Abstract
Rom2p is a GDP/GTP exchange factor for Rho1p and Rho2p GTPases; Rho proteins have been implicated in control of actin cytoskeletal rearrangements. ROM2 and RHO2 were identified in a screen for high-copy number suppressors of cik1Δ, a mutant defective in microtubule-based processes in Saccharomyces cerevisiae. A Rom2p::3XHA fusion protein localizes to sites of polarized cell growth, including incipient bud sites, tips of small buds, and tips of mating projections. Disruption of ROM2 results in temperature-sensitive growth defects at 11°C and 37°C. rom2Δ cells exhibit morphological defects. At permissive temperatures, rom2Δ cells often form elongated buds and fail to form normal mating projections after exposure to pheromone; at the restrictive temperature, small budded cells accumulate. High-copy number plasmids containing either ROM2 or RHO2 suppress the temperature-sensitive growth defects of cik1Δ and kar3Δ strains. KAR3 encodes a kinesin-related protein that interacts with Cik1p. Furthermore, rom2Δ strains exhibit increased sensitivity to the microtubule depolymerizing drug benomyl. These results suggest a role for Rom2p in both polarized morphogenesis and functions of the microtubule cytoskeleton.
INTRODUCTION
The cytoskeleton is the infrastructure of the cell; it aids in determining cell shape and participates in many dynamic cellular processes. Two major components of all eukaryotic cytoskeletons are microfilaments, composed of actin subunits, and microtubules, made up of tubulin heterodimers. The two systems are involved in distinct processes within the cell. This is particularly evident in the budding yeast Saccharomyces cerevisiae. The primary functions of the yeast actin cytoskeleton are in secretion, cell growth and polarized morphogenesis, resulting in bud and mating projection formation (Adams and Pringle, 1984; Kilmartin and Adams, 1984; Novick and Botstein, 1985; Read et al., 1992). Yeast microtubules are dispensable for secretion and cell growth; instead, they are required for nuclear positioning, chromosome segregation during mitosis and meiosis, and nuclear fusion during mating (karyogamy; Adams and Pringle, 1984; Kilmartin and Adams, 1984; Huffaker et al., 1988; Jacobs et al., 1988; Snyder et al., 1991; reviewed in Page and Snyder, 1993).
Studies of the regulation of cytoskeletal function and rearrangement in a wide variety of eukaryotes have provided insight into the mechanisms controlling the organization of microfilaments and, to a lesser degree, microtubules. A family of highly conserved ras-homologous small GTPases, known as Rho proteins, have emerged as key molecular switches that regulate organization of the actin cytoskeleton (reviewed in Hall, 1994; Ridley, 1995).
Rho proteins cycle between a GTP-bound active state and a GDP-bound inactive state with the assistance of several important classes of regulatory proteins. One type of Rho regulator is the GDP/GTP-exchange factor (GEF), which stimulates release of GDP from the Rho protein, allowing subsequent binding of GTP and thereby Rho activation (Hart et al., 1991). The Rho protein is returned to the GDP-bound inactive state by a Rho-specific GTPase-activating protein (GAP) that stimulates the inherent GTPase activity of the Rho protein (Ridley, 1995, and references therein). Additional Rho regulation involves Rho-GDP dissociation inhibitors that bind Rho proteins in their GDP-bound form and prevent exchange for GTP, thus locking them in the inactive state (Ohga et al., 1989; Fukumoto et al., 1990).
Five genes encoding members of the Rho family have been identified in S. cerevisiae, including CDC42, RHO1, RHO2, RHO3, and RHO4. All have been implicated in controlling polarized cell morphogenesis during bud and/or mating projection formation (Madaule et al., 1987; Adams et al., 1990; Johnson and Pringle, 1990; Matsui and Toh-e, 1992; Li et al., 1995; Imai et al., 1996). Cdc42p and Rho1p are essential and localize to sites of polarized cell growth, which are regions where actin patches accumulate (Ziman et al., 1993; Yamochi et al., 1994). Rho1p has recently been shown to regulate both cell wall biosynthesis and protein kinase C (Pkc1p) signaling (Drgonova et al., 1996; Qadota et al., 1996).
Members of the different classes of Rho regulators have also been identified in S. cerevisiae. CDC24 encodes a GEF for Cdc42p, and ROM1 and ROM2 encode GEFs for Rho1p (Zheng et al., 1994; Ozaki et al., 1996; Schmidt et al., 1997). BEM3 encodes a GAP for Cdc42p, and BEM2 and SAC7 encode GAPs for Rho1p (Zheng et al., 1993; Peterson et al., 1994; Schmidt et al., 1997). Genetic evidence suggests that Rom2p and Bem2p also regulate Rho2p activity (Kim et al., 1994; Ozaki et al., 1996; Schmidt et al., 1997; this study). Finally, a Rho-GDP dissociation inhibitor that binds Rho1p is encoded by RDI1 (Masuda et al., 1994).
Although the biochemical functions of Rho regulators have been well-characterized in both yeast and mammalian cells, much less is known about their cellular distribution and how they are controlled. The only direct Rho regulator whose subcellular distribution has been determined is S. cerevisiae Cdc24p, which localizes over the entire cell periphery (Pringle et al., 1995). This pattern contrasts that of the Cdc24p target Cdc42p, which concentrates at sites of polarized cell growth (Ziman et al., 1993). Whether other regulators localize and function at sites of Rho protein activity is an important but unresolved issue.
We have identified ROM2 in a screen for high-copy suppressors of cik1Δ, a mutant defective in microtubule-based processes (Page and Snyder, 1992). During the course of our work, ROM2 was also identified by two other groups, as a high-copy suppressor of both a cold-sensitive rho1 strain (Ozaki et al., 1996) and a temperature-sensitive tor2 strain (Schmidt et al., 1997). TOR2 encodes a phosphatidylinositol kinase homologue required for initiation of translation in response to nutrient concentrations (Barbet et al., 1996; DiComo and Arndt, 1996) and is also involved in polarization of the actin cytoskeleton (Schmidt et al., 1996). To further elucidate the function of ROM2, we localized its protein product and analyzed the phenotypes resulting from a rom2 null mutation. Our findings demonstrate that Rom2p is concentrated at sites of polarized growth in a cell-cycle–dependent manner, the first such localization for a regulator of Rho proteins. Phenotypic and genetic analysis suggests that Rom2p is involved in both cell morphogenesis and microtubule function. Possible mechanisms for such roles are discussed.
MATERIALS AND METHODS
Yeast Strains, Media, and Standard Methods
Yeast strains used in this study are listed in Table 1. Yeast genetic methods and growth media were as described in Guthrie and Fink (1991). Rich medium consisted of yeast extract, peptone, and dextrose, supplemented with adenine (YPDA). Yeast transformations were by the lithium acetate method of Ito et al. (1983). Where indicated, benomyl (DuPont, Wilmington, DE) dissolved in dimethyl sulfoxide was added to YPDA medium to final concentrations of 10, 20, and 30 μg/ml. For the ROM2 overexpression experiments, cells containing YEp24-ROM2 or YEp24 plasmids were first grown on SC medium lacking uracil prior to incubation on benomyl plates; this increases the benomyl sensitivity of these strains relative to those grown initially in YPDA medium.
Table 1.
Strain list
| Strain | Genotype |
|---|---|
| Y270 | MATa/MATα ura3-52/ura3-52 lys2-801/lys2-801 ade2-101/ade2-101 trp1-Δ1/trp1-Δ1 his3-Δ200/his3-Δ200 |
| Y930 | MATa ura3-52 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 bem2Δ::URA3 |
| Y1241 | MATa/MATα ura3-52/ura3-52 lys2-801/lys2-801 ade2-101/ade2-101 trp1-Δ1/trp1-Δ1 his3-Δ200/his3-Δ200 rom2Δ::HIS3/ROM2 |
| Y1242 | MATα ura3-52 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 rom2Δ::HIS3 |
| Y1243 | MATα ura3-52 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 |
| Y1244 | MATa ura3-52 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 |
| Y1245 | MATa ura3-52 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 rom2Δ::HIS3 |
| Y1693 | MATa/MATα ura3-52/ura3-52 lys2-801/lys2-801 ade2-101/ade2-101 trp1-Δ1/trp1-Δ1 his3-Δ200/his3-Δ200 rho2Δ::TRP1/RHO2 |
| Y1695 | MATa ura3-52 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 rho2Δ::TRP1 |
| Y1698 | MATa/MATα ura3-52/ura3-52 lys2-801/lys2-801 ade2-101/ade2-101 trp1-Δ1/trp1-Δ1 his3-Δ200/his3-Δ200 kar3Δ::TRP1/KAR3 |
| Y1700 | MATa ura3-52 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 kar3Δ::TRP1 |
| Y1703 | MATa/MATα ura3-52/ura3-52 lys2-801/lys2-801 ade2-101/ade2-101 trp1-Δ1/trp1-Δ1 cik1Δ::TRP1/CIK1 |
| Y1705 | MATa ura3-52 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 cik1Δ::TRP1 |
| Y1709a | MATa/MATα ura3-52/ura3-52 lys2-801/lys2-801 ade2-101/ade2-101 trp1-Δ1/trp1-Δ1 his3-Δ200/his3-Δ200 bem2Δ::URA3/BEM2 rom2Δ::HIS3/ROM2 |
| Y1709-1 | MATa ura3-52 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 bem2Δ::URA3 rom2Δ::HIS3 |
| Y1721 | MATa/MATα ura3-52/ura3-52 lys2-801/lys2-801 ade2-101/ade2-101 trp1-901/trp1-901 his3-Δ200/his3-Δ200 leu2-98/leu2-98 |
| Y1722 | MATa/MATα ura3-52/ura3-52 lys2-801/lys2-801 ade2-101/ade2-101 trp1-901/trp1-901 his3-Δ200/his3-Δ200 leu2-98/leu2-98 ROM2::3XHA/ROM2 |
| Y1723 | MATα ura3-52 lys2-801 ade2-101 trp1-901 his3-Δ200 leu2-98 ROM2::3XHA |
| Y1724 | MATa ura3-52 lys2-801 ade2-101 trp1-901 his3-Δ200 leu2-98 ROM2::3XHA |
| Y1725 | MATα ura3-52 lys2-801 ade2-101 trp1-901 his3-Δ200 leu2-98 |
| Y1726 | MATa ura3-52 lys2-801 ade2-101 trp1-901 his3-Δ200 leu2-98 |
| Y1727b | MATa/MATα ura3-52/ura3-52 lys2-801/lys2-801 ade2-101/ade2-101 trp1-Δ1/trp1-Δ1 his3-Δ200/his3-Δ200 rom2Δ::HIS3/rom2Δ::HIS3 |
| Y1728 | MATa ura3-52 lys2-801 ade2-101 his3-Δ200 leu2-98 cik1Δ::LEU2 |
Y930 × Y1242.
Y1242 × Y1245.
Construction of Epitope-tagged Rom2p Strains
A strain containing the ROM2::3XHA allele (Y1722) was constructed by using the polymerase chain reaction (PCR) epitope-tagging method of Schneider et al. (1995). The primers 5′-GCTACGAGGATTATCGCGGGTATGATACAGTTGCGTCGTTAGATTTCTGGGGTAGGGAACAAAAGCTGG-3′ and 5′-GCTTTTTTATTCTAAAGAAAATAAGGAAAGTCTATATACGTTGCTATCCTATAGGGCGAATTGG-3′ were used to amplify an ∼1.5-kb region of pMPY-3xHA. The PCR product contains the URA3 gene flanked by direct repeats encoding three copies of the hemagglutinin (HA) epitope and contains 53 bp of sequence from the 3′ end of the ROM2 gene at one end and the ROM2 translation termination codon and 45 bp of downstream sequence at the other end. This DNA fragment was used to transform yeast strain Y1721; transformants were selected on synthetic complete medium lacking uracil. For three strains, PCR analysis confirmed correct integration immediately upstream of the ROM2 stop codon. These strains were incubated on plates containing medium with 5-fluoroorotic acid to select for cells that had lost the URA3 gene through recombination between the two 3XHA coding regions. This event, as confirmed by PCR and immunoblot analysis, leaves a single in-frame copy of the 3XHA-encoding sequence at the carboxyl-terminal coding end of the ROM2 gene.
Yeast Immunoblot Analysis
Cells were grown in 10 ml of YPDA to early-logarithmic phase (OD600 = 0.5) and washed, and pellets were frozen at −70°C for 2 h. After thawing on ice, cells were lysed by using glass beads in 100 μl of lysis buffer (1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 150 mM NaCl, 50 mM Tris(hydroxymethyl)aminomethane hydrochloride, pH 7.5) containing protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin, 1 μg/ml pepstatin, 10 μg/ml soybean trypsin inhibitor, and 10 μg/ml l-1-tosylamide-2-phenylethyl chloromethyl ketone). Lysates were centrifuged for 10 min at 14,000 × g to remove unlysed cells and cell debris. Cell lysates were then mixed at a 4:1 (vol/vol) ratio with fivefold-concentrated Laemmli sample buffer (Sambrook et al., 1989) and boiled for 5 min before loading onto 8% polyacrylamide gels containing SDS. After electrophoretic separation, proteins were blotted onto Immobilon-P (Millipore, Bedford, MA) and probed with monoclonal anti-HA antibodies (12CA5 from BABCO, Richmond, CA); reactive bands were detected using alkaline phosphatase-conjugated anti-mouse antibodies (Ambersham, Arlington Heights, IL) and the CDP Star detection reagent (Boehringer Mannheim, Indianapolis, IN).
Fluorescence Microscopy
Indirect immunofluorescence was performed as described by Gehrung and Snyder (1990) and Pringle et al. (1991). Cells were fixed in 3.7% formaldehyde for 30 min, washed twice, and resuspended in 1.2 M sorbitol and 50 mM potassium phosphate buffer, pH 6.8 (solution A). Spheroplasts were prepared by incubating fixed cells in solution A containing 5 μg/ml Zymolyase 100T, 0.03% glusulase, and 0.2% 2-mercaptoethanol at 37°C for 30 min. Spheroplasts were washed, resuspended in solution A, and placed onto poly(l-lysine)-coated slides. Rom2p::3XHA was detected by primary antibody incubation overnight at 4°C with mouse 12CA5 antibodies diluted in 0.15 M NaCl, 0.05 M sodium phosphate, pH 7.4, and 0.1% bovine serum albumin (BSA; PBS/BSA), and secondary antibody incubation was for 2 h at room temperature with CY3-conjugated goat anti-mouse antibodies (Jackson ImmunoResearch, West Grove, PA) diluted in PBS/BSA. After both primary and secondary antibody incubations, slides were washed with PBS/BSA twice and PBS/BSA containing 0.1% Nonidet P-40 once. Finally, slides were mounted in 70% glycerol, 2% n-propyl gallate, and 0.25 μg/ml Hoechst 33258.
Cells with mating projections were prepared from midlogarithmic-phase cultures (OD600 = 1.2) of MATa stains grown in 5 ml of YPDA, washed twice with sterile double-distilled H20, and incubated for 2 h in 5 ml of YPDA containing 5 μg/ml α-factor (Sigma, St. Louis, MO) before formaldehyde fixation. Spheroplasts of cells with mating projections were prepared as described above but with incubation in spheroplast solution at 37°C for only 10 min.
The distribution of filamentous actin (F-actin) was analyzed by incubating fixed cells suspended in PBS with an equal volume of rhodamine- or fluorescein isothiocyanate-conjugated phalloidin (Molecular Probes, Eugene, OR; 3.3 μM, dissolved in methanol) for 2 h at room temperature in the dark. Stained cells were washed once, resuspended in PBS, and placed onto poly(l-lysine)-coated slide for fluorescence microscopy.
Disruption of ROM2 and RHO2
Deletion of the entire open reading frames (ORFs) of the ROM2 and RHO2 genes was performed by the PCR-based method described by Baudin et al. (1993). A heterozygous rom2Δ::HIS3/ROM2 strain (Y1241) was generated by using the primers 5′-CAGTGCTCTATTACTGCTGACTTAATTGGACAATTCATCTCTTTTCCTGCGGTTACTCTTGGCCTCCTCTAG-3′ and 5′-GTTATGCTTTTTATTCTAAAGAAAATAAGGAAAGTCTATATACGTTGCTATCGCCTCGTTCAGAATG-3′ to amplify the HIS3 gene from pRS303. The resulting PCR product, containing the HIS3 gene flanked on one side by 52 bp of sequence from the region directly upstream of the predicted ROM2 initiator codon and on the other side by 47 bp of sequence from the region directly downstream of the ROM2 termination codon, was used to transform Y270. Transformants with correct replacement of the ROM2 genomic locus with the HIS3 gene were confirmed by PCR analysis and sporulated, and tetrads were dissected and analyzed.
A heterozygous rho2Δ::TRP1/RHO2 strain (Y1693) was generated by using the primers 5′-GACATCAATTGCTGAAACGTTCTGCTTTGGTTGTGCTTTTGATCCCGTACTGGAGAGGGCCAAGAGGGAG-3′ and 5′-GTTTTTCCCTCCCTTGCTAAAAAGATAATGTATCATTTCAGTGTAAGTTTTTTGGCCTGCAGGCAAGTGCAC3′ to amplify the TRP1 gene from pRS304. The resulting PCR product, containing the TRP1 gene flanked on one side by 50 bp of sequence for the region directly upstream of the RHO2 initiator codon and on the other side by 52 bp of sequence from the region directly downstream of the RHO2 termination codon, was used to transform Y270. Transformants were confirmed and analyzed as described above.
Identification of Multicopy Suppressors of cik1Δts
A YEp24-based 2 μ S. cerevisiae genomic library (Carlson and Botstein, 1982) was transformed into a MATa cik1Δ::LEU2 strain (Y1728), spread onto plates containing synthetic complete medium lacking uracil (plasmid selection), and incubated for 12 h at the permissive temperature (room temperature) before shifting to the restrictive temperature (37°C) for 2 d. Of greater than 8000 total transformants, 34 that grew at the restrictive temperature were recovered. The plasmids from these strains were rescued into Escherichia coli, and 26 allowed growth at 37°C when reintroduced into Y1728 or a cik1Δ::TRP1 strain (Y1705). Based on restriction digest analysis, these plasmids contained eight distinct sequence groups. Primers flanking the YEp24 site of genomic DNA insertion were used to generate sequence from both ends of the insert. BLAST searches with these sequences identified regions from the S. cerevisiae genome that were encoded by these plasmids. Subcloning techniques were then employed to identify the cik1Δ suppressing (cis) gene from each group (see summary in Table 3). In each case, plasmids containing the single ORF indicated in Table 3 were competent for suppression; control plasmids lacking the ORF failed to suppress the temperature-sensitive growth defect.
Table 3.
Results of screen for 2 μ suppressors of cik1Δ
| Suppressor (no. of isolates) | Suppression ofa
|
Identityb—Homology | |
|---|---|---|---|
| cik1Δ | kar3Δ | ||
| CIS1 (1) | + | − | YDR022C—novel |
| CIS2 (1) | + | + | YLR299W—γ-glutamyltransferase |
| CIS3 (1) | + | + | YJL158C—Hsp150p homologue |
| CIS4 (2) | + | + | RHO2 |
| CIS5 (2) | + | + | ROM2 |
| CIS6 (1) | + | + | MID2 |
| CIS7 (9) | + | − | MATα,BUD5 ΔCc |
| CIS8 (9) | + | − | CIK1 |
Suppression was scored by growth of cik1Δ (Y1705) or kar3Δ (Y1700) strains containing YEp24 derived plasmids with the various CIS genes at 37°C on solid synthetic complete medium lacking uracil (see MATERIALS AND METHODS).
Saccharomyces Genomic Database ORF designation for previously uncharacterized genes.
Suppression is specific for cik1Δ MATa strains; isogenic MATα strains containing this plasmid fail to grow at the restrictive temperature. All other CIS gene encoding plasmids suppress the temperature sensitivity of both MATa and MATα cik1Δ strains. Smallest CIS7 plasmid tested contains the MATα locus and the amino-terminal end encoding two-thirds of BUD5. A CEN plasmid containing this region is sufficient for suppression.
RESULTS
ROM2
ROM2 was identified in a screen for high-copy suppressors of the temperature-sensitive growth defect of a cik1Δ strain. Cik1p complexes with the kinesin-related microtubule motor Kar3p, and together they are required for proper chromosome segregation and karyogamy (Meluh and Rose, 1990; Page and Snyder, 1992; Page et al., 1994; see below). ROM2 is predicted to encode a 1356-amino acid protein with a region of homology to the human oncogene-encoded dbl protein (Figure 1A), a GEF for human Cdc42 (Hart et al., 1991). This dbl-homology domain is common to Rho-GEFs and contains the GDP/GTP exchange activity (Hart et al., 1994; Zheng et al., 1994; Ozaki et al., 1996). The Rom2p amino acid sequence also predicts the presence of a pleckstrin-homology domain, also common to Rho-GEFs and implicated in protein–protein or protein–phosphatidylinositol derivative interactions (Musacchio et al., 1993; Lemmon et al., 1996). Rom2p has recently been identified as a GEF for the yeast Rho1 protein (Ozaki et al., 1996).
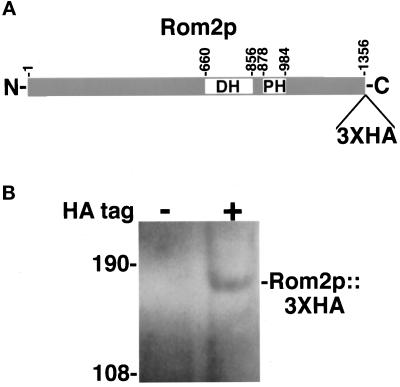
Figure 1.
(A) Schematic of Rom2p. (B) Immunoblot analysis of Rom2p::3XHA. (A) The Rom2 protein is predicted to be 1356 amino acids in length and contains a dbl-homology (DH) domain from amino acids 660 to 856 and a pleckstrin homology (PH) domain from amino acids 878 to 984. Epitope tagging of the protein results in the fusion of three tandem copies of the HA epitope to the carboxyl terminus of Rom2p (see MATERIALS AND METHODS). (B) Proteins from total cell extracts of an untagged ROM2 strain (Y1725; HA tag −) and an isogenic ROM2::3XHA strain (Y1723; HA tag +) were separated on an 8% polyacrylamide gel containing SDS. An immunoblot was prepared and probed with 12CA5 anti-HA monoclonal antibody (see MATERIALS AND METHODS). A protein of approximately 165 kDa is detected in extracts from ROM2::3XHA cells but not from ROM2 untagged cells. The mass of molecular weight markers are shown in kilodaltons.
Rom2p Localizes to Sites of Polarized Cell Growth
To gain further insight into the function of Rom2p, we determined its subcellular localization. The genomic locus of ROM2 was tagged with the HA epitope coding sequence (Schneider et al., 1995; see MATERIALS AND METHODS). A sequence encoding three copies of the HA epitope was integrated into the carboxyl-terminal coding sequence of ROM2, just prior to the translational stop codon. The resulting ROM2::3XHA allele is functional for ROM2 activity as growth rates at all temperatures, and bud morphology, are similar between ROM2::3XHA and wild-type cells (see below). Immunoblot analysis using anti-HA monoclonal antibodies detects a protein of approximately 165 kDa in cellular extracts from ROM2::3XHA strains but not from ROM2 untagged strains (Figure 1B). This corresponds well to the predicted 153-kDa size of the Rom2 protein plus the triple HA epitope.
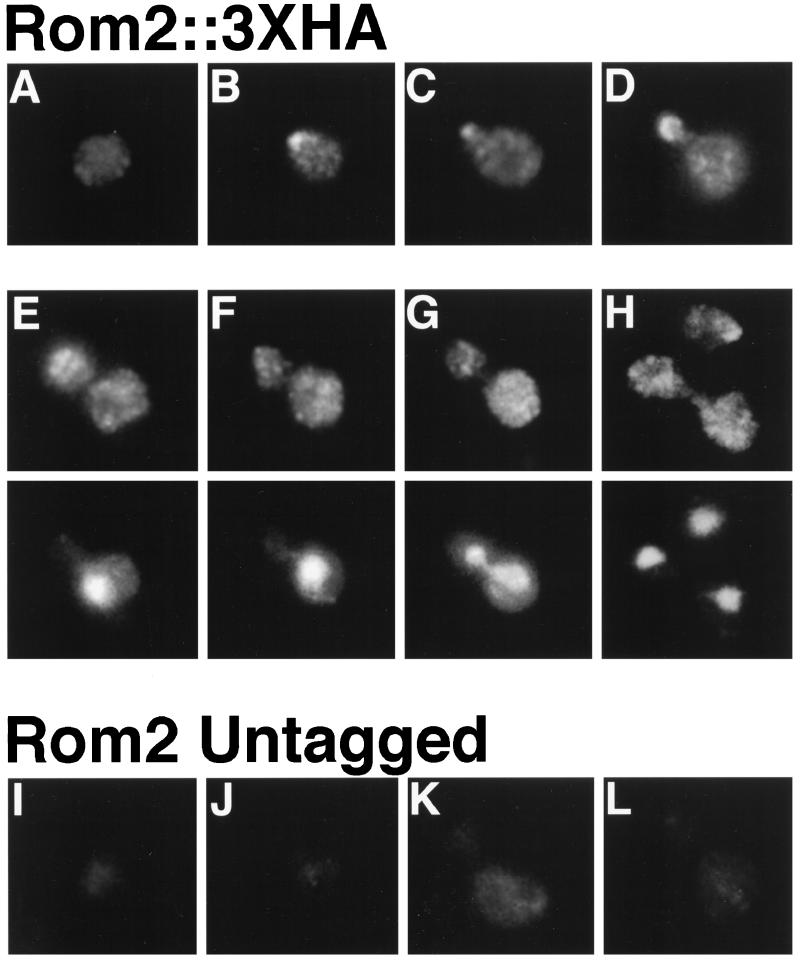
To determine the subcellular localization of the Rom2::3XHA fusion protein, indirect immunofluorescence using anti-HA monoclonal antibodies was performed (Pringle, et al., 1991; Santos and Snyder, 1997). As shown in Figure 2, Rom2p::3XHA exhibits a cell-cycle–dependent localization pattern. The majority of unbudded cells exhibit diffuse staining throughout the cell with a few small weakly stained patches (∼60% of unbudded cells, Figure 2A; total cells counted = 100). The remainder of unbudded cells contain a concentrated patch of bright staining at the cell cortex (Figure 2, B and H). Colocalization of Rom2p::3XHA and actin by using anti-HA antibodies and fluorescein isothiocyanate-conjugated phalloidin, respectively, indicates that this patch represents the presumptive site of bud emergence (our unpublished results). Rom2p::3XHA is concentrated at the bud tips of cells with emerging or small buds (>70% of tiny or small budded cells; Figure 2, C and D); some cytoplasmic patches are observed in these cells as well. In cells with larger buds, staining is more diffuse within the bud but is still concentrated toward the distal end (Figure 2E). Late in the cell cycle, when the nucleus has migrated to the bud neck (Figure 2F) and during anaphase (Figure 2G), Rom2p::3XHA localizes to cytoplasmic patches in both the mother and bud. This pattern continues through the end of mitosis and cytokinesis (Figure 2H). A strain containing Rom2p with three copies of the myc epitope fused to its carboxyl terminus exhibits identical cell-cycle–dependent staining patterns with anti-myc antibodies (our unpublished observation). Unlike F-actin and Rho1p, Rom2p::3XHA was not evident at the bud neck during cytokinesis (Adams and Pringle, 1984; Kilmartin and Adams, 1984; Yamochi et al., 1994). These staining patterns were not detected in ROM2 untagged cells at any stages of the cell cycle (Figure 2, I–L). Thus, the Rom2 protein localizes to sites of polarized bud growth, similarly to Rho1p and Cdc42p (Ziman et al., 1993; Yamochi et al., 1994).
Figure 2.
Cell-cycle–dependent distribution of Rom2p::3XHA. Indirect immunofluorescence with anti-HA antibodies (A–D; E–H, top; I–L) and DNA staining with Hoechst 33258 (E–H, bottom). Representative cells are pictured for a ROM2::3XHA strain (Y1723; A–H) and an untagged strain (Y1725; I–L). (A) An unbudded cell with diffuse cytoplasmic staining. (B) An unbudded cell with polarized cortical staining. (C) A cell with staining concentrated within an emerging bud. (D) Localization to the periphery of small buds. (E) A medium to large budded cell displaying cytoplasmic patches with staining strongest toward the distal end of the bud. (F) A large budded cell with nucleus positioned at the bud neck, and (G) a large budded cell in anaphase, demonstrating cytoplasmic patch staining. (H) A cell undergoing cytokinesis with bright patch staining in the cytoplasm and an unbudded cell with polarized cortical staining. Background staining of (I) an unbudded cell, (J and K) small to medium budded cells, (L) and a large budded cell is also shown.
Rom2p Localizes to the Tips of Mating Projections
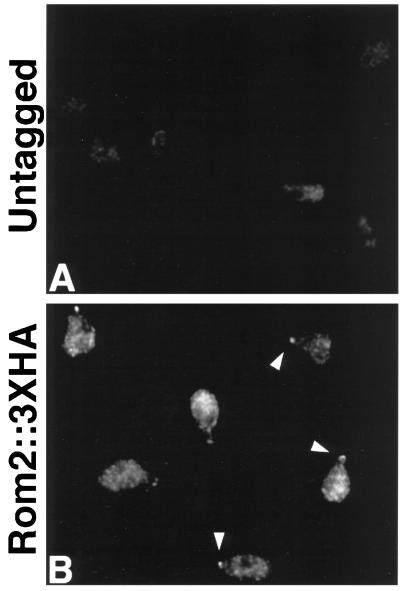
Cdc24p is required for mating projection formation and Cdc42p localizes to the projection tip (Field and Schekman, 1980; Ziman et al., 1993). As yet, the localization and function of other Rho proteins and their regulators in mating cells have not been determined. Concentration of Rom2p::3XHA at sites of vegetative polarized cell growth suggests the possibility that it may also localize to such sites in cells exposed to mating pheromone. Exponentially growing liquid cultures of ROM2 and ROM2::3XHA MATa strains were treated with α-factor (5 μg/ml, final concentration) for 2 h and then fixed for indirect immunofluorescence analysis. Staining of ROM2::3XHA cells with anti-HA antibodies reveals concentration of Rom2p::3XHA at the tips of mating projections (Figure 3B, arrowheads). In addition, these cells demonstrate a diffuse or granular cytoplasmic localization of Rom2p::3XHA; the cytoplasmic staining was stronger than that observed for unbudded vegetative cells. No staining above background was detected for untagged ROM2 cells treated with mating pheromone (Figure 3A). Therefore, Rom2p localizes to polarized cell growth sites both in vegetative and mating pheromone-treated cells.
Figure 3.
Mating projection localization of Rom2p::3XHA. A MATa untagged strain (Y1726; A) and a MATa ROM2::3XHA strain (Y1724; B) were treated with 5 μg/ml α-factor for 2 h before fixation and preparation for indirect immunofluorescence with anti-HA antibodies. Representative cells are pictured. (A) Untagged cells display very little background staining. (B) ROM2::3XHA cells demonstrate concentrated staining at the tips of mating projections (arrowheads) and diffuse cytoplasmic staining with some patches.
Rom2p Is Required for Proper Cell Growth and Bud Morphology
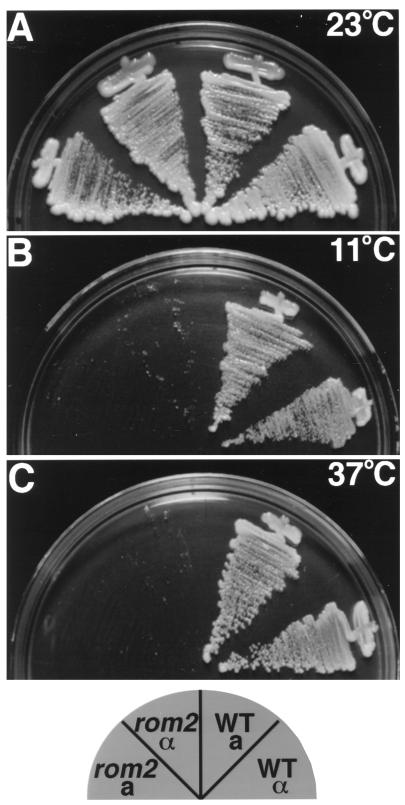
To further understand the function of Rom2p, the entire ROM2 genomic locus was replaced with a HIS3 marker (see MATERIALS AND METHODS). Two independent heterozygous rom2Δ::HIS3/ROM2 strains were sporulated and 20 tetrads were dissected for each. Tetrad analysis demonstrated 2:2 segregation for the HIS marker, and His+ segregants grew slower at 23°C. His+ segregants failed to grow at both 37°C and 11°C, but His− strains were viable at both temperatures (Figure 4). Therefore, although ROM2 is not essential at 23°C, it is required for growth at both elevated and lowered temperatures. High-copy plasmids containing either RHO1 or RHO2, but not CDC42, suppress the temperature-sensitive growth defect of rom2Δ strains at 37°C (our unpublished results). This is consistent with Rom2p serving as the GEF specific for Rho1p and Rho2p, as described independently by others (Ozaki et al., 1996; Schmidt et al., 1997).
Figure 4.
rom2Δ strains are temperature sensitive for growth at 11°C and 37°C. Haploid segregants from a rom2Δ::HIS3/ROM2 heterozygous diploid (Y1241) were streaked onto plates containing YPDA medium and incubated at room temperature for 3 d (A), 11°C for 10 d (B), or 37°C for 3 d (C). Pictured are MATa rom2Δ (Y1245), MATα rom2Δ (Y1242), MATa ROM2 (Y1244), MATα ROM2 (Y1243).
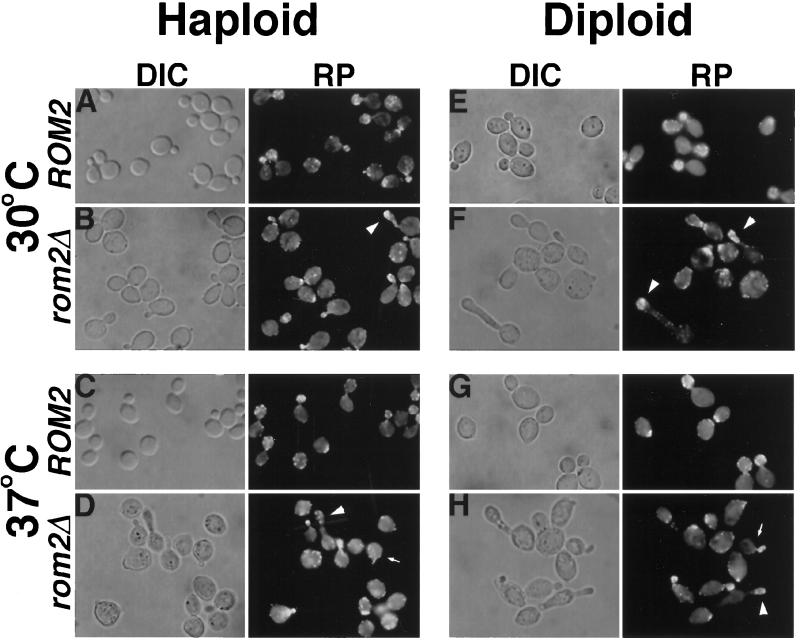
Cell morphology of rom2Δ strains was analyzed at both permissive and restrictive temperatures by using differential-interference contrast microscopy. The actin cytoskeleton was also examined in these cells by rhodamine-conjugated phalloidin staining and fluorescence microscopy (Figure 5). At the permissive temperature of 30°C, haploid rom2Δ strains differ in morphology from wild-type strains (four different rom2Δ segregants were analyzed; the results for one representative strain is quantified; Table 2). A significant number of cells (7%) with elongated buds are present (a cell with an elongated bud was scored as one in which the bud length is greater than twice the bud width, Table 2). Also, large-budded cells appear more oval or drop-shaped than those of ROM2 strains (Figure 5B). After shifting rom2Δ strains to 37°C for 3 h, the proportion of cells with small buds is substantially higher than wild-type strains (49% for rom2Δ cells as compared with 26% for isogenic wild-type cells) and many cells with elongated buds are again observed (Figure 5D and Table 2). Cells with elongated buds are not detected at either temperature in wild-type strains. F-actin polarization in most rom2Δ cells resembles that of ROM2 cells (Figure 5, A–D, rhodamine-phalloidin), in that actin patches accumulate at the incipient bud site and throughout the bud of small budded cells (Adams and Pringle, 1984; Kilmartin and Adams, 1984). However, in mutant cells with unusually elongated large buds, F-actin remains concentrated toward the bud tip instead of localizing throughout the bud (Figure 5B, arrowhead). At both temperatures, rom2Δ cells appear slightly larger than wild-type cells (Figure 5, A–D). All morphological defects of rom2Δ strains are suppressed by introduction of wild-type ROM2 on a plasmid (our unpublished results). The accumulation of small-budded cells and appearance of elongated buds at the restrictive temperature are similar to the results of Ozaki et al. (1996) describing a rom1Δ rom2Δ strain containing a PGAL1-ROM2 plasmid under Rom2p-depletion conditions at 30°C. The morphological defects of rom2Δ strains at the permissive temperature, as well as those of the homozygous rom2Δ/rom2Δ diploid discussed below, have not been described previously.
Figure 5.
Morphological defects of rom2Δ haploids and rom2Δ/rom2Δ diploids at permissive and restrictive temperatures. Cultures of a wild-type haploid strain (Y1244; A and C), rom2Δ haploid strain (Y1245; B and D), wild-type diploid strain (Y270; E and G), and rom2Δ homozygous diploid strain (Y1241; F and H) grown at 30°C were fixed with formaldehyde (A, B, E, and F) or shifted to 37°C for 3 h before fixation (C, D, G, and H). Fixed cells were then examined by differential-interference contrast microscopy (DIC; A–H, left), and F-actin distribution was analyzed with fluorescence microscopy by staining with rhodamine-conjugated phalloidin (RP; A–H, right). Representative fields are shown. Both haploid and diploid rom2Δ strains exhibit cells with elongated bud morphologies at restictive and permissive temperatures (B, D, F, and H; arrowheads). Upon shift to 37°C, wild-type cells bud normally (C and G), but rom2Δ strains produce an increased number of small budded cells, both normal and elongated, with F-actin concentrated within the bud (D and H, arrows; see Table 2).
Table 2.
Bud morphology of wild-type and rom2Δ strains
| Strain (no. of cells) | Temperature (°C)a | % of total cells
|
% elongatedc | ||
|---|---|---|---|---|---|
| Unbudded | Small buddedb | Medium–large budded | |||
| Haploid | |||||
| ROM2 (309) | 30 | 40 | 22 | 34 | 0 |
| rom2Δ (310) | 30 | 32 | 31 | 37 | 7 |
| ROM2 (307) | 37 | 42 | 26 | 32 | 0 |
| rom2Δ (304) | 37 | 27 | 49 | 24 | 7 |
| Diploid | |||||
| ROM2 (318) | 30 | 40 | 28 | 32 | 0 |
| rom2Δ (322) | 30 | 32 | 33 | 35 | 21 |
| ROM2 (319) | 37 | 44 | 33 | 22 | 0 |
| rom2Δ (327) | 37 | 25 | 46 | 29 | 28 |
ROM2 (Y1244), rom2Δ (Y1245), ROM2/ROM2 (Y270), and rom2Δ/rom2Δ (Y1727) strains were grown overnight in rich medium at 30°C then fixed with formaldehyde or shifted to 37°C for 3 h before fixation.
Small budded cells were scored as those with buds less than or equal to one-third the size of the mother. Medium to large budded cells were those with buds greater than one-third the size of the mother.
Cells with elongated buds were scored as those whose length from neck to bud tip was greater than twice the maximum bud width. This category includes both small and large budded cells.
The appearance of abnormal bud morphology is even more striking in rom2Δ/rom2Δ diploids (four homozygous diploid strains analyzed; the results for one representative strain are presented in Table 2). Approximately 30% of all budded cells at both 30°C and 37°C have elongated buds with F-actin staining concentrated toward the distal tip of the bud (Figure 5, F and H, arrowheads; Table 2). The number of small-budded cells increases upon shifting to 37°C for 3 h (46% compared with 33% for wild-type cells), but many of these are also elongated. Elongated buds are not detected for ROM2/ROM2 wild-type diploids (Figure 5, E and G; Table 2). Chains and clusters of cells are common in rom2Δ/rom2Δ cultures, suggesting a possible defect in cytokinesis. In summary, Rom2p is required to maintain proper polarized bud growth at the restrictive temperature, as evidenced by the accumulation of small-budded cells. The presence of cells with elongated buds suggests that Rom2p may also play a role in regulation of the switch from apical to isotropic bud growth (reviewed by Lew and Reed, 1995; see DISCUSSION).
Rom2 Is Important for Projection Formation during Mating
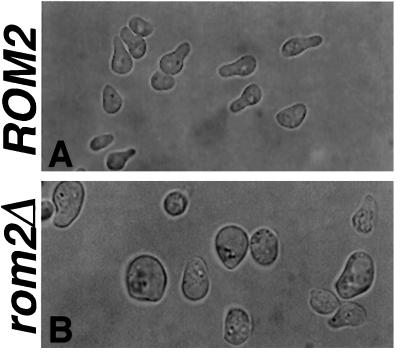
Mating projection formation was also analyzed in four different rom2Δ MATa cells. After a 2-h exposure to mating pheromone at 30°C, approximately 70% of wild-type MATa cells form mating projections of lengths greater than half the diameter of the cell (Figure 6A). In contrast, less than 20% of rom2Δ MATa cells under the same conditions form such projections. Mutant cells become much larger than wild-type cells and either form very small broad projections or do not form projections at all (Figure 6B). The actin cytoskeleton is polarized properly in the rom2Δ cells with detectable projections, as seen by rhodamine-phalloidin staining and fluorescence microscopy (our unpublished observations). Therefore, in addition to maintaining polarized cell growth during budding, Rom2p is also required for proper mating projection formation in response to pheromone.
Figure 6.
Mating-projection morphology defect of rom2Δ strains. Exponential phase cultures of a MATa ROM2 wild-type strain (Y1244; A) and a MATa rom2Δ strain (Y1245; B) were treated with 5 μg/ml α-factor for 2 h, fixed, and analyzed by differential interference contrast microscopy. ROM2 cells form normal mating projections (A), but rom2Δ cells become enlarged and form only short broad projections with abnormal morphology (B).
Involvement of ROM2 in the Microtubule Cytoskeleton
We originally identified ROM2 in a screen for high-copy suppressors of the temperature-sensitive growth defect of a cik1Δ strain (see MATERIALS AND METHODS; results summarized in Table 3). Interestingly, we also found RHO2 as a cik1 suppressing (CIS) gene, along with two other known loci, MID2 and MATα-BUD5 (see Chant et al., 1991; Herskowitz et al., 1992), and three previously uncharacterized genes, CIS1, CIS2, and CIS3. Mid2p is a transmembrane protein with a putative Ca2+ binding domain; mid2Δ mutants are sensitive to exposure to mating pheromone (Ono et al., 1994). CIS1 is predicted to encode a protein with no significant homology to known proteins. CIS2 is predicted to encode a protein with homology to γ-glutamyltransferases from a variety of organisms (22% amino acid identity to human γ-glutamyltransferase; Sakamuro et al., 1988). γ-Glutamyltransferases catalyze the transfer of glutamate residues to and from various peptide substrates, such as glutathione (Meister et al., 1981); because tubulin has been shown to be posttranslationally glutamylated (Edde et al., 1990), perhaps glutamylation of yeast tubulin by CIS2 affects microtubule function. Finally, CIS3 is predicted to encode a protein with homology to S. cerevisiae Hsp150p (29% amino acid identity; Russo et al., 1992). Further characterization of these genes will be described elsewhere.
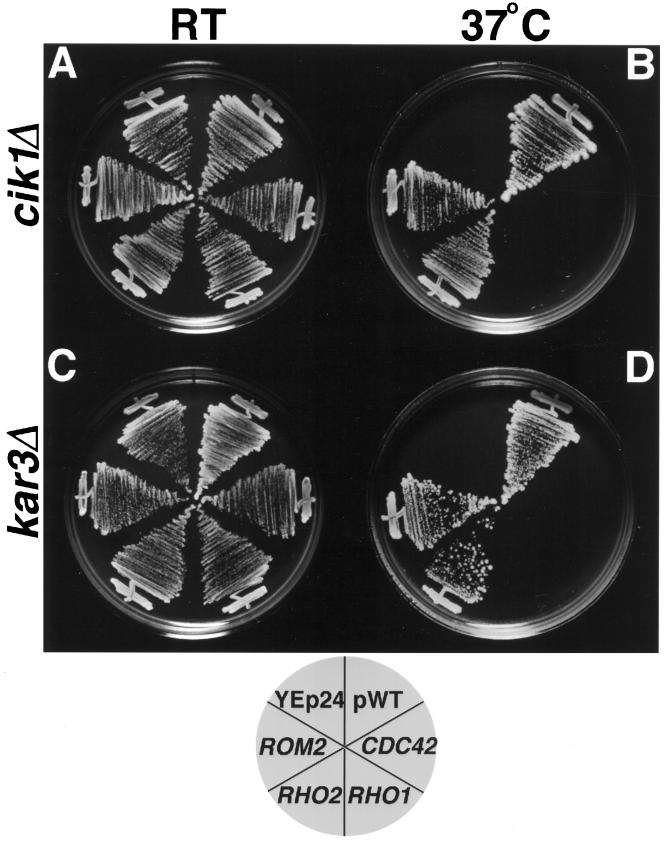
To determine whether the different suppressors are specific for cik1 mutants or whether they suppress a defect resulting from loss of function of the Kar3p–Cik1p complex, we tested the ability of 2 μ plasmids containing CIS genes to suppress the temperature sensitivity of a kar3Δ strain (Table 3). ROM2 and RHO2 plasmids allow both cik1Δ and kar3Δ strains to grow on solid medium at the restrictive temperature of 37°C; whereas vector alone or plasmids encoding one of two other S. cerevisiae Rho-related proteins, Cdc42p and Rho1p, do not (Figure 7). This genetic interaction of ROM2 and RHO2 with a microtubule–motor complex suggests involvement of these proteins in functions of the microtubule cytoskeleton.
Figure 7.
Increased gene dosage of ROM2 or RHO2 suppresses the temperature-sensitive growth defect of cik1Δ and kar3 Δ strains. A cik1Δ strain (Y1705; A and B) or a kar3 Δ strain (Y1700; C and D) containing YEp24-based 2 μ plasmids encoding the respective wild-type gene (pWT), CDC42, RHO1, RHO2, ROM2, or vector alone were plated to synthetic complete medium lacking uracil and incubated at either room temperature (RT; A and C) or 37°C (B and D). Strains containing ROM2, RHO2, or pWT were able to grow at the restrictive temperature of 37°C, but those containing CDC42, RHO1, or YEp24 alone failed to grow.
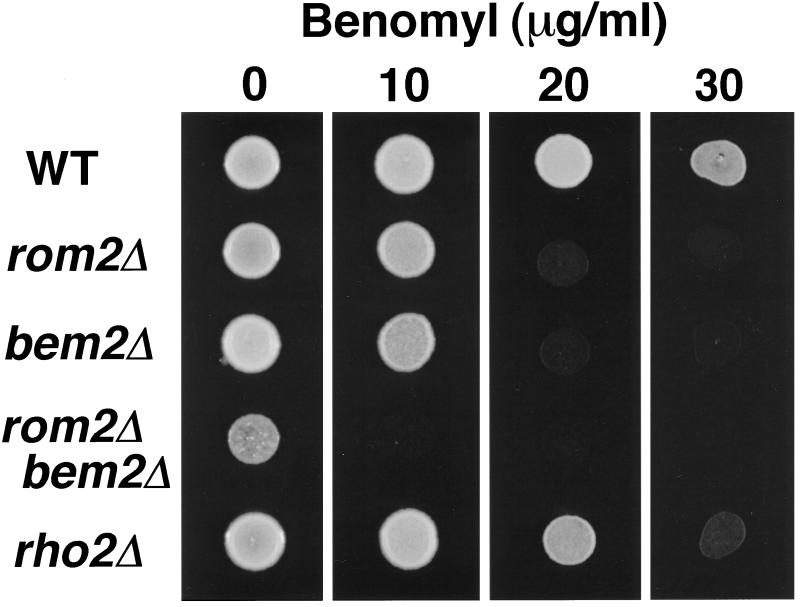
We analyzed whether strains lacking Rom2p or Rho2p displayed microtubule-related defects. rom2Δ and rho2Δ strains were tested for altered sensitivity to the microtubule-depolymerizing drug benomyl; failure to grow or enhanced growth in the presence of benomyl is a common characteristic of strains containing mutations in genes encoding structural or regulatory components of microtubules (e.g., Huffaker et al., 1988; Hoyt et al., 1991; Li and Murray, 1991; Reijo et al., 1994; Pellman et al., 1995; Saunders et al., 1997). Wild-type ROM2 strains grow on solid rich medium containing as much as 30 μg/ml of benomyl; in contrast, an isogenic rom2Δ strain fails to grow in the presence of benomyl concentrations of 20 and 30 μg/ml, and a rho2Δ strain displays a slight increase in benomyl sensitivity, failing to grow on plates containing 30 μg/ml (Figure 8; four mutant segregants were tested for each strain); these concentrations are similar to those that inhibit growth of several β-tubulin mutants (Reijo et al., 1994). This represents the first phenotype described for a RHO2 disruption strain. A strain containing the temperature-sensitive rho1–104 allele has been shown previously to exhibit wild-type levels of benomyl sensitivity (Wang and Bretscher, 1995). In addition, wild-type strains containing a high-copy ROM2 plasmid display increased resistance to benomyl compared with strains containing vector alone (the presence of the ROM2 plasmid allows colony formation in the presence 20 μg/ml benomyl at 102- to 103-fold lower cell concentrations than cells containing vector; see MATERIALS AND METHODS; our unpublished results). Thus, phenotypic analysis supports Rom2p involvement in microtubule stability and/or function.
Figure 8.
rom2Δ, bem2Δ, and rho2Δ strains exhibit increased sensitivity to benomyl. Approximately 5 × 105 cells from overnight cultures of a wild-type strain (Y1244), rom2Δ strain (Y1245), bem2Δ strain (Y930), rom2Δ bem2Δ strain (Y1709–1), and rho2Δ strain (Y1695) were plated in 5-μl spots to YPDA medium containing benomyl at 0, 10, 20, or 30 μg/ml. Wild-type cells grow in medium containing 30 μg/ml benomyl; cells of rom2Δ and bem2Δ mutants fail to grow in medium with 20 and 30 μg/ml benomyl; cells of a rom2Δ bem2Δ double mutant fail to grow in medium with 10 μg/ml benomyl; finally, rho2Δ strains are impaired for growth in medium containing 30 μg/ml benomyl.
Interestingly, it has been reported that strains lacking Bem2p, a GAP for Rho1p and Rho2p, also display increased sensitivity to benomyl (Kim et al., 1994; Wang and Bretscher, 1995; Figure 8). Because the GEF activity of Rom2p should oppose the GAP activity of Bem2p, we tested whether a BEM2 deletion could partially suppress the growth defect of rom2Δ on benomyl. A double rom2Δ bem2Δ mutant was constructed by mating a rom2Δ::HIS3 strain (Y1242) with a bem2Δ::URA3 strain (Y930) and sporulating the resulting double heterozygous diploid. Tetrad dissection yielded His+ Ura− and His− Ura+ segregants that grew slowly as compared with His− Ura− wild-type segregants. His+ Ura+ segregants grew even slower, often taking several days to form a colony at room temperature (20 tetrads analyzed; our unpublished results). These rom2Δ bem2Δ double mutants fail to grow on the lowest concentration of benomyl tested (10 μg/ml; Figure 8; 4 His+Ura+ segregants tested). Therefore, disrupting both a GEF and a GAP for the Rho1p/Rho2p GTPase cycle exacerbates the defect seen in the single mutants (see DISCUSSION).
Microtubule structures from strains containing either a ROM2 deletion or a 2 μ ROM2 plasmid were also examined. Exponentially growing cells, as well as those from cultures treated with 5 μg/ml α-factor (which arrests cells in G1 phase) or 100 mM hydroxyurea (which arrests cells in S phase with a short spindle and the nucleus migrated to the bud neck), were analyzed by indirect immunofluorescence with anti-tubulin antibodies. No significant difference in microtubule staining pattern, length, or number was detected at any temperature compared with wild-type or vector control cells for any of the samples (our unpublished observations). Additionally, nuclear migration and spindle orientation were analyzed in the different cells by 4,6-diamidino-2-phenylindole and anti-tubulin staining, respectively. Again, no significant defect in these processes was observed (our unpublished observation). Therefore, although genetic and phenotypic analysis of ROM2 suggests that, in addition to a role in actin-mediated events, Rom2p participates in microtubule-related functions, immunofluorescence techniques fail to detect differences in microtubule structure or cytoplasmic microtubule function (see DISCUSSION).
DISCUSSION
A Rho Regulator Localizes to Sites of Rho Activity
Despite its importance in understanding the molecular and spatial control of Rho-protein function, data on the subcellular localization of Rho regulators, in any system, have been limited. We demonstrate that the yeast Rho-GEF Rom2p localizes to sites of polarized cell growth, both during budding and mating projection formation. The cell-cycle–dependent localization of Rom2p is very similar to that of Rho1p (Yamochi et al., 1994) and represents the first example of a Rho regulator concentrating at sites of Rho function. Unlike Rom2p, another S. cerevisiae Rho-GEF, Cdc24p, is distributed around the entire cell periphery (Pringle et al., 1995), although its target, Cdc42p, localizes to discrete sites of polarized cell growth (i.e., incipient bud sites and the tips of small buds; Ziman et al., 1993). Therefore, the regulation of Cdc42p and Rho1p may differ through the spatial organization of their GEFs. One model for control of Cdc42p function is that Cdc24p is only active at sites of Cdc42p accumulation, specified by cortical cues (Pringle et al., 1995). Another possibility is that Cdc24p is not regulated in a spatial manner at all; instead it may be competent for GEF activity throughout the cell periphery, and Cdc42p localization would, therefore, determine where the GTPase switch is activated.
The accumulation of both Rho1p and Rom2p at cell growth sites suggests a different mechanism for Rho1p localized activation. These proteins may localize to polarized growth sites independently by recognizing polarity establishment components, such as Cdc42p. Presumably, activation and translocation of Rom2p and release of Rho1p from cytoplasmic Rdi1p would occur at the G1–S transition prior to bud emergence. While this manuscript was in preparation, Schmidt et al. (1997) found that Tor2p directly or indirectly activates the Rho1p-GEF activity of Rom2p. Perhaps the putative phosphatidylinositol kinase activity of Tor2p leads to a local accumulation of phosphatidylinositol derivatives that interact with the pleckstrin-homology domain of Rom2p (Harlan et al., 1994), resulting in both polarized localization and activation of Rom2p. Rho1p may independently localize to sites of cell wall synthesis through its interaction with the transmembrane subunits of 1,3-β-glucan synthase (Qadota et al., 1996). Alternatively, it is possible that Rom2p and Rho1p are interdependent for this localization or that there exists a hierarchy, perhaps with activated Rom2p responsible for recruiting Rho1p. Future localization studies should determine which of these models is correct.
Although the distributions of Rho1p and Rom2p overlap, the Rom2p::3XHA cytoplasmic patch distribution in large-budded cells and absence from the bud neck during cytokinesis differs from that reported for HA-Rho1p. In large-budded cells, HA-Rho1p staining was very faint and diffuse but reappeared at the bud neck in cells undergoing cytokinesis (Yamochi et al., 1994). These differences may be due to the sensitivity of detection of each protein or could represent actual differences in the subcellular distribution of these proteins during the later stages of the cell cycle.
Rom2p Is Required for Proper Cell Growth and Polarized Morphogenesis
We have found that disruption of the ROM2 gene in our strain background results in slow growth at room temperature and failure to grow at both 11°C and 37°C. Two independent groups have found somewhat different phenotypes for rom2Δ strains: Ozaki et al. (1996) report that along with slow growth at 20°C, these strains fail to grow at temperatures above 33°C; Schmidt et al. (1997) report normal growth at 37°C and impaired growth at temperatures of 30°C and lower. Presumably, these discrepancies are the result of differences in strain backgrounds.
We demonstrate that both haploid and homozygous diploid rom2Δ strains have various morphological defects at both restrictive and permissive temperatures. Upon shift to 37°C, rom2Δ strains accumulate small-budded cells with the actin cytoskeleton polarized to the bud. A strain with a temperature-sensitive allele of RHO1 also arrests with small-budded cells and a polarized actin cytoskeleton (Yamochi et al., 1994). It is likely that the cessation of bud growth observed in rho1 and rom2 strains at the restrictive temperature is due to loss of 1,3-β-glucan synthase activity, which has been shown to require GTP-bound Rho1p (Qadota et al., 1996), and/or altered regulation of the protein kinase C signalling pathway (Drgonova et al., 1996). In addition to the small-budded phenotype, many rom2Δ cells have elongated buds, even at the permissive temperature. The phenotype is most striking in homozygous diploid cells, with bud lengths often exceeding five times the length of its mother. This is suggestive of a defect in the switch from apical to isotropic bud growth (see Lew and Reed, 1995). It is possible that Rom2p is involved in redirecting growth from the bud tip to regions throughout the bud; regulation of Rom2p localization or function by cell cycle regulatory components might mediate this switch. Finally, although rom2Δ cells exposed to mating pheromone arrest normally, they become enlarged and fail to form typical wild-type mating projections. This phenotype, along with the localization of Rom2p to projection tips, demonstrates that Rom2p-mediated cellular morphogenesis extends to mating-pheromone differentiated cells as well.
Rom2p May Play a Role in Functions of the Microtubule Cytoskeleton
CIS5, a gene identified as a high-copy suppressor of the temperature-sensitive growth defect of a cik1Δ strain, was found to be identical to ROM2. Cik1p is involved in microtubule functions, such as chromosome segregation and karyogamy (Page and Snyder, 1992). Cik1p complexes with the kinesin-related Kar3 protein, a minus-end–directed microtubule motor (Meluh and Rose, 1990; Endow et al., 1994; Page et al., 1994). These proteins localize to spindle pole bodies and microtubules and are involved in spindle assembly and/or stability in vegetative cells (Meluh and Rose, 1990; Page et al., 1994). A high-copy plasmid containing ROM2 also suppressed a kar3Δts strain, indicating that Rom2p suppresses a defect resulting from loss of the motor complex function, rather than just loss of Cik1p. Interestingly, CIS4 was found to encode Rho2p and also suppresses the kar3Δts strain. This suppression was specific for RHO2, as no other yeast Rho-related gene, including RHO1, on a 2 μ plasmid demonstrated this activity.
Similar to mutants defective in microtubule stability and/or function, rom2Δ strains exhibit increased sensitivity to microtubule-depolymerizing growth conditions (i.e., cold temperature and the presence of benomyl). Strains with disruptions in the BEM2 gene also exhibit growth defects in the presence of benomyl (Kim et al., 1994; Wang and Bretscher, 1995; this study). Wang and Bretscher (1995) have also observed that a bem2Δ strain displays synthetic lethality with a tub2 allele defective in nuclear microtubules. Surprisingly, disruption of both ROM2, encoding a Rho1/Rho2-GEF (Ozaki et al., 1996), and BEM2, encoding a Rho1/Rho2-GAP (Zheng et al., 1993; Peterson et al., 1994), results in an additive defect for both growth rate and benomyl sensitivity. This implies that either Rho protein cycling between GDP- and GTP-bound form is important for its function or that Rom2p and Bem2p have downstream targets of their own involved in microtubule function. Interestingly, the temperature-sensitive growth defect of rom2Δ was partially suppressed by disruption of SAC7, encoding a Rho1p-specific GAP (Schmidt et al., 1997); it will be interesting to test whether rom2Δ sac7Δ double mutants also exhibit enhanced growth defects in the presence of benomyl.
There are at least two mechanisms by which Rom2p may participate in microtubule function. One possibility is that Rho proteins or their regulators affect assembly or stability of contact sites between cytoplasmic microtubules and the actin cytoskeleton. Putative microtubule-capture sites have been implicated in proper positioning of mitotic spindles prior to asymmetric cell divisions in such diverse systems as Caenorhabditis elegans embryogenesis and yeast budding (Hyman 1989; Snyder et al., 1991; Palmer et al., 1992; Cheng et al., 1994). In yeast these sites are thought to lie, at least in part, at sites of polarized growth (Snyder et al., 1991; Page and Snyder, 1993). However, we detected no significant difference in nuclear migration or spindle orientation in strains lacking Rom2p function. Thus, if actin–microtubule contact sites are affected in rom2Δ cells, it does not result in significant defects in these particular processes.
Another possibility, not mutually exclusive from the first, is that Rom2p or Rho proteins may be directly or indirectly important for microtubule stability. The differences in growth observed in rom2Δ strains under conditions that destablize microtubules is consistent with this possibility. In mammalian cells, it has recently been reported that Rho activation is required to stabilize wound-oriented microtubules in cultured fibroblasts and that this stabilization occurs independently of the actin cytoskeleton (Cook and Gundersen, 1996). Furthermore, it has been demonstrated that microtubule depolymerization, by treatment of fibroblasts with nocodazole, activates Rho (Liu et al., 1996). The molecular basis for these events is unknown, but it is likely that they are controlled by Rho regulators, such as GEFs.
Conclusion
We have both cytologically and genetically characterized the Rho-GEF Rom2p. The localization of Rom2p to sites of Rho1p activity and the phenotypic similarities between rho1 and rom2 mutants provide further understanding of the mechanisms by which Rho proteins are spatially regulated. The function of Rom2p in mating-projection formation and its localization to projection tips supports evidence that components required for polarized morphogenesis are shared between the budding cycle and the mating pheromone-induced differentiation program in S. cerevisiae (reviewed in Madden et al., 1992; Chenevert, 1994). This conservation of function may apply to polarized cell growth events in other eukaryotic systems as well. Finally, we have demonstrated genetic interactions between a Rho GTPase cycle and elements of the microtubule cytoskeleton. These and future studies should provide insight into the ever expanding functions of Rho proteins and their regulators.
ACKNOWLEDGMENTS
We thank J. Barrett, N. Burns, and S. Erdman for critical comments on the manuscript, and J. Barrett for strains and constructs. D. Johnston and Y. Takai provided the CDC42 and RHO1 plasmids, respectively. B.D.M. was supported by an NIH training grant. This research was supported by NIH grants GM52197 and GM36494.
REFERENCES
- Adams A, Johnson DI, Longnecker RM, Sloat BF, Pringle JR. CDC42 and CDC43, two additional genes involved in budding and the establishment of cell polarity in Saccharomyces cerevisiae. J Cell Biol. 1990;111:131–142. doi: 10.1083/jcb.111.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams AEM, Pringle JR. Relationship of actin and tubulin distribution to bud growth in wild-type and morphogenetic-mutant Saccharomyces cerevisiae. J Cell Biol. 1984;98:934–945. doi: 10.1083/jcb.98.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbet NC, Schneider U, Helliwell SB, Stansfield I, Tuite MF, Hall MN. TOR controls translation initiation and early G1 progression in yeast. Mol Biol Cell. 1996;7:25–42. doi: 10.1091/mbc.7.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M, Botstein D. Two differentially regulated mRNAs with different 5′ ends encode secreted and intracellular forms of yeast invertase. Cell. 1982;28:145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- Chant J, Corrado K, Pringle JR, Herskowitz I. The yeast BUD5 gene, which encodes a putative GDP-GTP exchange factor, is necessary for bud-site selection and interacts with bud-formation gene BEM1. Cell. 1991;65:1213–1224. doi: 10.1016/0092-8674(91)90016-r. [DOI] [PubMed] [Google Scholar]
- Chenevert J. Cell polarization directed by extracellular cues in yeast. Mol Biol Cell. 1994;5:1169–1175. doi: 10.1091/mbc.5.11.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng N, Kirby C, Kemphues K. Control of cleavage spindle orientation in Caenorhabditis elegans: the role of the genes par-2 and par-3. Genetics. 1994;139:549–559. doi: 10.1093/genetics/139.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook, T.A., and Gundersen, G.G. (1996). Rho regulates microtubule stability in fibroblasts. Mol. Biol. Cell 7(suppl), 574a (Abstract).
- DiComo CJ, Arndt KT. Nutrients, via the TOR proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev. 1996;10:1904–1916. doi: 10.1101/gad.10.15.1904. [DOI] [PubMed] [Google Scholar]
- Drgonova J, Drgon T, Tanaka K, Kollar R, Chen G, Ford RA, Chan CSM, Takai Y, Cabib E. Rho1p, a yeast protein at the interface between cell polarization and morphogenesis. Science. 1996;272:277–279. doi: 10.1126/science.272.5259.277. [DOI] [PubMed] [Google Scholar]
- Edde B, Rossier J, Caer JL, Desbruyeres E, Gros F, Denoulet P. Posttranslational glutamylation of α-tubulin. Science. 1990;247:83–85. doi: 10.1126/science.1967194. [DOI] [PubMed] [Google Scholar]
- Endow SA, Kang SJ, Satterwhite LL, Rose MD, Skeen VP, Salmon ED. Yeast Kar3 is a minus-end microtubule motor protein that destabilizes microtubules preferentially at the minus ends. EMBO J. 1994;13:2708–2713. doi: 10.1002/j.1460-2075.1994.tb06561.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field C, Schekman R. Localized secretion of acid phosphatase reflects the pattern of cell surface growth in Saccharomyces cerevisiae. J Cell Biol. 1980;86:123–128. doi: 10.1083/jcb.86.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumoto Y, Kaibuchi K, Hori Y, Fujioka H, Araki S, Ueda T, Kikuchi A, Takai Y. Molecular cloning and characterization of a novel type of regulatory protein (GDI) for the rho proteins, ras p21-like small GTP-binding proteins. Oncogene. 1990;5:1321–1328. [PubMed] [Google Scholar]
- Gehrung S, Snyder M. The SPA2 gene of Saccharomyces cerevisiae is important for pheromone-induced morphogenesis and efficient mating. J Cell Biol. 1990;111:1451–1464. doi: 10.1083/jcb.111.4.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C, Fink GR. Guide to yeast genetics and molecular biology. Methods Enzymol. 1991;194:1–933. [PubMed] [Google Scholar]
- Hall A. Small GTP-binding proteins and the regulation of the actin cytoskeleton. Annu Rev Cell Biol. 1994;10:31–54. doi: 10.1146/annurev.cb.10.110194.000335. [DOI] [PubMed] [Google Scholar]
- Harlan JE, Hajduk PJ, Yoon HS, Fesik SW. Pleckstrin homology domains bind to phosphatidylinositol-4,5-bisphosphate. Nature. 1994;371:168–170. doi: 10.1038/371168a0. [DOI] [PubMed] [Google Scholar]
- Hart MJ, Eva A, Evans T, Aaronson SA, Cerione RA. Catalysis of guanine nucleotide exchange on the CDC42Hs protein by the dbl oncogene product. Nature. 1991;354:311–314. doi: 10.1038/354311a0. [DOI] [PubMed] [Google Scholar]
- Hart MJ, Eva A, Zangrilli D, Aaronson SA, Evans T, Cerione RA, Zheng Y. Cellular transformation and guanine nucleotide exchange activity are catalyzed by a common domain on the dbl oncogene product. J Biol Chem. 1994;269:63–65. [PubMed] [Google Scholar]
- Herskowitz I, Rine J, Strathern J. Mating-type determination and mating-type interconversion in Saccharomyces cerevisiae. In: Jones EW, Pringle JR, Broach JR, editors. The Molecular and Cellular Biology of the Yeast Saccharomyces. II. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1992. pp. 583–656. [Google Scholar]
- Hoyt MA, Toti L, Roberts BT. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66:507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- Huffaker TC, Thomas JH, Botstein D. Diverse effects of β-tubulin mutations on microtubule formation and function. J Cell Biol. 1988;106:1997–2010. doi: 10.1083/jcb.106.6.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman AA. Centrosome movement in the early divisions of Caenorhabditis elegans: a cortical site determining centrosome position. J Cell Biol. 1989;109:1185–1193. doi: 10.1083/jcb.109.3.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai J, Toh-e A, Matsui Y. Genetic analysis of the Saccharomyces cerevisiae RHO3 gene, encoding a Rho-type small GTPase, provides evidence for a role in bud formation. Genetics. 1996;142:359–369. doi: 10.1093/genetics/142.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Fukada Y, Murata K, Kimura A. Transformation of intact yeast cells with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs CW, Adams AE M, Szaniszlo PJ, Pringle JR. Functions of microtubules in the Saccharomyces cerevisiae cell cycle. J Cell Biol. 1988;107:1409–1426. doi: 10.1083/jcb.107.4.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DI, Pringle JR. Molecular characterization of CDC42 a Saccharomyces cerevisiae gene involved in the development of cell polarity. J Cell Biol. 1990;111:143–152. doi: 10.1083/jcb.111.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin JV, Adams AEM. Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces. J Cell Biol. 1984;98:922–933. doi: 10.1083/jcb.98.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Francisco L, Chen G, Marcotte E, Chan CS M. Control of cellular morphogenesis by the Ipl2/Bem2 GTPase-activating protein: possible role of protein phosphorylation. J Cell Biol. 1994;127:1381–1394. doi: 10.1083/jcb.127.5.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA, Ferguson KM, Schlessinger J. PH domains: diverse sequences with a common fold recruit signaling molecules to the cell surface. Cell. 1996;85:621–624. doi: 10.1016/s0092-8674(00)81022-3. [DOI] [PubMed] [Google Scholar]
- Lew D, Reed S. Cell cycle control of morphogenesis in budding yeast. Curr Opin Genet Dev. 1995;5:17–23. doi: 10.1016/s0959-437x(95)90048-9. [DOI] [PubMed] [Google Scholar]
- Li R, Murray AW. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- Li R, Zheng Y, Drubin D. Regulation of cortical actin cytoskeleton assembly during polarized cell growth in budding yeast. J Cell Biol. 1995;128:599–615. doi: 10.1083/jcb.128.4.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, B.P., Chrzanowska-Wodnicka, M., and Burridge, K. (1996). Rho mediates nocodazole-induced stress fiber and focal adhesion formation. Mol. Biol. Cell 7(suppl), 525a. (Abstract).
- Madaule P, Axel R, Myers AM. Characterization of two members of the rho gene family from the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1987;84:779–784. doi: 10.1073/pnas.84.3.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madden K, Costigan C, Snyder M. Cell polarity and morphogenesis in Saccharomyces cerevisiae. Trends Cell Biol. 1992;2:22–29. doi: 10.1016/0962-8924(92)90140-i. [DOI] [PubMed] [Google Scholar]
- Masuda T, Tanaka K, Nonaka H, Yamochi W, Maeda A, Takai Y. Molecular cloning and characterization of yeast rho GDP dissociation inhibitor. J Biol Chem. 1994;269:19713–19718. [PubMed] [Google Scholar]
- Matsui Y, Toh-e A. Yeast RHO3 and RHO4 ras super-family genes are necessary for bud growth, and their defect is suppressed by a high dose of bud formation genes CDC42 and BEM1. Mol Cell Biol. 1992;12:5690–5699. doi: 10.1128/mcb.12.12.5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister A, Tate SS, Griffith OW. γ-Glutamyl transpeptidase. Methods Enzymol. 1981;77:237–253. doi: 10.1016/s0076-6879(81)77032-0. [DOI] [PubMed] [Google Scholar]
- Meluh PB, Rose MD. KAR3, a kinesin-related gene required for yeast nuclear fusion. Cell. 1990;60:1029–1941. doi: 10.1016/0092-8674(90)90351-e. [DOI] [PubMed] [Google Scholar]
- Musacchio A, Gibson T, Rice P, Thompson J, Saraste M. The PH domain: a common piece in the structural patchwork of signalling proteins. Trends Biochem Sci. 1993;18:343–348. doi: 10.1016/0968-0004(93)90071-t. [DOI] [PubMed] [Google Scholar]
- Novick P, Botstein D. Phenotypic analysis of temperature-sensitive yeast actin mutants. Cell. 1985;40:405–416. doi: 10.1016/0092-8674(85)90154-0. [DOI] [PubMed] [Google Scholar]
- Ohga N, Kikuchi A, Ueda T, Yamamoto J, Takai Y. Rabbit intestine contains a protein that inhibits the dissociation of GDP from and the subsequent binding of GTP to rhoB p20, a ras p21-like GTP-binding protein. Biochem Biophys Res Commun. 1989;163:1523–33. doi: 10.1016/0006-291x(89)91153-4. [DOI] [PubMed] [Google Scholar]
- Ono T, Suzuki T, Anraku Y, Iida H. The MID2 gene encodes a putative integral membrane protein with a Ca2+-binding domain and shows mating pheromone-stimulated expression in Saccharomyces cerevisiae. Gene. 1994;151:203–208. doi: 10.1016/0378-1119(94)90657-2. [DOI] [PubMed] [Google Scholar]
- Ozaki K, Tanaka K, Imamura H, Hihara T, Kameyama T, Nonaka H, Hirano H, Matsuura Y, Takai Y. Rom1p and Rom2p are GDP/GTP exchange proteins (GEPs) for the Rho1p small GTP binding protein in Saccharomyces cerevisiae. EMBO J. 1996;15:2196–2207. [PMC free article] [PubMed] [Google Scholar]
- Page B, Snyder M. CIK1: a developmentally regulated spindle pole body-associated protein important for microtubule functions in Saccharomyces cerevisiae. Genes Dev. 1992;6:1414–1429. doi: 10.1101/gad.6.8.1414. [DOI] [PubMed] [Google Scholar]
- Page B, Snyder M. Chromosome segregation in yeast. Annu Rev Microbiol. 1993;47:231–261. doi: 10.1146/annurev.mi.47.100193.001311. [DOI] [PubMed] [Google Scholar]
- Page BD, Satterwhite LL, Rose MD, Snyder M. Localization of the KAR3 kinesin heavy chain-like protein requires the CIK1 interacting protein. J Cell Biol. 1994;124:507–519. doi: 10.1083/jcb.124.4.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer RE, Sullivan DS, Huffaker T, Koshland D. Role of astral microtubules and actin in spindle orientation and migration in the budding yeast, Saccharomyces cerevisiae. J Cell Biol. 1992;119:583–593. doi: 10.1083/jcb.119.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellman D, Bagget M, Tu H, Fink GR. Two microtubule-associated proteins required for anaphase spindle movement in Saccharomyces cerevisiae. J Cell Biol. 1995;130:1373–1385. doi: 10.1083/jcb.130.6.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson J, Zheng Y, Bender L, Myers A, Cerione R, Bender A. Interactions between the bud emergence proteins Bem1p and Bem2p and Rho-type GTPases in yeast. J Cell Biol. 1994;127:1395–1406. doi: 10.1083/jcb.127.5.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle J, Adams AEM, Drubin DG, Haarer BK. Immunofluorescence methods for yeast. In: Guthrie C, Fink GR, editors. Guide to Yeast Genetics and Molecular Biology. San Diego: Academic Press; 1991. pp. 565–601. [Google Scholar]
- Pringle JR, Bi E, Harkins HA, Zahner JE, Virgilio CD, Chant J, Corrado K, Fares H. Establishment of cell polarity in yeast. Cold Spring Harbor Symp Quant Biol. 1995;60:729–744. doi: 10.1101/sqb.1995.060.01.079. [DOI] [PubMed] [Google Scholar]
- Qadota H, Python CP, Inoue SB, Arisawa M, Anraku Y, Zheng Y, Watanabe T, Levin DE, Ohya Y. Identification of yeast Rho1 GTPase as a regulatory subunit of 1,3-β-glucan synthase. Science. 1996;272:279–281. doi: 10.1126/science.272.5259.279. [DOI] [PubMed] [Google Scholar]
- Read EB, Okamura HH, Drubin DG. Actin- and tubulin-dependent functions during Saccharomyces cerevisiae mating projection formation. Mol Biol Cell. 1992;3:429–444. doi: 10.1091/mbc.3.4.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijo RA, Cooper EM, Beagle GJ, Huffaker TC. Systematic mutational analysis of the yeast β-tubulin gene. Mol Biol Cell. 1994;5:29–43. doi: 10.1091/mbc.5.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley A. Rho-related proteins: actin cytoskeleton and cell cycle. Curr Opin Genet Dev. 1995;5:24–30. doi: 10.1016/s0959-437x(95)90049-7. [DOI] [PubMed] [Google Scholar]
- Russo P, Kalkkinen N, Sareneva H, Paakkola J, Makarow M. A heat shock gene from Saccharomyces cerevisiae encoding a secretory glycoprotein. Proc Natl Acad Sci USA. 1992;89:3671–3675. doi: 10.1073/pnas.89.9.3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamuro D, Yamazoe M, Matsuda Y, Kangawa K, Taniguchi N, Matsuo H, Yoshikawa H, Ogasowara N. The primary structure of human gamma-glutamyl transpeptidase. Gene. 1988;73:1–9. doi: 10.1016/0378-1119(88)90307-1. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd ed. Plainview, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Santos B, Snyder M. Targeting of chitin synthase 3 to polarized growth sites in yeast requires Chs5p and Myo2p. J Cell Biol. 1997;136:95–110. doi: 10.1083/jcb.136.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders W, Hornack D, Lengyel V, Deng C. The Saccharomyces cerevisiae kinesin-related motor Kar3p acts at preanaphase spindle poles to limit the number and length of cytoplasmic microtubules. J Cell Biol. 1997;137:417–431. doi: 10.1083/jcb.137.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Bickle M, Beck T, Hall MN. The yeast phosphatidylinositol kinase homolog TOR2 activates RHO1 and RHO2 via the exchange factor ROM2. Cell. 1997;88:531–542. doi: 10.1016/s0092-8674(00)81893-0. [DOI] [PubMed] [Google Scholar]
- Schmidt A, Kunz J, Hall MN. TOR2 is required for organization of the actin cytoskeleton in yeast. Proc Natl Acad Sci USA. 1996;93:13780–13785. doi: 10.1073/pnas.93.24.13780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider BL, Seufert W, Steiner B, Yang QH, Futcher AB. Use of PCR epitope tagging for protein tagging in Saccharomyces cerevisiae. Yeast. 1995;11:1265–1274. doi: 10.1002/yea.320111306. [DOI] [PubMed] [Google Scholar]
- Snyder M, Gehrung S, Page BD. Temporal and genetic control of cell polarity in Saccharomyces cerevisiae. J Cell Biol. 1991;114:515–532. doi: 10.1083/jcb.114.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Bretscher A. The rho-GAP encoded by BEM2 regulates cytoskeletal structure in budding yeast. Mol Biol Cell. 1995;6:1011–1024. doi: 10.1091/mbc.6.8.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamochi W, Tanaka K, Nonaka H, Maeda A, Musha T, Takai Y. Growth site localization of Rho1 small GTP-binding protein and its involvement in bud formation in Saccharomyces cerevisiae. J Cell Biol. 1994;125:1077–1093. doi: 10.1083/jcb.125.5.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Cerione R, Bender A. Control of the yeast bud-site assembly GTPase Cdc42. Catalysis of guanine nucleotide exchange by Cdc24 and stimulation of GTPase activity by Bem3. J Biol Chem. 1994;269:2369–2372. [PubMed] [Google Scholar]
- Zheng Y, Hart MJ, Shinjo K, Evans T, Bender A, Cerione RA. Biochemical comparisons of the Saccharomyces cerevisiae Bem2 and Bem3 proteins. Delineation of a limit Cdc42 GTPase-activating protein domain. J Biol Chem. 1993;268:24629–24634. [PubMed] [Google Scholar]
- Ziman M, Preuss D, Mulholland J, O’Brien JM, Botstein D, Johnson DI. Subcellular localization of Cdc42p, a Saccharomyces cerevisiae GTP-binding protein involved in the control of cell polarity. Mol Biol Cell. 1993;4:1307–1316. doi: 10.1091/mbc.4.12.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]