). However, from the deduced amino acid sequence, the murine receptor appears to possess an additional N-glycosylation site and lacks one of the cysteine residues in the stalk region. Given these differences to the human receptor, we characterized the structural features of murine CLEC9A expressed in transduced NIH3T3 fibroblasts. As for the human receptor, mCLEC9A was expressed at the cell surface, but Western blot analysis indicated that mCLEC9A was not glycosylated and that the receptor was expressed as a monomer (supplemental Fig. S2).
Expression of CLEC9A on Primary Cells and Tissues—We first examined the expression of CLEC9A by RT-PCR, using primers, which amplified the entire ORF from cDNA isolated from a variety of tissues (Fig. 3A). Human CLEC9A was detected in most tissues, with highest expression apparent in the brain, thymus, and spleen. Only a single band was detected, suggesting that there were no alternative splice variants. The murine orthologue was expressed in most tissues, but a number of bands were detected, which upon sequencing, were shown to be due to alternative splicing (supplemental Fig. S3). While most of these variants lack one or more exons, as found in many other C-type lectins, there was an isoform containing a 7th exon, which extended the polypeptide by 26 amino acids.
To characterize CLEC9A further, we generated a monoclonal antibody (mAb) by immunizing mice with a soluble Fc-hCLEC9A fusion protein. One mAb (9A11, IgG1) was chosen for further analysis, as this mAb specifically recognized human CLEC9A, as demonstrated by flow cytometry and Western blotting, under nonreducing conditions (supplemental Fig. S4, A and B). 9A11 could not detect CLEC9A by Western blotting under reduced conditions, or in paraffin-fixed tissues (data not shown).
We then examined expression of CLEC9A by multicolor live-cell flow cytometry (Fig. 3B). In total human peripheral blood, expression of CLEC9A could be detected on only a very small number of PBMCs (supplemental Fig. S4C), consistent with the low levels of expression detected by RT-PCR in PBL. To characterize these cells further, we purified CLEC9A+ leukocytes by cell sorting, and verified that the isolated cells expressed CLEC9A by RT-PCR (supplemental Fig. S4). By single staining with selected markers, we were able to demonstrate that all sorted CLEC9A+ cells expressed CD4, CD11c, and HLA-DR (Fig. 3B). The majority of these cells expressed BDCA-3 (21), but we also detected subpopulations of 9A11+ cells expressing CD11b, CD64, and CD14. Double staining with these markers revealed that CLEC9A is expressed on three populations of cells, BDCA3+ dendritic cells (DC), CD11b+CD64+CD14+ CD16- monocytes, and a CD11b+CD64+CD14-CD16- population of cells, which we were unable to identify further. These cells were all negative for expression of CD1a, CD2, CD8, BDCA-1, BDCA-2, DC-SIGN, CD83 as well as lineage specific markers including CD3, CD19, and CD56 (data not shown). Further analysis of PBMCs indicated that CLEC9A was expressed on all BDCA3+ cells, 5% (± 2%) of all CD11b+ cells, 2% (± 1%) of all CD14+ cells, and 6% (± 3%) of all CD64+ cells (data not shown). In vitro cultured, peripheral blood-derived, macrophages, and dendritic cells did not express CLEC9A (data not shown). Thus CLEC9A is predominantly expressed by BDCA3+ DCs, but also on a minor subset of CD14+CD16- monocytes as well as a population of CD14-CD11b+CD64+ cells, which remain to be identified.
CLEC9A Is an Endocytic Receptor—Given the limitation in obtaining primary CLEC9A+ cells for analysis, we resorted to determining the function of this receptor in transduced cell lines. Many DC-expressed molecules are involved in antigen uptake (22) and as the cytoplasmic tail of this molecule contains potential internalization motifs (Fig. 1), we examined the ability of CLEC9A to mediate endocytosis (Fig. 4). For this analysis, we cross-linked CLEC9A on the surface of transduced NIH3T3 fibroblasts and monitored the disappearance of this receptor from the cell surface over time by flow cytometry (Fig. 4A). Cross-linking of CLEC9A led to a rapid reduction of the receptor at the cell surface, compared with cells maintained at 4 °C, indicating that the receptor was mediating internalization of the bound antibodies. As before, Dectin-1, which is also known to mediate endocytosis (19), was included as a positive control. Similar results were obtained in transduced RAW264.7 macrophages (supplemental Fig. S5), in which we could also demonstrate the formation of intracellular CLEC9A+ endocytic vesicles, some of which co-stained with the late endosomal/lysosomal marker, LAMP-1 (Fig. 4B). These data therefore indicate that CLEC9A is an endocytic receptor.
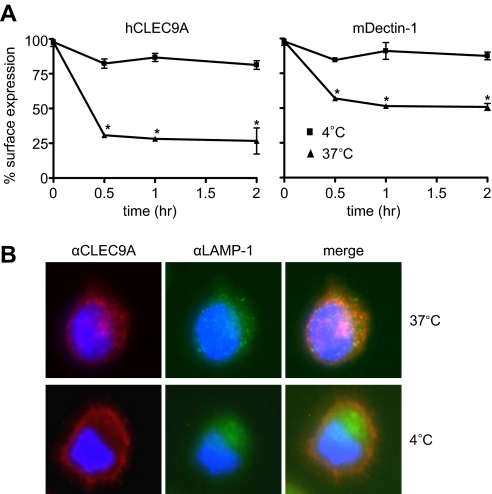
FIGURE 4.
hCLEC9A is an endocytic receptor. A, internalization of CLEC9A expressed in NIH3T3 fibroblasts, following antibody cross-linking, was determined by flow cytometry. The analysis, performed by comparing cells incubated at 37 versus 4 °C (to prevent internalization), demonstrates that hCLEC9A is rapidly removed from the cell surface following cross-linking, indicative of receptor endocytosis. Dectin-1, previously shown to be endocytic (19) was included as a control. The data shown are mean ± S.D. of triplicates, and representative of three independent experiments. *, p < 0.05 versus control (Student's t test). B, fluorescent microscopy demonstrating internalization of CLEC9A (red) into LAMP1+(green) vesicles, as indicated following cross-linking and incubation at 37 °C versus 4 °C. Nuclei are stained with DAPI (blue).
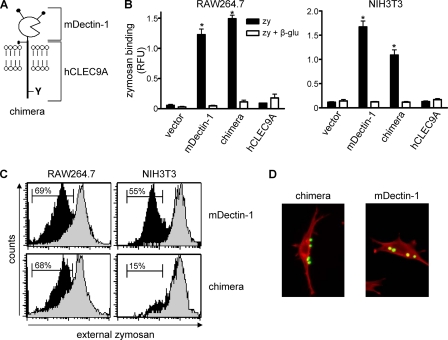
CLEC9A Does Not Mediate Phagocytosis—CLEC9A contains a tyrosine-based sequence, which is similar to that shown to mediate phagocytosis in Dectin-1 (19), so we investigated the possibility that CLEC9A could also mediate particle uptake. As the ligand for CLEC9A is unknown, we constructed a chimeric receptor expressing an HA-tagged extracellular domain of Dectin-1, coupled to the cytoplasmic tail and transmembrane regions of CLEC9A (Fig. 5A). This monomeric chimera would allow us to trigger CLEC9A signaling using zymosan, a defined particulate ligand of Dectin-1 (18).
FIGURE 5.
hCLEC9A is not a phagocytic receptor. A, schematic representation of the chimeric Dectin-1/CLEC9A receptor. B, fluorometric quantitation of zymosan (zym) binding by the transduced RAW264.7 macrophages or NIH3T3 fibroblasts, in the presence or absence of soluble β-glucan (β-glu), as indicated. RFU, relative fluorescence units. Shown are the mean ± S.D. of one representative experiment of three. C, flow cytometric quantitation of zymosan uptake (black histograms) by the chimera or Dectin-1 expressing RAW264.7 macrophages or NIH3T3 fibroblasts, as indicated. Cytochalasin D (gray-filled histograms) was used to inhibit actin polymerization and served as a control in this assay. The histograms shown are representative of at least three independent experiments, and the bars indicate the percentage of cells with internalized particles. *, p < 0.05 versus control (Student's t test). D, representative confocal images demonstrating FITC-zymosan (green) uptake in NIH3T3 fibroblasts transduced with Dectin-1, but not the chimeric receptor. Actin was stained with TRITC-phallodin, and is shown in red.
We then expressed the chimeric receptor, as well as full-length HA-tagged CLEC9A and Dectin-1, in RAW264.7 macrophages and examined the ability of these transduced cells to recognize and ingest zymosan. The expression of these receptors at the cell surface was confirmed by flow cytometry (supplemental Fig. S6). As expected (18, 23), the expression of Dectin-1 conferred the ability to bind zymosan in a β-glucan-dependent fashion, and similar levels of β-glucan-dependent zymosan binding were also obtained with cells expressing the chimeric receptor (Fig. 5B). In contrast, cells transduced with vector only or hCLEC9A were unable to recognize zymosan.
We next determined the ability of these transduced cells to internalize zymosan, by incubating the cells at 37 °C and measuring the amount of internalized particles by flow cytometry and microscopy, as described under “Experimental Procedures” (Fig. 5C and supplemental Fig. S7). Where indicated, some cells were also treated with cytochalasin D to inhibit actin polymerization, and hence particle uptake, and were used as controls in these experiments. As expected, Dectin-1 mediated the internalization of zymosan, in an actin-dependent manner (19), but we also observed that macrophages expressing the chimeric receptor were able to mediate zymosan uptake.
The phagocytosis of complex particles can involve several macrophage receptors, and to prove the phagocytic ability of a receptor requires demonstration of this activity when expressed in normally non-phagocytic cells lines, such as NIH3T3 cells (19, 24–26). We therefore expressed the chimeric receptor in these fibroblasts and examined the ability of these cells to ingest zymosan. As before, expression of the chimera and Dectin-1, but not vector only or full-length CLEC9A, conferred the ability on these cells to bind zymosan in a β-glucan-dependent fashion (Fig. 5B and supplemental Fig. S6). As previously demonstrated (19), Dectin-1 mediated particle internalization in these cells, whereas the chimeric receptor, despite mediating zymosan binding, did not induce particle uptake (Fig. 5, C and D). Thus, hCLEC9A is not a phagocytic receptor.
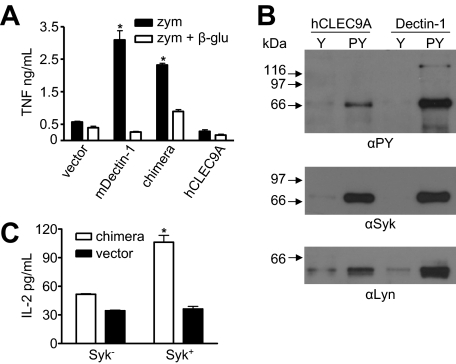
CLEC9A Can Induce Inflammatory Cytokine Production—In addition to phagocytosis, the tyrosine-based motif of Dectin-1 can induce a variety of cellular responses, including the production of proinflammatory cytokines in response to zymosan (27). So we next explored the possibility that signaling via CLEC9A could similarly induce TNF production in the RAW264.7 macrophages transduced with the chimeric receptor. As described above, the expression of Dectin-1 and the chimeric receptor resulted in increased levels of zymosan binding in these cells (Fig. 5). Furthermore, both Dectin-1 (23) and the chimeric receptor induced high levels of TNF in response to zymosan (Fig. 6A). As we have previously shown that Dectin-1 lacking a cytoplasmic tail can mediate zymosan binding, but not the induction of TNF (23), our results indicate that the cellular response induced by the chimeric receptor is due to the presence of the CLEC9A cytoplasmic tail. Thus these data suggest that, like Dectin-1, CLEC9A signaling can induce a proinflammatory response.
FIGURE 6.
hCLEC9A can induce pro-inflammatory cytokine production and can recruit and signal via Syk kinase. A, quantitation of zymosan (zym) induced TNF production by transduced RAW264.7 macrophages, in the presence or absence of soluble β-glucan (β-glu), as indicated, demonstrating the ability of the chimeric receptor to induce proinflammatory cytokine production. Shown are the mean ± S.D. of one representative experiment of three. B, Western blotting of immunoprecipitates using phosphorylated (pY) and unphosphorylated peptides (Y) corresponding to the cytoplasmic tail of hCLEC9A or Dectin-1 from RAW264.7 cell lysates. Blots were probed with anti-phosphotyrosine (αPY), anti-Syk, and anti-Lyn, as indicated. C, IL-2 production following zymosan stimulation of Syk-deficient (Syk-) and Syk-sufficient (Syk+) cells, transduced with the chimeric receptor or vector-only control, as indicated, showing that cytokine production induced by the chimeric receptor in response to zymosan requires Syk. The data shown are the mean ± S.D. and are representative of three independent experiments. *, p < 0.05 versus control (Student's t test).
CLEC9A Can Associate and Signal via Syk Kinase—As cytokine induction mediated by Dectin-1 involves signaling via Syk (27, 28), we examined the possibility that this kinase was involved in the intracellular signaling mediated by CLEC9A. For this, we generated tyrosine-phosphorylated and unphosphorylated forms of peptides corresponding to the cytoplasmic tails of CLEC9A and Dectin-1. These peptides were then used to immunoprecipitate associated signaling molecules from RAW264.7 cell lysates, which were subsequently analyzed by Western blotting with specific antibodies (Fig. 6B). Probing with anti-phosphotyrosine antibodies indicated a predominant band of ∼70 kDa in immunoprecipitates with phosphorylated peptides of both CLEC9A and Dectin-1, which we confirmed to be Syk by probing with anti-Syk antibodies. We were also able to show an association with the Src kinase, Lyn, which is thought to be involved in tyrosine phosphorylation of ITAM motifs upon ligand binding (29).
To confirm that CLEC9A could induce intracellular signaling via Syk kinase, we expressed the chimeric receptor in Syk-sufficient and Syk-deficient B-cell lines and monitored their response to zymosan (Fig. 6C). This approach was similar to that we had used previously to demonstrate the Syk-dependent activity of Dectin-1 (11). The chimeric receptor was expressed at similar levels in both cell types, as determined by flow cytometry (supplemental Fig. S8A). The addition of zymosan to the Syk-sufficient B-cells expressing the chimeric receptor induced the production of IL-2. In contrast, this response was absent in vector control-transduced cells and in the transduced Syk-deficient B-cell lines. Furthermore, we could also inhibit the IL-2 response in the Syk-sufficient cells by including the Syk inhibitor piceatannol (supplemental Fig. S8B). Thus, CLEC9A can mediate intracellular signaling via Syk kinase.
DISCUSSION
We describe here the first characterization of human CLEC9A and demonstrate that it functions as an activation receptor, capable of triggering intracellular signaling via Syk kinase. This interaction is mediated by the cytoplasmic tail of the CLEC9A that contains a single tyrosine-based (EXYXXL) sequence, which can be considered to be an ITAM-like motif (19). These sequences were first identified in two other receptors of the Dectin-1 cluster, Dectin-1 itself (11), and more recently CLEC-2 (12). While the mechanism of Syk signaling via these motifs is unknown, it requires both SH2 domains of this kinase (13) and may occur via bridging of two receptor chains (11).
In myeloid cells, ITAM-like mediated signaling through Syk has largely been studied with Dectin-1. Signaling via this kinase occurs through a novel pathway involving CARD9 (30) and can induce a variety of cellular responses including the induction of cytokines (such as TNF, IL-6, IL-10, IL-23, and IL-2), the respiratory burst, and the production of arachidonic acid (27, 31). In addition, signaling via this pathway was recently shown to induce T-helper type 17 (TH17) adaptive responses in vivo (31). As CLEC9A can signal via this pathway, it is possible that this receptor may be able to induce many, if not all, of these responses, which may be influenced by its ability to dimerize.
The ITAM-like motif of Dectin-1 also mediates phagocytosis (19). Although we could demonstrate that CLEC9A could induce particle uptake in macrophages, it was unable to do so when expressed in NIH3T3 cells. As the definition of a phagocytic receptor is its ability to confer phagocytic activity to normally non-phagocytic cell lines, we conclude that CLEC9A is not directly able to mediate particle uptake. In the RAW264.7 macrophages, it is possible that the residual low levels of endogenous Dectin-1 were contributing to this process (32). In addition Dectin-1-mediated phagocytosis occurs through novel mechanisms (19), which are still undefined and requires additional tyrosine-proximal residues (33) that are absent in CLEC9A.
The functional characterization of CLEC9A on primary cells was limited by its expression on extremely rare populations of peripheral blood leukocytes. The receptor is predominantly expressed by BDCA3+ DCs, which represent less than 0.05% of PBMC (34). Very little is known about this cell type, but they are thought to be immature precursors of interstitial DCs, and were recently shown to express multiple Toll-like receptors (34–36). CLEC9A is also expressed on small subsets of CD14+CD16- monocytes and on CD64+CD11b+ CD14- cells we were unable to identify further. CD14+CD16- monocytes, the so called “inflammatory monocytes,” normally constitute around 14% of the PBMC and are thought to migrate to sites of inflammation where they can differentiate into dendritic cells (37, 38). The expression of CLEC9A on a minor subset of these cells is suggestive of further differentiation of this monocyte population, and this receptor could potentially be used as a marker to characterize this subset in future analyses.
Although the ligand and physiological role of CLEC9A is unknown, its limited expression on peripheral blood leukocytes suggests that it is unlikely to function as a pattern recognition receptor. However, the high levels of expression in the thymus and spleen, as determined by RT-PCR analysis (Fig. 2), are suggestive of a role in immunity. Furthermore, as an endocytic receptor, CLEC9A may facilitate antigen uptake and presentation, and may provide a suitable target for antibody-mediated antigen delivery, as has been demonstrated for a variety of other C-type lectins, including Dectin-1 (39).
Despite being conserved between species (20), the murine CLEC9A orthologue appears to be structurally dissimilar to human CLEC9A. Murine, but not human, receptor is alternatively spliced into a number isoforms. Furthermore, murine CLEC9A is not glycosylated nor does the receptor dimerize. While the significance of these differences is unclear, they may reflect some degree of functional divergence of this molecule between the various species. However, given the rarity of cells expressing this receptor in human blood, it may still be practical for further study of this receptor to be performed in mice.
Supplementary Material
Acknowledgments
We thank the Western Province Blood Transfusion service for providing the buffy coats, and Andrew Marshall, John Davies, Elwira Pyz, Ronald Dreyer, Vicky Tsoni, and Ann Kerrigan for reagents and advice. We also thank Delphine Le Roux and Ana-Maria Lennon-Duménil (Institut Curie, Paris) for generously providing the Syk-sufficient and deficient B-cells.
This work was supported by the National Research Foundation (South Africa), the Wellcome Trust and scholarships from Harry Crossley, KW Johnston Research, and Marion Beatrice. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Author's Choice—Final version full access.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S8.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBank™/EBI Data Bank with accession number(s) EU339276, EU339277, EU339278, EU339279, EU339280, and EU339281.
The abbreviations used are: CTLD, C-type lectin domain; HA, hemagglutinin; PBS, phosphate-buffered saline; BSA, bovine serum albumin; FITC, fluorescein isothiocyanate; FACS, fluorescent-activated cell sorting; NK, natural killer cells; TNF, tumor necrosis factor; IL, interleukin; DC, dendritic cells; PBMC, peripheral blood mononuclear cells; TRITC, tetramethylrhodamine isothiocyanate; ORF, open reading frame.
References
- 1.Weis, W. I., Taylor, M. E., and Drickamer, K. (1998) Immunol. Rev. 163 19-34 [DOI] [PubMed] [Google Scholar]
- 2.Zelensky, A. N., and Gready, J. E. (2005) Febs J. 272 6179-6217 [DOI] [PubMed] [Google Scholar]
- 3.Yokoyama, W. M., and Plougastel, B. F. (2003) Nat. Rev. Immunol. 3 304-316 [DOI] [PubMed] [Google Scholar]
- 4.Ito, M., Maruyama, T., Saito, N., Koganei, S., Yamamoto, K., and Matsumoto, N. (2006) J. Exp. Med. 203 289-295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iizuka, K., Naidenko, O. V., Plougastel, B. F., Fremont, D. H., and Yokoyama, W. M. (2003) Nat. Immunol. 4 801-807 [DOI] [PubMed] [Google Scholar]
- 6.Ravetch, J. V., and Lanier, L. L. (2000) Science 290 84-89 [DOI] [PubMed] [Google Scholar]
- 7.Sobanov, Y., Bernreiter, A., Derdak, S., Mechtcheriakova, D., Schweighofer, B., Duchler, M., Kalthoff, F., and Hofer, E. (2001) Eur. J. Immunol. 31 3493-3503 [DOI] [PubMed] [Google Scholar]
- 8.Pyz, E., Marshall, A. S., Gordon, S., and Brown, G. D. (2006) Ann. Med. 38 242-251 [DOI] [PubMed] [Google Scholar]
- 9.Robinson, M. J., Sancho, D., Slack, E. C., LeibundGut-Landmann, S., and Reis e Sousa, C. (2006) Nat. Immunol. 7 1258-1265 [DOI] [PubMed] [Google Scholar]
- 10.Taylor, P. R., Martinez-Pomares, L., Stacey, M., Lin, H. H., Brown, G. D., and Gordon, S. (2005) Annu. Rev. Immunol. 23 901-944 [DOI] [PubMed] [Google Scholar]
- 11.Rogers, N. C., Slack, E. C., Edwards, A. D., Nolte, M. A., Schulz, O., Schweighoffer, E., Williams, D. L., Gordon, S., Tybulewicz, V. L., Brown, G. D., and Reis e Sousa, C. (2005) Immunity 22 507-517 [DOI] [PubMed] [Google Scholar]
- 12.Suzuki-Inoue, K., Fuller, G. L., Garcia, A., Eble, J. A., Pohlmann, S., Inoue, O., Gartner, T. K., Hughan, S. C., Pearce, A. C., Laing, G. D., Theakston, R. D., Schweighoffer, E., Zitzmann, N., Morita, T., Tybulewicz, V. L., Ozaki, Y., and Watson, S. P. (2006) Blood 107 542-549 [DOI] [PubMed] [Google Scholar]
- 13.Fuller, G. L., Williams, J. A., Tomlinson, M. G., Eble, J. A., Hanna, S. L., Pohlmann, S., Suzuki-Inoue, K., Ozaki, Y., Watson, S. P., and Pearce, A. C. (2007) J. Biol. Chem. 282 12397-12409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Willment, J. A., Marshall, A. S., Reid, D. M., Williams, D. L., Wong, S. Y., Gordon, S., and Brown, G. D. (2005) Eur. J. Immunol. 35 1539-1547 [DOI] [PubMed] [Google Scholar]
- 15.Yokozeki, T., Adler, K., Lankar, D., and Bonnerot, C. (2003) J. Immunol. 171 1328-1335 [DOI] [PubMed] [Google Scholar]
- 16.Marshall, A. S., Willment, J. A., Lin, H. H., Williams, D. L., Gordon, S., and Brown, G. D. (2004) J. Biol. Chem. 279 14792-14802 [DOI] [PubMed] [Google Scholar]
- 17.Graham, L. M., Tsoni, S. V., Willment, J. A., Williams, D. L., Taylor, P. R., Gordon, S., Dennehy, K., and Brown, G. D. (2006) J. Immunol. Methods 314 164-169 [DOI] [PubMed] [Google Scholar]
- 18.Brown, G. D., and Gordon, S. (2001) Nature 413 36-37 [DOI] [PubMed] [Google Scholar]
- 19.Herre, J., Marshall, A. J., Caron, E., Edwards, A. D., Williams, D. L., Schweighoffer, E., Tybulewicz, V. L., Reis e Sousa, C., Gordon, S., and Brown, G. D. (2004) Blood 104 4038-4045 [DOI] [PubMed] [Google Scholar]
- 20.Hao, L., Klein, J., and Nei, M. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 3192-3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dzionek, A., Fuchs, A., Schmidt, P., Cremer, S., Zysk, M., Miltenyi, S., Buck, D. W., and Schmitz, J. (2000) J. Immunol. 165 6037-6046 [DOI] [PubMed] [Google Scholar]
- 22.Steinman, R. M., and Banchereau, J. (2007) Nature 449 419-426 [DOI] [PubMed] [Google Scholar]
- 23.Brown, G. D., Herre, J., Williams, D. L., Willment, J. A., Marshall, A. S. J., and Gordon, S. (2003) J. Exp. Med. 197 1119-1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le Cabec, V., Emorine, L. J., Toesca, I., Cougoule, C., and Maridonneau-Parini, I. (2005) J. Leukoc. Biol. 77 934-943 [DOI] [PubMed] [Google Scholar]
- 25.Ezekowitz, R. A., Sastry, K., Bailly, P., and Warner, A. (1990) J. Exp. Med. 172 1785-1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Indik, Z. K., Park, J. G., Hunter, S., and Schreiber, A. D. (1995) Blood 86 4389-4399 [PubMed] [Google Scholar]
- 27.Brown, G. D. (2006) Nat. Rev. Immunol. 6 33-43 [DOI] [PubMed] [Google Scholar]
- 28.Dennehy, K. M., Ferwerda, G., Faro-Trindade, I., Pyz, E., Willment, J. A., Taylor, P. R., Kerrigan, A., Tsoni, S. V., Gordon, S., Meyer-Wentrup, F., Adema, G. J., Kullberg, B. J., Schweighoffer, E., Tybulewicz, V., Mora-Montes, H. M., Gow, N. A., Williams, D. L., Netea, M. G., and Brown, G. D. (2008) Eur. J. Immunol. 38 500-506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strzelecka-Kiliszek, A., Kwiatkowska, K., and Sobota, A. (2002) J. Immunol. 169 6787-6794 [DOI] [PubMed] [Google Scholar]
- 30.Gross, O., Gewies, A., Finger, K., Schafer, M., Sparwasser, T., Peschel, C., Forster, I., and Ruland, J. (2006) Nature 442 651-656 [DOI] [PubMed] [Google Scholar]
- 31.Leibundgut-Landmann, S., Gross, O., Robinson, M. J., Osorio, F., Slack, E. C., Tsoni, S. V., Schweighoffer, E., Tybulewicz, V., Brown, G. D., Ruland, J., and Reis, E. S. C. (2007) Nat. Immunol. 8 630-638 [DOI] [PubMed] [Google Scholar]
- 32.Brown, G. D., Taylor, P. R., Reid, D. M., Willment, J. A., Williams, D. L., Martinez-Pomares, L., Wong, S. Y. C., and Gordon, S. (2002) J. Exp. Med. 296 407-412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Underhill, D. M., Rossnagle, E., Lowell, C. A., and Simmons, R. M. (2005) Blood 106 2543-2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sato, K., and Fujita, S. (2007) Allergol. Int. 56 183-191 [DOI] [PubMed] [Google Scholar]
- 35.MacDonald, K. P., Munster, D. J., Clark, G. J., Dzionek, A., Schmitz, J., and Hart, D. N. (2002) Blood 100 4512-4520 [DOI] [PubMed] [Google Scholar]
- 36.Piccioli, D., Tavarini, S., Borgogni, E., Steri, V., Nuti, S., Sammicheli, C., Bardelli, M., Montagna, D., Locatelli, F., and Wack, A. (2007) Blood 109 5371-5379 [DOI] [PubMed] [Google Scholar]
- 37.Geissmann, F., Jung, S., and Littman, D. R. (2003) Immunity 19 71-82 [DOI] [PubMed] [Google Scholar]
- 38.Passlick, B., Flieger, D., and Ziegler-Heitbrock, H. W. (1989) Blood 74 2527-2534 [PubMed] [Google Scholar]
- 39.Carter, R. W., Thompson, C., Reid, D. M., Wong, S. Y., and Tough, D. F. (2006) J. Immunol. 177 2276-2284 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.