Abstract
Electron transfer dissociation (ETD) has become increasingly used in proteomic analyses due to its complementarity to collision-activated dissociation (CAD) and its ability to sequence peptides with post-translation modifications (PTMs). It was previously unknown, however, whether ETD would be compatible with a commonly employed quantification technique, isobaric tags for relative and absolute quantification (iTRAQ), since the fragmentation mechanisms and pathways of ETD differ significantly from CAD. We demonstrate here that ETD of iTRAQ labeled peptides produces c- and z•-type fragment ions as well as reporter ions that are unique from those produced by CAD. Exact molecular formulas of product ions were determined by ETD fragmentation of iTRAQ-labeled synthetic peptides followed by high mass accuracy orbitrap mass analysis. These experiments revealed that ETD cleavage of the N – Cα bond of the iTRAQ tag results in fragment ions that could be used for quantification. Synthetic peptide work demonstrates that these fragment ions provide up to three channels of quantification and that the quality is similar to that provided by beam-type CAD. Protein standards were used to evaluate peptide and protein quantification of iTRAQ labeling in conjunction with ETD, beam-type CAD, and pulsed Q dissociation (PQD) on a hybrid ion trap-orbitrap mass spectrometer. For reporter ion intensities above a certain threshold all three strategies provided reliable peptide quantification (average error <10%). Approximately 36%, 8%, and 16% of scans identified fall below this threshold for ETD, HCD, and PQD respectively. At the protein level, average errors were 2.3%, 1.7%, and 3.6% for ETD, HCD, and PQD respectively.
Protein quantification has become an important and, in many cases, critical component of modern mass spectrometry-based proteomic research[1-9]. Over the past decade, numerous quantification strategies have evolved – nearly all of them rely on the incorporation of stable isotopes for subsequent mass spectrometric sorting and relative quantification[10-19]. Time and method of isotope integration distinguish these approaches. Whether introduced metabolically through heavy amino acids or chemically with differentially labeled tags at the peptide or protein level, mixing of the light and heavy (e.g., control and treated) peptides results in co-eluting peptide pairs with subtle, but measurably, different masses[10-12, 14-16, 20, 21].
In a clever departure from this paradigm, Pappin et al. described the concept of amine-reactive isobaric tagging[13]. Here differentially isotopically labeled, but isobaric amine-reactive tags (up to four) are embedded into peptides from as many as four separate peptide pools (e.g., control and three treatment time points). Once labeled, the four samples are combined and peptides are sequenced individually by tandem mass spectrometry using collision-based dissociation methods (i.e., beam-type collision-activated dissociation (CAD) or pulsed Q dissociation (PQD)). Identical peptides arising from each of the four samples co-elute and have equivalent m/z values. During MS/MS, however, vibrational excitation induces cleavage of both the peptide backbone and the isobaric tag. Dissociation of the backbone gives rise to fragment ions characteristic of the peptide sequence; dissociation of the tag generates low mass product ions where each of the four labels creates a unique m/z reporter peak. Because it allows for the simultaneous quantification of up to four samples, iTRAQ has become an important and powerful protein quantification methodology.
Due to the loss of low mass ions during resonant excitation (low mass cutoff), the use of iTRAQ labeling in conjunction with ion trap and ion trap hybrid mass spectrometers has been limited. That is, ion trap CAD, often results in the inability to detect iTRAQ reporter ions due to the loss of low mass ions during precursor fragmentation[22]. However, the rapid scanning, excellent sensitivity, and ability to couple with other analyzers such as the orbitrap and Fourier transform ion cyclotron resonance (FT-ICR) have made ion traps among the most useful devices for protein and peptide identification[23]. Beam-type CAD is now available on hybrid ion trap-orbitrap mass spectrometers, but these systems only permit detection of product ions in the orbitrap mass analyzer which is inherently slower and less sensitive that ion trap mass analysis[24]. PQD, a form of ion trap CAD designed to eliminate low mass cutoff, does allow for detection of low mass-to-charge fragment ions and is available on a some ion trap and ion trap hybrid mass spectrometers[25]. Griffin et al. have demonstrated that PQD is compatible with iTRAQ labeling and have characterized the quantitative merits of this approach[26, 27].
Electron transfer dissociation is complementary to CAD and can be especially useful for sequencing peptides containing post-translational modification (PTM).[28-36] ETD allows for rapid peptide sequencing, with speeds similar to ion trap CAD, but is operated such that ions of mass lower than 100 m/z are detected regardless of precursor m/z (i.e., no low mass cutoff). Work by us and others demonstrate that ion trap CAD and ETD are complementary; however, ETD, and other electron-based dissociation methods, rely on free radical-initiated peptide backbone cleavage and hence are not obviously compatible with the iTRAQ tagging strategy. Here we demonstrate that ETD produces iTRAQ reporter ions, unique from those produced by CAD, and that these reporters allow for peptide quantification of up to three different samples. Fragmentation of iTRAQ labeled peptides with ETD results in c- and z•-type fragment ions and two fragment ions resulting from cleavage of the iTRAQ tag. One of the cleavages results in reporter ions that allow for quantitative comparison of up to three different samples. Synthetic peptides as well as digests of protein standards were used to evaluate the quality of iTRAQ based quantification in conjunction with ETD in an ETD enabled hybrid linear ion trap-orbitrap mass spectrometer. Peptide and protein quantification was compared using ETD, PQD, and beam-type CAD (HCD).
Materials and Methods
Sample Preparation
Synthetic peptides were obtained at the University of Wisconsin-Madison Biotechnology Center, standard proteins were purchased from Sigma-Adrich (St. Louis, MO), and the iTRAQ labeling reagent was purchased from Applied Biosystems (Foster City, CA). Bovine serum albumin, beta-casein, horse cytochrome C, beta-lactoglobin, rabbit phosphorylase B, and carbonic anhydrase were reduced, alkylated, and digested as previously described[37]. iTRAQ labeling was performed according to the manufacturer supplied protocol in approximately 70% ethanol and 0.15 M triethylammonium bicarbonate at room temperature for 1 hour. Samples were subsequently mixed, desalted using solid phase extraction, dried to completion, and resuspended in 100 mM acetic acid prior to LC-MS/MS analysis.
Liquid Chromatography and Mass Spectrometry
Synthetic peptides were resuspended in 30% acetonitrile with 100 mM acetic acid and infused in the mass spectrometer via static nanospray Econotips (New Objective, Woburn MA). The six protein digest was separated on-line using nanoflow reversed phase high performance liquid chromatography (nRP-HPLC) as previously described[38]. Briefly, the sample was bomb loaded onto a 5 cm × 75 μm ID precolumn packed with 5 μm C18 reversed-phase packing material (Alltech, Nicholasville, KY). This precolumn was then butt connected to a 7 cm × 50 μm ID analytical column with Teflon® tubing. The sample was eluted into the mass spectrometer using a 60 minute linear gradient from 100 mM acetic acid to 100 mM acetic acid 70% acetonitrile at a flow rate of approximately 60 nL/min.
ETD reactions and mass analysis were carried out in a hybrid linear ion trap-orbitrap mass spectrometer (Orbitrap, Thermo Fisher Scientific, Bremen, Germany) which was modified as previously described to perform ETD reactions[39]. A negative chemical ionization (NCI) source was fitted to the back of the instrument and connected to the back of the C-trap via a multipole. The linear ion trap was modified to enable charge-sign independent trapping (CSIT). Radical fluoranthene ions are generated in the NCI source and transported down the added multipole, through the C-trap and second multipole, and finally into the linear ion trap where ETD reactions proceed exactly as they would in a commercially available linear ion trap mass spectrometer. After fragmentation, product ions can either be analyzed by the linear ion trap or sent to the orbitrap mass analyzer for high mass accuracy detection. All ETD reactions were performed for 85 ms. Precursor cation target values of 40,000 for ion trap mass analysis and 300,000 for orbitrap mass analysis were used. PQD and HCD collision energies were optimized on iTRAQ labeled synthetic peptides prior to LC-MS/MS runs. Normalized collision energies of 45 and 31 were used for HCD and PQD respectively. Spectra from infused samples were averaged for 100 scans. LC-MS/MS experiments comprised of 10 scan events; an MS1 scan with orbitrap mass analysis followed by HCD (beam-type CAD in the collision cell followed by orbitrap mass analysis), PQD, and ETD of the 3 most abundant precursors. Fragment ions generated by PQD and ETD were analyzed in the ion trap mass analyzer while those produced by HCD were detected in the orbitrap mass analyzer.
Data Analysis
MS2 spectra were searched using OMSSA (Open Mass Spectrometry Search Algorithm)[40]. The database searched consisted of the six proteins sequences for our standard peptides as well as a reversed human IPI database which allowed false positive rate filtering[41, 42]. To limit false positive identifications, the results were filtered by precursor mass error and OMSSA e-value such that no reversed database entries were included. Software was written in-house to extract quantitative information from the .dta files. For each scan, peak intensities within +/- 0.5 daltons (0.01 for HCD scans) of expected reporter ion m/z ratios were summed. The software also grouped peptides identifications into protein identifications, calculated protein ratios by averaging peptide ratios, and provided standard deviation calculations for each protein. As shown in Figure 2, each channel was extremely pure so no corrections were made for isotopic purity.
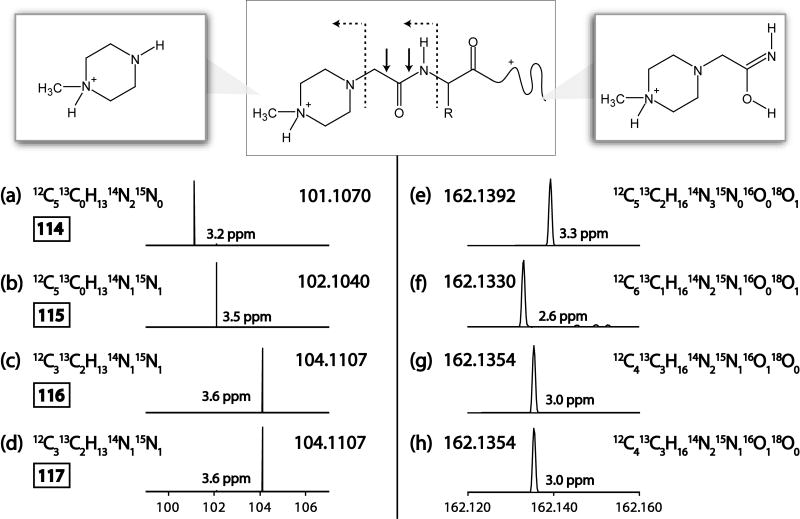
Figure 2.
Proposed structures of ETD-generated iTRAQ reporter ions. The structure of an iTRAQ labeling reagent as attached to a peptide is shown (Top middle). Bonds cleaved by CAD are indicated by solid arrows. Bonds cleaved by ETD are indicated by dotted arrows. Possible structures for cleavage products are shown in the top left and top right. Panels A-H show mass spectrum of cleavage products generated by ETD of iTRAQ labeled peptides. Measured masses and error in ppm are shown.
Synthetic peptide impurities and unequal sample loss during preparation required normalization. To accomplish this the synthetic peptide SSAAKAAAK was labeled with three iTRAQ tags (114, 115, and 116), mixed in 1:1:1 ratio, and infused into a hybrid linear ion trap-orbitrap mass spectrometer. The precursor population was then fragmented by beam-type CAD in the collision cell and analyzed in the orbitrap mass analyzer. The relative peak heights observed for the 114, 115, and 116 peaks were used to normalize all subsequent ETD and HCD scans of labeled synthetic peptide mixtures. No normalization was performed on the quantitative data from LC-MS/MS analyses.
Results
Fragmentation pathways
To determine the effect of iTRAQ labeling upon ETD fragmentation a synthetic peptide with the sequence HAAAHAAAH, no joke, was labeled with each of the iTRAQ tags (i.e. 114, 115, 116, and 117). Peptides from each group were separately ionized via nano electrospray (infusion) and sampled by an ETD-enabled linear ion trap-orbitrap hybrid mass spectrometer. Following the ETD reaction the product ions were injected into the orbitrap for m/z analysis. Orbitrap mass analysis revealed that numerous c- and z•-type ions were generated. In this case, only the N-terminus of the peptide contains an isobaric tag; thus, the entire c-type product ion series is increased by the exact mass of the intact isobaric tag (144.1059, 144.0996, or 144.1021 Da depending on the tag used). Further, we find no cases wherein c-type ions have lost the isobaric tag.
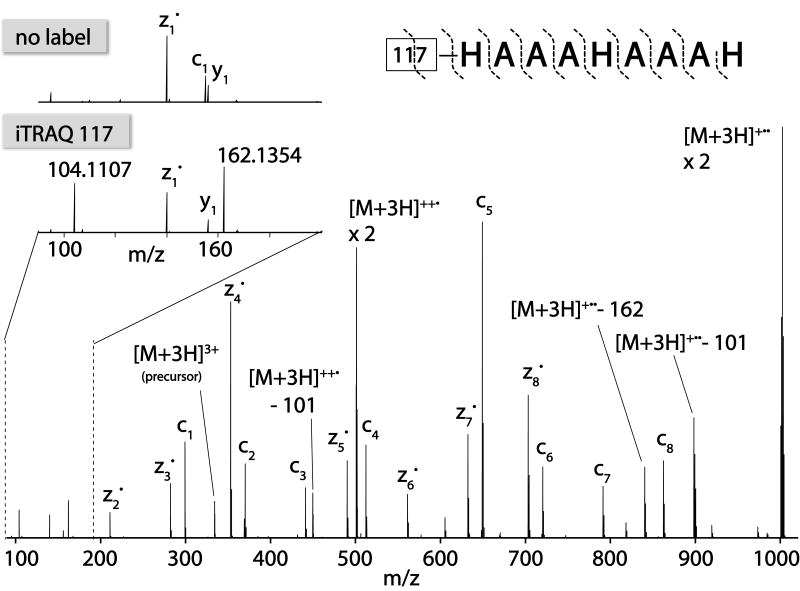
A comparison of the low m/z region produced following ETD tandem MS of unlabeled and labeled (117) precursors, however, reveals the presence of two new m/z peaks – 104.1107 and 162.1354 (Figure 1). Note CAD cleavage of the peptide generates a reporter tag having a nominal m/z value of 117 and this peak was not observed in the ETD spectrum. To identify the composition of the ETD-generated low m/z peaks we examined these mass spectral regions from the product ion spectra of each iTRAQ tagged peptide (e.g., 114, 115, 116, and 117). Panels A – D of Figure 2 display the lower m/z iTRAQ-specific ion from each of the iTRAQ tagged species. From the accurate masses of these respective peaks we deduced the best-fit molecular formulas for each: 101.1073 - 12C514N2H13; 102.1044 - 12C515N114N1H13; 104.1110 - 13C212C315N114N1H13. Although two molecular formulas were possible for the latter two of these masses within an error of 5 ppm, only one for each peak could be explained by the known structure of the iTRAQ tagging reagent. The theoretical m/z values of each of these formulas fits the measured m/z value to within 4 ppm and is consistent with cleavage of the N-methylpiperazine reporter region between the N – Cα of the iTRAQ tag. Note the intended CAD cleavage site of the reporter group is one methylene group downstream (i.e., between the carbonyl C and Cα). High mass accuracy measurements of the 162 peak were consistent with cleavage between the N – Cα of the first amino acid (panels E-H of Figure 2).
Figure 1.
ETD MS/MS spectra with orbitrap mass analysis of synthetic peptide HAAAHAAAH labeled with the 117 iTRAQ tag. The pullout in the upper left depicts the low mass region observed upon ETD MS/MS of unlabeled HAAAAHAAAH. The lower frame is the same reaction but with HAAAAHAAAH labeled with the iTRAQ 117 tag.
From these data we can identify where the heavy atoms are located in the iTRAQ tag. The 114 tag contains two 13C atoms (at the Cα and the carbonyl) and an 18O; 115 incorporates an 15N within the piperazine ring system, a 13C at the Cα, and an 18O; 116 and 117 each have two 13C atoms and one 15N within the piperazine ring system – they differ only in the placement of a third 13C atom – 116 places it at the carbonyl C while 117 moves it to the Cα. Because ETD cleaves the N – Cα bond product ion spectra from the 116 and 117 tags generate identical reporter peaks, 104.1107 m/z. Therefore, only three unique reporter ions are available for relative quantification. Our results suggest slightly different chemical compositions from those originally reported by Pappin et al.[13]. However, our results agree with the chemical compositions provided in the manufacturer supplied reference guide.
ETD-generated reporter ions are quantitative
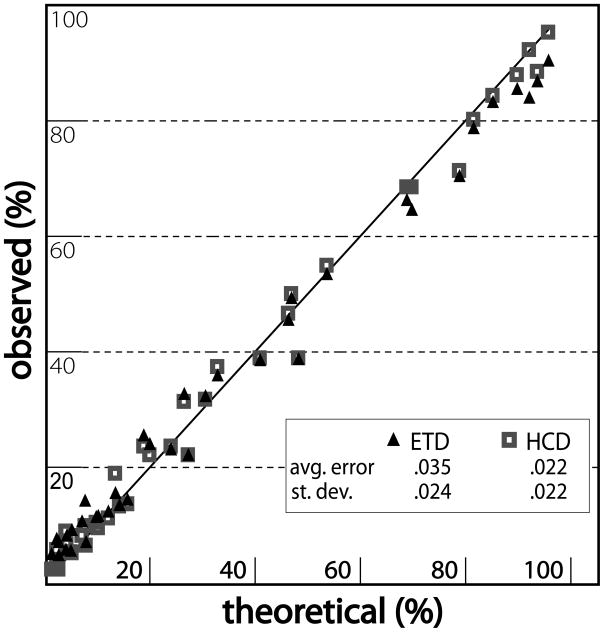
To determine the quantitative capability of these ETD-generated reporter ions, a synthetic peptide SSAAKAAAK was labeled with three different iTRAQ tags (114, 115, and 116). Samples were mixed in known ratios, infused into an ETD-enabled orbitrap mass spectrometer, fragmented by ETD, and analyzed using the linear ion trap mass analyzer. For comparison to beam-type CAD, each mixture was also fragmented by HCD with product ion m/z detection performed in the orbitrap mass analyzer. Peptide ratios were calculated as previously described based on relative peaks areas of the reporter ions using the following formula; area(reporter ion of interest)/(area(all reporter ions))[26]. Thus all values fall between 0 and 1 with a 1:1:1 ratio ∼ 0.33 for all three reporter ions. Relative peak areas of the 101, 102, and 104 reporter ions corresponded well with peptide abundance for ratios ranging from 1:1 to 1:68. Figure 3 depicts observed versus expected percentage of reporter ion intensity. Least squares fit yielded an equation of y=0.8973x + 0.0342 (R2 =0.99). This compares favorably with our beam-type CAD control; y=0.9588x + 0.0137 (R2 =0.99) (See Figure 3).
Figure 3.
Theoretical versus observed ratios for both HCD and ETD fragmentation of an iTRAQ labeled peptide standard. Samples of labeled SSAAKAAAK were mixed in ratios ranging from 1:1 to 1:68. Ratios after ETD fragmentation and ion trap mass analysis are depicted as solid triangles. Ratios after beam-type CAD (HCD) fragmentation and orbitrap mass analysis are depicted as squares.
Comparison of protein qunatification using ETD, PQD, and HCD
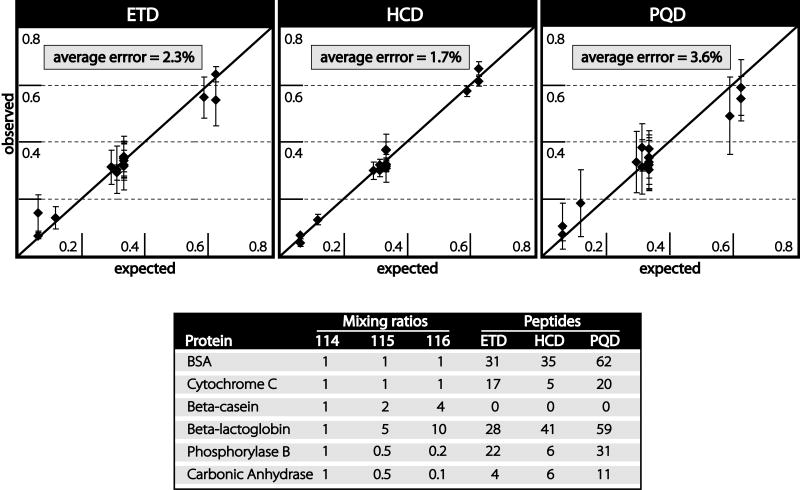
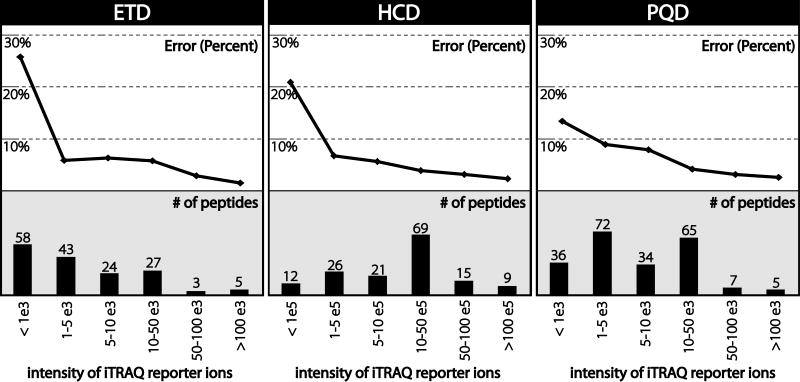
Quality of protein level quantification was determined using protein standards. Six protein standards were digested by trypsin, labeled with the same three iTRAQ labels (114, 115, and 116), mixed in known ratios, and analyzed via LC-MS/MS with consecutive HCD, ETD, and PQD scans. Mixing ratios (summarized in Figure 5) are; BSA 1:1:1, carbonic anhydrase 1:0.5:0.1, beta-lactoglobin 1:5:10, beta-casein 1:2:4, phosphorylase B 1:0.5:0.2, and cytochrome C 1:1:1. After peptide identification by database searching, average absolute error (absolute value of expected – observed) was compared to the sum total intensity of iTRAQ reporter ions in that scan. Figure 4 shows the number of scans that produced confident peptide identifications as a function of sum total reporter ion intensity. Scans that produced confident peptide identifications were binned according to the sum of all three reporter ion intensities. The bar graphs indicate how many scans fall into each bin while the line graphs above indicate the average quantitative error. For all scan types, average error decreased as reporter intensity increased. For ion trap scans (ETD and PQD), scans that exhibited reporter ion intensities summing to less than 1,000 exhibited poor quantification; absolute error = 26% for ETD, and 14% for PQD. For HCD scans with orbitrap detection scans with reporter ion intensities summing to less than 100,000 resulted in an error of 21%. However, above these values all three scan types resulted in average errors of less than 10%. Thus, each mass analyzer appears to have a threshold intensity for reporter ions below which quantitative results are unreliable.
Figure 5.
Protein level quantification using iTRAQ labeling in conjunction with ETD, PQD, and HCD. (top) Each diamond denotes the ratio of one iTRAQ channel averaged across all identified peptides corresponding to one protein. Error bars indicate standard deviation. (bottom) The table describes the ratios in which the proteins were mixed as well as the number of peptides used to quantify each protein. Note, although beta-casein was identified by all fragmentation techniques, none of the peptides exhibited reporter ion ratios above the minimum threshold required for accurate quantification.
Figure 4.
Quantitative error as a function of total reporter ion intensity. Scans were binned according to the sum of all three reporter ion intensities. Bar graphs depict the number of scans that fall into each bin. The line graph above depicts the average error for each bin. All three fragmentation methods exhibit high error (>10%) below a certain intensity threshold; however, ETD generated a large number of scans that fall below this threshold.
Interestingly, approximately 36% of all ETD scans that produced a confident identification exhibited low reporter ion intensities compared to only 8% of HCD scans and 16% of PQD scans. One possible explanation for the high proportion of scans that exhibit low intensity reporter ion populations is that ETD does not result in preferential cleavage of the N-Cα bond of the iTRAQ tag. CAD is known to preferentially cleave the weakest bonds in the peptide backbone. iTRAQ tagging reagents were designed specifically so that CAD would preferentially cleave this bond producing very intense reporter ion peaks. ETD cleaves randomly along the peptide backbone and is largely unaffected by amino acid composition and/or chemical modification. Therefore, ETD does not always produce intense iTRAQ reporter ions.
For the evaluation of protein quantification, only scans with sum total reporter ion intensities of 1,000 or greater were included for ion trap measurements and 100,000 for spectra acquired in the orbitrap. The observed ratios were averaged across all scans for each of the proteins identified. Figure 5 depicts expected versus observed ratios with standard deviations shown as error bars. The average difference between expected and observed was similar for all three dissociation methods (ETD = 2.2%, PQD = 3.6%, and HCD = 1.7%). This compares favorably with the < 6% error originally reported for protein level quantification using iTRAQ labeling and beam-type CAD fragmentation[13].
ETD is limited by poor fragmentation efficiency of doubly charged peptides as well as peptides with high amino acid-to-charge ratios[30, 37]. These caveats can be overcome by the use of supplemental activation of the non-dissociated electron transfer products[34, 37]. However, supplemental activation, much like CAD, results in the loss of low mass ions and is therefore incompatible with iTRAQ quantification. More tractable approaches include the use different enzymes (e.g. Lys-C) or modified digestion conditions (incomplete tryptic digests) that result in more highly charged precursors. Chemical approaches may also be used to increase precursor charge states. Recently Kjeldsen et al. have demonstrated that addition of m-nitrobenzl alcohol to the liquid chromatography mobile phase increases the average charge states of precursors[32]. The more highly charged precursors exhibited increased ETD fragmentation efficiency. Introduction of basic moieties to the peptide through chemical modification prior to mass spectrometry may also provide enhancement of ETD fragmentation efficiency. And though we have not yet measured it in depth, the addition of the basic iTRAQ group appears to broadly elevate peptide charge. This could provide an added benefit for ETD fragmentation and is a subject of current investigation in our laboratory.
Conclusions
We have demonstrated that ETD of iTRAQ labeled peptides produces c- and z•-type fragment ions and generates unique reporter ions that allow for peptide and protein quantification. Since the 116 and 117 tags produce the same reporter ion after fragmentation by ETD, the user is limited to only three channels of relative quantification. Synthetic peptides were used to evaluate the quality of quantification provided by ETD fragmentation of iTRAQ labeled peptides and the results are comparable with those published using beam-type CAD as well as with our own beam-type CAD controls. Furthermore, we have compared protein quantification using iTRAQ and three different fragmentation strategies. All strategies provided reliable quantitative information when reporter ions exceeded a certain intensity threshold. However, compared to HCD and PQD, a high percentage (∼36%) of the scans identified by ETD exhibited low reporter ion intensities. Taking efforts to increase precursor charge states may help reduce the percentages of identified peptide that exhibit low intensity iTRAQ reporter ions.
A major advantage of this approach is that it enables multiplexed quantification to be performed on all mass spectrometers capable of ETD, regardless of their ability to perform PQD or beam-type CAD. Furthermore, iTRAQ labeling can now be used in conjunction with ETD for the quantitative analysis of phosphorylated peptides and other post-translationally modified peptides that can be difficult to sequence by CAD. Lastly, decision tree based mass spectrometry approaches have recently been shown to enhance peptide and protein identification[43]. By combining iTRAQ labeling with ETD and either PQD or beam-type CAD it may be possible to maximize peptide identifications while retaining the ability to perform multiplexed quantification.
A version of iTRAQ has recently been released that allows for up to eight channels of quantification. Since the reporter ion structure remains the same it seems likely that ETD will be compatible with these reagents. Further experiments should confirm this supposition and determine how many channels of quantification it can provide.
Acknowledgments
We thank Graeme McAlister for instrument support. The University of Wisconsin-Madison, Thermo Scientific, the Beckman Foundation, the American Society of Mass Spectrometry, Eli Lilly, the National Science Foundation (0701846; 0747990 both to JJC), and the NIH (1R01GM080148 to JJC) provided financial support for this work. DP gratefully acknowledges support from NIH pre-doctoral fellowship – (the Genomic Sciences Training Program, NIH 5T32HG002760).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ong SE, Foster LJ, Mann M. Mass spectrometric-based approaches in quantitative proteomics. Methods. 2003;29:124–130. doi: 10.1016/s1046-2023(02)00303-1. [DOI] [PubMed] [Google Scholar]
- 2.Bantscheff M, Schirle M, Sweetman G, Rick J, Kuster B. Quantitative mass spectrometry in proteomics: a critical review. Analytical and Bioanalytical Chemistry. 2007;389:1017–1031. doi: 10.1007/s00216-007-1486-6. [DOI] [PubMed] [Google Scholar]
- 3.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 4.Heck AJR, Krijgsveld J. Mass spectrometry-based quantitative proteomics. Expert Review of Proteomics. 2004;1:317–326. doi: 10.1586/14789450.1.3.317. [DOI] [PubMed] [Google Scholar]
- 5.Righetti PG, Campostrini N, Pascali J, Hamdan M, Astner H. Quantitative proteomics: a review of different methodologies. European Journal of Mass Spectrometry. 2004;10:335–348. doi: 10.1255/ejms.600. [DOI] [PubMed] [Google Scholar]
- 6.Ong SE, Mann M. Mass spectrometry-based proteomics turns quantitative. Nature Chemical Biology. 2005;1:252–262. doi: 10.1038/nchembio736. [DOI] [PubMed] [Google Scholar]
- 7.Julka S, Regnier F. Quantification in proteomics through stable isotope coding: A review. Journal of Proteome Research. 2004;3:350–363. doi: 10.1021/pr0340734. [DOI] [PubMed] [Google Scholar]
- 8.Leitner A, Lindner W. Chemistry meets proteomics: The use of chemical tagging reactions for MS-based proteomics. Proteomics. 2006;6:5418–5434. doi: 10.1002/pmic.200600255. [DOI] [PubMed] [Google Scholar]
- 9.Conrads TP, Issaq HJ, Veenstra TD. New tools for quantitative phosphoproteome analysis. Biochemical and Biophysical Research Communications. 2002;290:885–890. doi: 10.1006/bbrc.2001.6275. [DOI] [PubMed] [Google Scholar]
- 10.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Molecular & Cellular Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 11.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nature Biotechnology. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 12.Oda Y, Huang K, Cross FR, Cowburn D, Chait BT. Accurate quantitation of protein expression and site-specific phosphorylation. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:6591–6596. doi: 10.1073/pnas.96.12.6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ross PL, Huang YLN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Molecular & Cellular Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Goshe MB, Conrads TP, Panisko EA, Angell NH, Veenstra TD, Smith RD. Phosphoprotein isotope-coded affinity tag approach for isolating and quantitating phosphopeptides in proteome-wide analyses. Analytical Chemistry. 2001;73:2578–2586. doi: 10.1021/ac010081x. [DOI] [PubMed] [Google Scholar]
- 15.Mirgorodskaya OA, Kozmin YP, Titov MI, Korner R, Sonksen CP, Roepstorff P. Quantitation of peptides and proteins by matrix-assisted laser desorption/ionization mass spectrometry using O-18-labeled internal standards. Rapid Communications in Mass Spectrometry. 2000;14:1226–1232. doi: 10.1002/1097-0231(20000730)14:14<1226::AID-RCM14>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 16.Yao XD, Freas A, Ramirez J, Demirev PA, Fenselau C. Proteolytic O-18 labeling for comparative proteomics: Model studies with two serotypes of adenovirus. Analytical Chemistry. 2001;73:2836–2842. doi: 10.1021/ac001404c. [DOI] [PubMed] [Google Scholar]
- 17.Pasa-Tolic L, Jensen PK, Anderson GA, Lipton MS, Peden KK, Martinovic S, Tolic N, Bruce JE, Smith RD. High throughput proteome-wide precision measurements of protein expression using mass spectrometry. Journal of the American Chemical Society. 1999;121:7949–7950. [Google Scholar]
- 18.Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, Rappsilber J, Mann M. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Molecular & Cellular Proteomics. 2005;4:1265–1272. doi: 10.1074/mcp.M500061-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Zhou HL, Ranish JA, Watts JD, Aebersold R. Quantitative proteome analysis by solid-phase isotope tagging and mass spectrometry. Nature Biotechnology. 2002;20:512–515. doi: 10.1038/nbt0502-512. [DOI] [PubMed] [Google Scholar]
- 20.Faca V, Coram M, Phanstiel D, Glukhova V, Zhang Q, Fitzgibbon M, McIntosh M, Hanash S. Quantitative analysis of acrylamide labeled serum proteins by LC-MS/MS. Journal of Proteome Research. 2006;5:2009–2018. doi: 10.1021/pr060102+. [DOI] [PubMed] [Google Scholar]
- 21.Wu CC, MacCoss MJ, Howell KE, Matthews DE, Yates JR. Metabolic labeling of mammalian organisms with stable isotopes for quantitative proteomic analysis. Analytical Chemistry. 2004;76:4951–4959. doi: 10.1021/ac049208j. [DOI] [PubMed] [Google Scholar]
- 22.Schwartz JC, Senko MW, Syka JEP. A two-dimensional quadrupole ion trap mass spectrometer. Journal of the American Society for Mass Spectrometry. 2002;13:659–669. doi: 10.1016/S1044-0305(02)00384-7. [DOI] [PubMed] [Google Scholar]
- 23.Domon B, Aebersold R. Review - Mass spectrometry and protein analysis. Science. 2006;312:212–217. doi: 10.1126/science.1124619. [DOI] [PubMed] [Google Scholar]
- 24.Olsen JV, Macek B, Lange O, Makarov A, Horning S, Mann M. Higher-energy C-trap dissociation for peptide modification analysis. Nature Methods. 2007;4:709–712. doi: 10.1038/nmeth1060. [DOI] [PubMed] [Google Scholar]
- 25.Schwartz JC, Syka JP, Quarmby ST. Improving the Fundamentals of Msn on 2D Ion Traps: New Ion Activation and Isolation Techniques. 53rd ASMS Conference on Mass Spectrometry; San Antonio, Texas. 2005. [Google Scholar]
- 26.Griffin TJ, Xie HW, Bandhakavi S, Popko J, Mohan A, Carlis JV, Higgins L. iTRAQ reagent-based quantitative proteomic analysis on a linear ion trap mass spectrometer. Journal of Proteome Research. 2007;6:4200–4209. doi: 10.1021/pr070291b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meany DL, Xie HW, Thompson LV, Arriaga EA, Griffin TJ. Identification of carbonylated proteins from enriched rat skeletal muscle mitochondria using affinity chromatography-stable isotope labeling and tandem mass spectrometry. Proteomics. 2007;7:1150–1163. doi: 10.1002/pmic.200600450. [DOI] [PubMed] [Google Scholar]
- 28.Chi A, Huttenhower C, Geer LY, Coon JJ, Syka JEP, Bai DL, Shabanowitz J, Burke DJ, Troyanskaya OG, Hunt DF. Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:2193–2198. doi: 10.1073/pnas.0607084104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Syka JEP, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Good DM, Wirtala M, McAlister GC, Coon JJ. Performance characteristics of electron transfer dissociation mass spectrometry. Molecular & Cellular Proteomics. 2007;6:1942–1951. doi: 10.1074/mcp.M700073-MCP200. [DOI] [PubMed] [Google Scholar]
- 31.Molina H, Horn DM, Tang N, Mathivanan S, Pandey A. Global proteomic profiling of phosphopeptides using electron transfer dissociation tandem mass spectrometry. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:2199–2204. doi: 10.1073/pnas.0611217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kjeldsen F, Giessing AMB, Ingrell CR, Jensen ON. Peptide sequencing and characterization of post-translational modifications by enhanced ion-charging and liquid chromatography electron-transfer dissociation tandem mass spectrometry. Analytical Chemistry. 2007;79:9243–9252. doi: 10.1021/ac701700g. [DOI] [PubMed] [Google Scholar]
- 33.Mikesh LM, Ueberheide B, Chi A, Coon JJ, Syka JEP, Shabanowitz J, Hunt DF. The utility of ETD mass spectrometry in proteomic analysis. Biochimica Et Biophysica Acta-Proteins and Proteomics. 2006;1764:1811–1822. doi: 10.1016/j.bbapap.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu SL, Huehmer AFR, Hao ZQ, Karger BL. On-line LC-MS approach combining collision-induced dissociation (CID), electron-transfer dissociation (ETD), and CID of an isolated charge-reduced species for the trace-level characterization of proteins with post-translational modifications. Journal of Proteome Research. 2007;6:4230–4244. doi: 10.1021/pr070313u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gunawardena HP, Emory JF, McLuckey SA. Phosphopeptide anion characterization via sequential charge inversion and electron- transfer dissociation. Analytical Chemistry. 2006;78:3788–3793. doi: 10.1021/ac060164j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hogan JM, Pitteri SJ, Chrisman PA, McLuckey SA. Complementary structural information from a tryptic N-linked glycopeptide via electron transfer ion/ion reactions and collision-induced dissociation. Journal of Proteome Research. 2005;4:628–632. doi: 10.1021/pr049770q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swaney DL, McAlister GC, Wirtala M, Schwartz JC, Syka JEP, Coon JJ. Supplemental activation method for high-efficiency electron-transfer dissociation of doubly protonated peptide precursors. Analytical Chemistry. 2007;79:477–485. doi: 10.1021/ac061457f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McAlister GC, Phanstiel D, Good DM, Berggren WT, Coon JJ. Implementation of electron-transfer dissociation on a hybrid linear ion trap-orbitrap mass spectrometer. Analytical Chemistry. 2007;79:3525–3534. doi: 10.1021/ac070020k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McAlister GC, Berggren WT, Horning S, Makarov A, Phanstiel D, Griep-Raming J, Stafford G, Swaney DL, Syka JEP, Zabrouskov V, Coon JJ. A Proteomics Grade Electron Transfer Dissociation-enabled Hybrid Linear Ion Trap-orbitrap Mass Spectrometer. 2008 doi: 10.1021/pr800264t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Geer LY, Markey SP, Kowalak JA, Wagner L, Xu M, Maynard DM, Yang XY, Shi WY, Bryant SH. Open mass spectrometry search algorithm. Journal of Proteome Research. 2004;3:958–964. doi: 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
- 41.Peng JM, Elias JE, Thoreen CC, Licklider LJ, Gygi SP. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: The yeast proteome. Journal of Proteome Research. 2003;2:43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- 42.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nature Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 43.McAlister GC, Swaney D, Griep-Raming J, Makarov A, Lange O, Schwartz JC, Syka JEP, Horning S, Stafford G, Coon JJ. A Probabilistic Decision Tree-Driven Tandem Mass Spectrometer for Shotgun Proteomics. 2008 [Google Scholar]