Abstract
Our previous results showed that the polycomb protein mel-18 binds to a protein called HSF2 and inhibits HSF2 sumoylation, thereby functioning as an anti-SUMO E3 factor. This study also suggested that mel-18 regulates the sumoylation of other cellular proteins, but the identities of these other proteins were unknown. Here we show that mel-18 interacts with the RanGAP1 protein and inhibits its sumoylation, and that these activities do not require the RING domain of mel-18. The results also show that RanGAP1 sumoylation is decreased during mitosis, and that this is associated with increased interaction between RanGAP1 and mel-18 during this stage of the cell cycle. Intriguingly, this regulatory relationship is the opposite of that found for mel-18 and HSF2, in which the interaction between these two proteins decreases during mitosis, resulting in elevated HSF2 sumoylation. The results of this study strengthen the conclusion that mel-18 functions as an anti-SUMO E3 factor, and extend its targets to include regulation of the sumoylation of the important cellular protein RanGAP1.
Keywords: RanGAP1, mel-18, polycomb, SUMO, SUMO-1, mitosis
Introduction
Covalent attachment of Small Ubiquitin-like Modifier (SUMO) proteins to lysine residues in target proteins, or sumoylation, is an important regulator of protein functional properties [1–5]. SUMO proteins are covalently attached to target lysine residues by the SUMO E2 enzyme, ubc9, and these substrate lysines are typically found within the consensus sequence ΨKXE (Ψ represents hydrophobic amino acids) [6–9].
SUMO E3 proteins have been identified that enhance the efficiency of sumoylation by interacting with both ubc9 (SUMO E2) and the target protein, thereby acting as bridging factors to increase the rate of the sumoylation reaction [1–3, 5, 10–14]. However, the results of a recent study in our laboratory revealed the surprising finding that a member of the polycomb group of proteins, called mel-18, function like an anti-SUMO E3 protein [15]. In this study it was found that mel-18 binds to and inhibits the sumoylation of a protein called HSF2 by interacting with and inhibiting the activity of ubc9 enzymes in the vicinity of HSF2. These results also indicated that mel-18 may function to inhibit the sumoylation of other cellular proteins, but the identities of these other proteins was unknown.
RanGAP1, a Ran GTPase-activating protein that plays a critical role in nuclear transport, was the first identified substrate for SUMO modification [16, 17]. The results presented in this paper reveal that mel-18 interacts with RanGAP1 and inhibits its sumoylation, and that these effects do not require the RING domain of mel-18. The results also show that RanGAP1 sumoylation decreases during mitosis and that this is correlated with increased interaction between RanGAP1 and mel-18 during this stage of the cell cycle, which is the reverse of the regulatory relationship between mel-18 and HSF2 sumoylation with respect to mitosis. These results strengthen support for the function of the polycomb protein mel-18 as an anti-SUMO-E3 factor, and indicate that it is an important regulator of the sumoylation of a number of vital proteins in the cell.
Materials and Methods
Cell culture
HEK293T cells were grown at 37°C in DMEM supplemented with 10% FBS.
Mutagenesis of Plasmids
pEGFP-mel-18 plasmid mutations, including the Δ17–56 mutant of mel-18 (ring finger deletion referred to as RINGΔ) and the C53G and C56G substitution mutants, were generated using the QuickChange mutagenesis method (Stratagene) according to the manufacturer’s protocol. Mutations were confirmed by DNA sequencing. A T7-RanGAP1 Δ419 mutant of RanGAP1 was generated using the QuickChange mutagenesis kit (Stratagene) according to the manufacturer’s protocol by using pQE-RanGAP1-NΔ419 plasmid (C-terminus of RanGAP1 sequence from amino acid 420 to end 589). Mutations were confirmed by DNA sequencing.
Immunoprecipitation analysis
For co-immunoprecipitation experiments, HEK 293 cells were transfected with GFP-mel-18 expression constructs (wild type, ring finger deletion (RINGΔ), C53G, or C56G mutations) using jetPEI reagent according to the manufacturer’s instructions (polyplus-transfection), and then blocked in mitosis by treatment with 400 ng/ml nocodazole for 16 hours. After 48 hours transfected cells were extracted on ice with NP-40 lysis buffer (1% NP-40, 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1 mM dithiothreitol, and complete protease inhibitor cocktail (Roche Applied Science)) for 20 minutes. Lysates were then cleared by centrifugation at 13,000 rpm for 10 minutes at 4°C. Supernatants were precleared by incubation with goat (control) IgG and protein G-sepharose beads for 2 hours at 4°C with gentle rotation. Precleared extracts were then incubated with primary goat polyclonal RanGAP1 antibody or control IgG and 50% slurry of protein G-sepharose for 4 hours at 4°C with rotation. After washing beads 6 times for 5 minutes each at 4°C with NP-40 buffer, bound proteins were released by boiling in SDS-PAGE sample dye and analyzed by Western blot using the GFP mouse monoclonal antibody (JL-8 clone, Invitrogen). For immunoprecipitation analysis of RanGAP1 sumoylation, HEK 293T cells were transfected with pEGFP-mel-18 (wild type or mutants) or pEGFP-C1 along with the myc-sumo-1 expression plasmid using jetPEI reagent according to the manufacturer’s instructions (polyplus-transfection). After 48 hours cell extracts were prepared using NP-40 Buffer (50 mM Tris–HCl, (pH 8.0), 150 mM NaCl, 1% Nonidet P-40, 1 mM dithiothreitol, and complete protease inhibitor cocktail (Roche Applied Science), 10 mM N-ethylmaleimide) before adding RanGAP1 polyclonal antibodies or non-specific IgG to proceed as described above, followed by Western blot assay using anti-myc monoclonal antibody (Invitrogen) or anti-SUMO-1 antibodies (Bethyl).
In vitro sumoylation assay
T7-RanGAP1 Δ419 (gift of Mike Matunis) was in vitro translated in a rabbit reticulocyte lysates (TNT, Promega) and then subjected to in vitro SUMO-1 modification assay as previously described [18] in the presence or absence of purified recombinant GST-mel-18 or GST.
Results
Our previous study showed that mel-18 interacts with HSF2 and inhibits its sumoylation by binding to and inhibiting the activity of ubc9 enzymes in the vicinity of HSF2 [15]. The results also suggested that mel-18 inhibits the sumoylation of other cellular proteins. RanGAP1 is a very important cellular SUMO substrate protein, in fact the first identified substrate for SUMO-1 conjugation [16, 17]. Therefore, in the present study we sought to determine whether the sumoylation of RanGAP1 is regulated by mel-18, and if so whether this involves interaction between these two proteins.
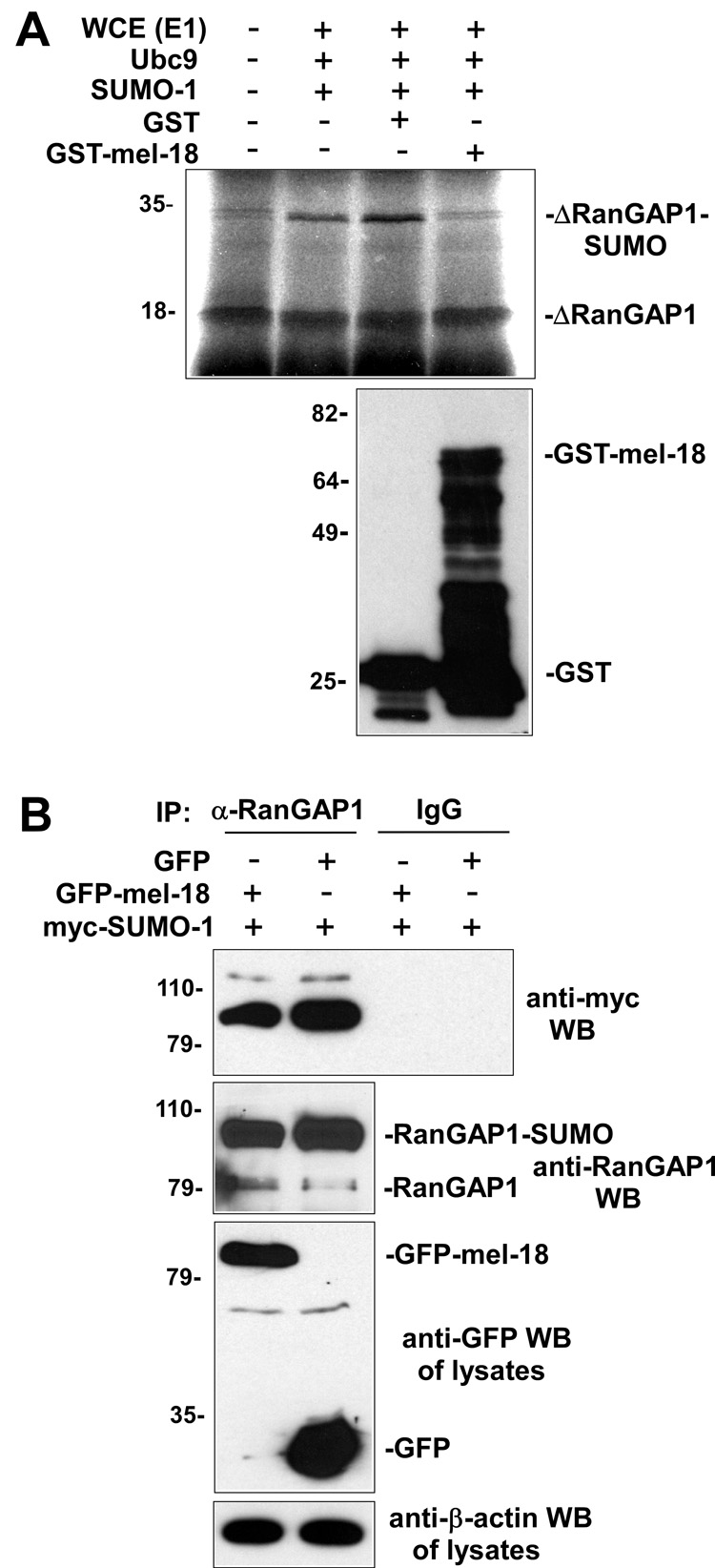
As a first test of this hypothesis, we determined whether adding purified recombinant GST-mel-18 to would affect the SUMO modification of RanGAP1 in an in vitro sumoylation assay. The results of this experiment, shown in Figure 1A, indicate that the addition of purified GST-mel-18 is indeed associated with decreased sumoylation of RanGAP1. Next, we wanted to examine whether mel-18 can inhibit the sumoylation of RanGAP1 expressed in cells. To test this, GFP-mel-18 or GFP expression constructs, along with a myc-SUMO-1 expression plasmid, were transfected into cells and then extracts of these cells were subjected to immunoprecipitation using anti-RanGAP1 antibodies or non-specific IgG (negative control), followed by anti-myc Western blot to detect the sumoylated forms of the RanGAP1 protein. The immunoprecipitates were also subjected to Western blot using anti-RanGAP1 antibodies in order to provide an additional way to visualize the sumoylated form of RanGAP1. The results of this experiment, shown in Figure 1B, indicate that expression of GFP-mel-18, but not GFP, is associated with decreased RanGAP1 sumoylation. The results of the experiments shown in Figure 1 demonstrate the ability of mel-18 to inhibit RanGAP1 sumoylation.
Fig. 1. Mel-18 inhibits RanGAP1 sumoylation.
(A) Purified mel-18 inhibits in vitro sumoylation of RanGAP1. 35S-labeled in vitro translated T7-RanGAP1 Δ419 RanGAP1 fragment was subjected to in vitro sumoylation in the absence of any additional purified proteins (lane 2), or in the presence of purified GST (lane 3) or GST-mel-18 (lane 4). Samples were then analyzed by SDS-PAGE and autoradiography (top panel). The anti-GST Western blot (lower panel) shows the relative amounts of GST-mel-18 or GST that were added to the reactions in lanes 3 and 4 of the top panel, respectively. (B) Mel-18 inhibits sumoylation of RanGAP1 in vivo. HEK 293 cells were transfected with GFP-mel-18 or GFP expression constructs along with myc-SUMO-1 expression plasmid, and then extracts of the cells were subjected to immunoprecipitation with anti-RanGAP1 antibodies or non-specific IgG (negative control), followed by anti-myc Western blot to detect the sumoylated forms of RanGAP1, and to Western blot using anti-RanGAP1 antibodies. The cell lysates were subjected to anti-GFP Western blot to analyze expression levels of GFP-mel-18 and GFP, and to anti-β-actin Western as a loading control
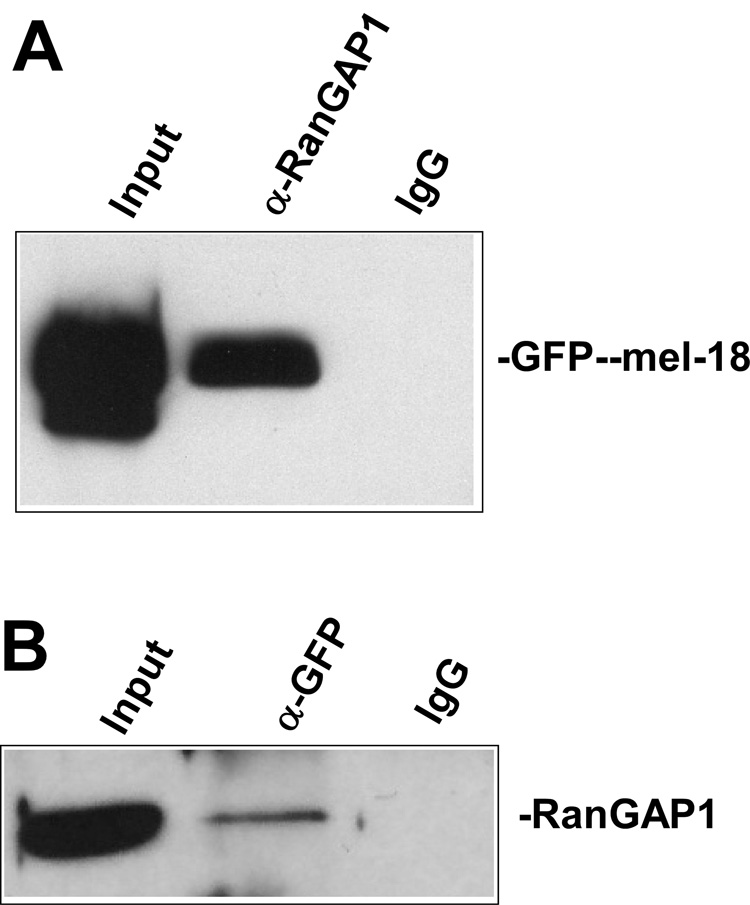
As described above, our previous work demonstrated that mel-18 binds to HSF2 and inhibit its sumoylation [15]. Therefore, we next investigated whether the inhibition of RanGAP1 sumoylation demonstrated by the data in Figure 1 above could also involve interaction between RanGAP1 and mel-18. To examine this, HEK 293 cells were transfected with GFP-mel-18 expression constructs and then extracts of the cells were subjected to immunoprecipitation with anti-RanGAP1 antibodies or non-specific IgG (negative control), followed by anti-GFP antibody Western blot (Fig. 2A). As a complementary approach, we preformed the reverse approach of subjecting extracts of these transfected cells to immunoprecipitation with anti-GFP antibodies or non-specific IgG (negative control), followed by Western blot using anti-RanGAP1 antibodies (Figure 2B). The results of these experiments both indicate that RanGAP1 and mel-18 proteins expressed in cells do interact.
Fig. 2. RanGAP1 interacts with mel-18.
(A) HEK 293 cells were transfected with GFP-mel-18 expression constructs, and then extracts of the cells were subjected to immunoprecipitation with anti-RanGAP1 antibodies or non-specific IgG (negative control), followed by anti-GFP Western blot to detect the interaction between mel-18 and RanGAP1. (B) A similar experiment to that described in panel A was performed, except that here the cell extracts were subjected to immunoprecipitation with anti-GFP antibodies or non-specific IgG (negative control), followed by anti-RanGAP1 Western blot.
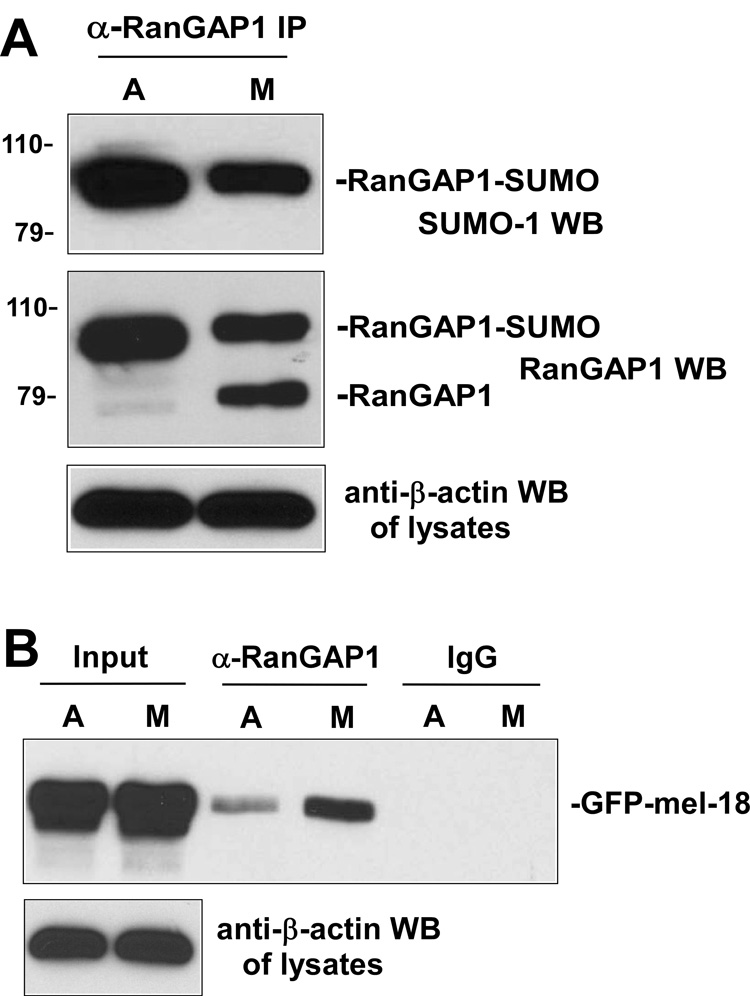
Although it has been described that RanGAP1 remains SUMO modified during mitosis [19], it is still unclear whether the SUMO level is changed during this part of the cell cycle. Our previous finding showed that increased sumoylation of HSF2 in mitosis is coupled with decreased interaction between HSF2 and mel-18, so we hypothesized that RanGAP1 may also exhibit mitotic-dependent regulation of its sumoylation, which might be associated with different interaction level with mel-18 during this stage of the cell cycle. As a first test of this hypothesis, extracts of asynchronous or mitotic HEK 293 cells were immunoprecipitated using anti-RanGAP1 antibodies or non-specific IgG and the immunoprecipitates were subjected to Western blot using anti-SUMO-1 antibodies (top panel) or anti-RanGAP1 antibodies (middle panel) (Figure 3A). The results of this experiment indicate that sumoylation of RanGAP1 is decreased during mitosis.
Fig. 3. Decreased RanGAP1 sumoylation and increased interaction between RanGAP1 and mel-18 during mitosis.
(A) Sumoylation of RanGAP1 decreases during mitosis. Extracts of asynchronous or mitotic HEK 293 cells were immunoprecipitated using anti-RanGAP1 (goat polyclonal) antibodies and the immunoprecipitates subjected to Western blot using anti-SUMO-1 antibodies (top panel) or anti-RanGAP1 antibodies (middle panel). The cell lysates were subjected to anti-β-actin Western blot as a loading control (bottom panel). (B) Interaction between RanGAP1 and mel-18 increases during mitosis. HEK 293 cells were transfected with GFP-mel-18 expression constructs, and then extracts of asynchronous or mitotic transfected cells were immunoprecipitated using anti-RanGAP1 antibodies or non-specific IgG and the immunoprecipitates subjected to Western blot using anti-GFP antibodies (top panel). The cell lysates were subjected to anti-β-actin Western blot as a loading control (bottom panel).
Next, we wanted to determine whether this mitotic-dependent decrease in RanGAP1 sumoylation is associated with increased interaction between mel-18 and RanGAP1 during this part of the cell cycle, as predicted by our hypothesis. To test this, HEK 293 cells were transfected with GFP-mel-18 expression constructs, and then extracts of asynchronous or mitotic transfected HEK 293 cells were immunoprecipitated using anti-RanGAP1 antibodies and the immunoprecipitates subjected to Western blot using anti-GFP antibodies. The results of this experiment indicate that higher levels of interaction between RanGAP1 and mel-18 are indeed observed in extracts of mitotic cells compared to those of asynchronous cells (Figure 3B).
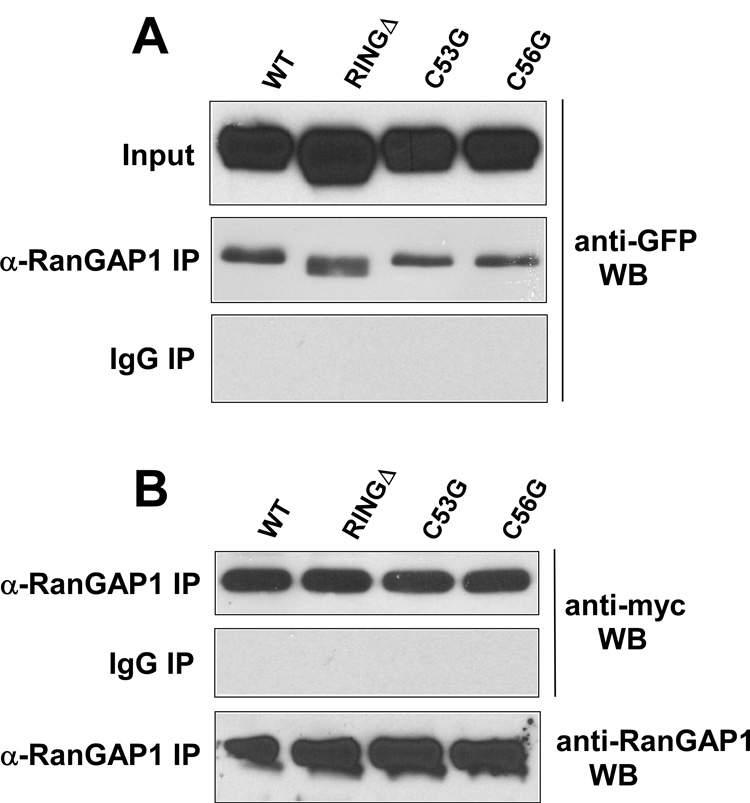
The mel-18 protein contains a RING finger domain in its N-terminal region [20–22]. To test whether this RING domain is important for the ability of mel-18 to interact with RanGAP1 or inhibit RanGAP1 sumoylation, we made mutant GFP-mel-18 plasmids with a RING finger deletion (RINGΔ) or cysteine-to-glycine substitutions at key cysteine residues 53 and 56 of the RING domain (C53G, C56G). First, to test the importance of the RING finger for mel-18 interaction with RanGAP1, HEK 293 cells were transfected with the wild type or mutant (RINGΔ, C53G, or C56G) GFP-mel-18 constructs along with the myc-SUMO-1 expression plasmid, and then extracts of the transfected cells were subjected to immunoprecipitation using anti-RanGAP1 antibodies or non-specific IgG followed by Western blot of the immunoprecipitates with anti-GFP antibodies (Fig. 4A). The results indicate that none of the RING finger mutations appear to affect mel-18 interaction with RanGAP1. To examine their effect on RanGAP1 sumoylation, HEK 293 cells transfected with these constructs along with the myc-SUMO-1 expression plasmid were subjected to immunoprecipitation using anti-RanGAP1 antibodies or non-specific IgG, followed by Western blot of the immunoprecipitates with anti-myc antibodies to detect sumoylated RanGAP1. The results, shown in Figure 4B, indicate that deletion of the RING domain or mutation of cysteines 53 or 56 also has no effect on RanGAP1 sumoylation.
Fig. 4. Ring finger domain of mel-18 is not required for its interaction with RanGAP1 or inhibition of RanGAP1 sumoylation.
(A) HEK 293 cells were transfected with GFP-mel-18 expression constructs (wild type, ring finger deletion, C53G, or C56G mutations), along with the myc-SUMO-1 expression plasmid. Extracts of the transfected cells were immunoprecipitated using anti-RanGAP1 antibodies or non-specific IgG and the immunoprecipitates subjected to Western blot using anti-GFP antibodies (top panel). (B) As in panel A, except that here the anti-RanGAP1 or non-specific IgG immunoprecipitates were subjected to Western blot using anti-myc antibodies to examine the levels of sumoylated RanGAP1. The anti-RanGAP1 immunoprecipitates were also subjected to anti-RanGAP1 Western blot to normalize for levels of RanGAP1.
Discussion
The results presented in this paper show that mel-18 not only acts as an anti-SUMO E3 factor for the HSF2 protein, but for RanGAP1 as well. This finding increases the likelihood that mel-18 regulates the sumoylation of other, as-yet-undiscovered cellular proteins. Indeed, our previous results indicated that there are many sumoylated bands on SDS-PAGE gels whose amounts change significantly in response to changes in mel-18 level, indicating that mel-18 may in fact regulate the sumoylation of a large number of proteins. Thus, one important goal of future studies is to identify these other targets whose SUMO modification is modulated by mel-18. Mel-18 is known to act as a tumor suppressor, and so it would be particularly exciting if it was found to regulate the sumoylation of cellular proteins involved in the control of cell proliferation.
Another intriguing finding of this paper is that the cell cycle-dependence of the interaction between mel-18 and HSF2 vs. RanGAP1 are exactly the opposite of each other: HSF2 interaction with mel-18 decreases during mitosis, resulting in elevated HSF2 sumoylation, in contrast to higher interaction of RanGAP1 and mel-18 and less RanGAP1 sumoylation during this stage of the cell cycle. This suggests that there is a mechanism or mechanisms for differentially controlling mel-18 interaction with partners in a mitosis-dependent manner. Future studies into this area would likely reveal valuable insight into the regulation of the anti-SUMO E3 function of mel-18.
Finally, our results indicate that the conserved RING finger motif of mel-18, found in its N-terminal region [20–22], is not required for its interaction with RanGAP1 or its ability to inhibit RanGAP1 sumoylation. Since the results of our previous study suggested that mel-18 interaction with the SUMO E2 enzyme ubc9 is likely important for the sumoylation-inhibitory function of mel-18 [15], this suggests that other regions of the mel-18 protein are involved in binding RanGAP1 and ubc9. Future studies to identify these regions would reveal important new functional domains of the mel-18 protein.
Acknowledgments
We would like to thank Mike Matunis for the RanGAP1 construct, and to other members of the laboratory for insightful discussions. This research was supported by NIH grant GM64606 to K.D.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- 2.Mukhopadhyay D, Dasso M. Modification in reverse: the SUMO proteases. Trends Biochem Sci. 2007;32:286–295. doi: 10.1016/j.tibs.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 3.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 4.Gill G. Something about SUMO inhibits transcription. Curr Opin Genet Dev. 2005;15:536–541. doi: 10.1016/j.gde.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 5.Hay RT. SUMO: a history of modification. Mol Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 6.Desterro JM, Thomson J, Hay RT. Ubch9 conjugates SUMO but not ubiquitin. FEBS Lett. 1997;417:297–300. doi: 10.1016/s0014-5793(97)01305-7. [DOI] [PubMed] [Google Scholar]
- 7.Johnson ES, Blobel G. Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J Biol Chem. 1997;272:26799–26802. doi: 10.1074/jbc.272.43.26799. [DOI] [PubMed] [Google Scholar]
- 8.Rodriguez MS, Dargemont C, Hay RT. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J Biol Chem. 2001;276:12654–12659. doi: 10.1074/jbc.M009476200. [DOI] [PubMed] [Google Scholar]
- 9.Sampson DA, Wang M, Matunis MJ. The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J Biol Chem. 2001;276:21664–21669. doi: 10.1074/jbc.M100006200. [DOI] [PubMed] [Google Scholar]
- 10.Johnson ES, Gupta AA. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell. 2001;106:735–744. doi: 10.1016/s0092-8674(01)00491-3. [DOI] [PubMed] [Google Scholar]
- 11.Takahashi Y, Kahyo T, Toh EA, Yasuda H, Kikuchi Y. Yeast Ull1/Siz1 is a novel SUMO1/Smt3 ligase for septin components and functions as an adaptor between conjugating enzyme and substrates. J Biol Chem. 2001;276:48973–48977. doi: 10.1074/jbc.M109295200. [DOI] [PubMed] [Google Scholar]
- 12.Kahyo T, Nishida T, Yasuda H. Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol Cell. 2001;8:713–718. doi: 10.1016/s1097-2765(01)00349-5. [DOI] [PubMed] [Google Scholar]
- 13.Pichler A, Gast A, Seeler JS, Dejean A, Melchior F. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell. 2002;108:109–120. doi: 10.1016/s0092-8674(01)00633-x. [DOI] [PubMed] [Google Scholar]
- 14.Kagey MH, Melhuish TA, Wotton D. The polycomb protein Pc2 is a SUMO E3. Cell. 2003;113:127–137. doi: 10.1016/s0092-8674(03)00159-4. [DOI] [PubMed] [Google Scholar]
- 15.Zhang J, Goodson ML, Hong Y, Sarge KD. MEL-18 interacts with HSF2 and the SUMO E2 UBC9 to inhibit HSF2 sumoylation. J Biol Chem. 2008;283:7464–7499. doi: 10.1074/jbc.M707122200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- 17.Matunis MJ, Wu J, Blobel G. SUMO-1 modification and its role in targeting the Ran GTPase-activating protein, RanGAP1, to the nuclear pore complex. J Cell Biol. 1998;140:499–509. doi: 10.1083/jcb.140.3.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hilgarth RS, Sarge KD. Analysis of protein sumoylation. Curr Protoc Protein Sci Chapter. 2006;14 doi: 10.1002/0471140864.ps1408s44. Unit 14 8. [DOI] [PubMed] [Google Scholar]
- 19.Joseph J, Tan SH, Karpova TS, McNally JG, Dasso M. SUMO-1 targets RanGAP1 to kinetochores and mitotic spindles. J Cell Biol. 2002;156:595–602. doi: 10.1083/jcb.200110109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tagawa M, Sakamoto T, Shigemoto K, Matsubara H, Tamura Y, Ito T, Nakamura I, Okitsu A, Imai K, Taniguchi M. Expression of novel DNA-binding protein with zinc finger structure in various tumor cells. J Biol Chem. 1990;265:20021–20026. [PubMed] [Google Scholar]
- 21.Asano H, Ishida A, Hasegawa M, Ono T, Yoshida MC, Taniguchi M, Kanno M. The mouse Mel-18 "RING-finger" gene: genomic organization, promoter analysis and chromosomal assignment. DNA Seq. 1993;3:369–377. doi: 10.3109/10425179309020838. [DOI] [PubMed] [Google Scholar]
- 22.Ishida A, Asano H, Hasegawa M, Koseki H, Ono T, Yoshida MC, Taniguchi M, Kanno M. Cloning and chromosome mapping of the human Mel-18 gene which encodes a DNA-binding protein with a new 'RING-finger' motif. Gene. 1993;129:249–255. doi: 10.1016/0378-1119(93)90275-8. [DOI] [PubMed] [Google Scholar]