Summary
The Mycobacterium tuberculosis PE_PGRS multigene family is thought to be involved in antigenic variation, which can be generated by differential regulation of expression and a high frequency of genetic polymorphism. PE_PGRS16 and PE_PGRS26 are inversely regulated during persistent M. tuberculosis infection, suggesting that differential regulation of the expression of these two PE_PGRS genes may have a role in latency. To understand how genetic diversity, in addition to differential regulation, contributes to antigenic variability, we investigated the sequence variations in the PE_PGRS16 and PE_PGRS26 genes among 200 clinical M. tuberculosis strains, in comparison to the sequenced laboratory strain H37Rv, using PCR and DNA sequencing. Among the 200 strains, 102 (51%) and 100 (50%) had sequence variations within the PE_PGRS16 gene and the PE_PGRS26 gene, respectively. In-frame insertions and deletions, frameshifts, and SNPs were observed in both the PE_PGRS16 gene and the PE_PGRS26 gene. However, the frequency of frameshifts and in-frame deletions differed between the two PE_PGRS genes. Examining the profile of the PE_PGRS16, PE_PGRS26, and the previously investigated PE_PGRS33 amino acid sequences for each of the 200 strains, 72 different profiles were observed with frequencies ranging from 0.5% to 13%. In conclusion, a remarkable level of genetic diversity exists in the PE_PGRS16 and PE_PGRS26 genes of M. tuberculosis clinical strains. The significant sequence variations in the two PE_PGRS genes observed in this study could impact the function of these two PE_PGRS proteins and be associated with differences in the ability of the tubercle bacilli to remain persistent within the host.
Introduction
Mycobacterium tuberculosis, killing approximately 2 million people worldwide each year, has been called the most successful human pathogen. Controlling tuberculosis will require a better understanding of the mechanisms which allow M. tuberculosis to evade the immune system and remain persistent in the host. Sequencing of the genomes of M. tuberculosis strains has provided important insights into possible mechanisms of persistence, including the discovery of the multigene family of ~ 60 genes named PE_PGRS that is thought to be involved in antigenic variation. The PE domain of the PE_PGRS protein has a proline-glutamic acid sequence near the amino terminus and the PGRS domain of the protein varies in size and contains many repeats of alanine and glycine 1. There is evidence that at least some members of this gene family are expressed on the cell surface during M. tuberculosis infection and recognized by the host immune system 2 – 5. The maintenance of this large multigene family in the M. tuberculosis genome suggests that these genes are important to the success of the organism, perhaps because the variability of the PE_PGRS protein surface antigens may contribute to the ability of M. tuberculosis to persist in the face of the host immune system.
The regulation of gene expression is one mechanism for generating antigenic diversity. There is evidence that M. tuberculosis PE_PGRS genes are variably expressed in different conditions and during different time points of infection 6 – 8. In a study of persistent M. tuberculosis infection in a mouse model, PE_PGRS16 and PE_PGRS26 were inversely regulated, with expression of PE_PGRS16 being significantly up-regulated and expression of PE_PGRS26 being significantly down-regulated, suggesting that differential regulation of these two PE_PGRS genes may have a role in latency and that the inverse expression of these two genes could potentially serve as a marker of latent infection 7.
Antigenic variation of an organism can also be generated by a high level of genetic variability of the genes that encode antigens. Thus, to understand the full scope of surface antigen variability generated by the PE_PGRS gene family, it is important to investigate the genetic diversity of these genes among clinical isolates. The sequence variations in one member of this gene family, PE_PGRS33, have been characterized for 123 clinical M. tuberculosis strains and included single nucleotide polymorphisms (SNPs), insertions, deletions, and a frameshift mutation. These sequence variations were observed in different combinations resulting in 23 different PE_PGRS33 alleles 9. Furthermore, in a population-based study of 649 clinical M. tuberculosis isolates, patients infected with M. tuberculosis isolates having large changes to the PE_PGRS33 protein were 1.9 times more likely to belong to a cluster of tuberculosis cases, defined by M. tuberculosis genotyping, and 1.6 times more likely to lack cavitations in the lungs than were patients infected with M. tuberculosis isolates having no or minimal change to the PE_PGRS33 protein. This suggests that PE_PGRS33 may have an important role in M. tuberculosis persistence 10.
To extend our knowledge of the genetic diversity of M. tuberculosis generated by the PE_PGRS genes and to understand how genetic diversity, in addition to differential regulation, contributes to antigenic variability, we investigated the sequence variations within the PE_PGRS16 and PE_PGRS26 genes among 200 clinical M. tuberculosis strains. The frequency of different types of sequence variations was compared between the PE_PGRS16 gene and the PE_PGRS26 gene and the potential antigenic diversity of the 200 strains generated by sequence variations in three PE_PGRS genes, the PE_PGRS16 and PE_PGRS26 genes and the previously investigated PE_PGRS33 gene, was examined.
Materials and Methods
M. tuberculosis strains
A study sample of 200 M. tuberculosis strains was selected from 705 isolates collected in Arkansas between 1996 and 2000. Strains were selected based on the isolate genotyping data that were available from the Mycobacteriology Research Laboratory at the Central Arkansas Veterans Healthcare Center to represent the broad range of strains found in this population-based isolate collection. The IS6110 copy numbers of each strain ranged from one to 22. The sample included 94 strains representing 94 clusters and 106 unique strains based on a combination of IS6110 and pTBN12 fingerprinting data 11 – 13. The sample contained strains from all three Principal Genetic Groups (PGG) 14 with 20 (10%), 130 (65%), and 50 (25%) of the 200 strains belonging to PGG1, PGG2, and PGG3, respectively. The PE_PGRS33 gene sequences for these 200 strains were previously determined 9.
PCR of the PE_PGRS16 and PE_PGRS26 genes
The PE_PGRS16 gene and the PE_PGRS26 gene of each strain were PCR amplified for DNA sequencing. The primers used for the PCR amplification of the PE_PGRS16 gene were PE_PGRS16-F and PE_PGRS16-R and the primers used for the PCR amplification of the PE_PGRS26 gene were PE_PGRS26-F and PE_PGRS26-R (Table 1). The primers were located in the regions flanking the respective target gene and were determined to be specific for the amplification of the target gene using the BLAST program of the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/BLAST). The PE_PGRS16 gene was PCR amplified using the EPICENTER MasterAmp™ Extra-Long PCR Kit (EPICENTER, Madison, WI). Each standard 50 µl reaction consisted of 25 µl of 2X Premix #7 (containing PCR buffer and dNTPs) from the kit, 20 pmol of each primer in 2 µl, 2.5 U MasterAmp™ Extra-Long DNA Polymerase in 1 µl, 50 ng DNA template in 5 µl, and 15 µl PCR grade water. The thermocycling program used was 1 cycle at 94°C for 3 minutes, 31 cycles of 94°C for 1 minute, 68°C for 1 minute, and 72°C for 14.5 minutes, and one final cycle at 72°C for 10 minutes. The PE_PGRS26 gene was PCR amplified using the BD Advantage™-GC 2 PCR Kit (BD Biosciences Clontech, Palo Alto, CA). Each standard 50 µl reaction consisted of 10 µl of 5X reaction buffer, 5 µl of GC Melt, 20 pmol of each primer in 2 µl, 1 µl of a 50X dNTP mix, 1 µl of 50X BD Advantage™ 2 Polymerase Mix, 2 µl DNA solution containing 20 ng DNA template, and 27 µl PCR grade water. The thermocycling program used was 1 cycle at 94°C for 1 minute, 30 cycles of 94°C for 30 seconds, 61°C for 30 seconds, and 72°C for 2.5 minutes, and a final cycle at 72°C for 10 minutes. The PCR products were examined by 0.8% (w/v) agarose gel electrophoresis performed in 1X TBE buffer.
Table 1.
Primers used in this study for the PCR amplification and DNA sequencing of the M. tuberculosis PE_PGRS16 and PE_PGRS26 genes
| Primer | Sequence | Positiona |
|---|---|---|
| PE_PGRS16-F | 5' - GGG ATC GGC GAC GCT ACC AAC CAA - 3' | 164 bp left of start |
| PE_PGRS16-R | 5' - GCC CGC TGC AGA CGC CCC TTC C - 3' | 47 bp right of end |
| PE_PGRS16-F2 | 5' - CGG GCT GCT ATT CGG CAA CGG T - 3' | 633 |
| PE_PGRS16-R2 | 5' - TCG CGT TGG TCA ACG TGG TGC TG - 3' | 1972 |
| PE_PGRS16-SeqR | 5' - ATC ACT TCC CGC GGT GGC CAT - 3' | 1209 |
| PE_PGRS16-SeqR2 | 5' - GGC ACC ACC GTT GCC GAA TAG C - 3' | 660 |
| PE_PGRS16-SeqF | 5' - TCG GCG GAA TTA ACG GCA CGT TCG - 3' | 1832 |
| PE_PGRS26-F | 5' - GCT GTT CGT TAC CGG CAT CTG - 3' | 206 bp left of end |
| PE_PGRS26-R | 5' - AGA CCT GCA TTT GCA GCA GTC - 3' | 117 bp right of start |
| PE_PGRS26-SeqF | 5' - GCT GCT GCC ATT GCC GAT GGT C - 3' | 1167 |
| PE_PGRS26-SeqR | 5' - ACG GCG GGG ATG CCG GAA TGT T - 3' | 476 |
Positions of primers are based on the M. tuberculosis strain H37Rv genome sequence (NC_000962) and are in relation to the coding strand for the PE_PGRS16 gene (rv0977) and the PE_PGRS26 gene (rv1441c). The positions of the primers used for PCR (PE_PGRS16-F, PE_PGRS16-R, PE_PGRS26-F, and PE_PGRS26-R) are outside of the target gene. The positions of all other primers are within the target gene sequence.
Automated DNA sequencing
The PCR products were sequenced to identify any insertions, deletions, or SNPs in the DNA sequences of the PE_PGRS16 and the PE_PGRS26 genes. The PCR products used for DNA sequencing were purified using QIAquick® PCR Purification Kit following the manufacturer’s instructions (QIAGEN Inc., Stanford, Valencia, CA). DNA sequencing of the PE_PGRS16 gene was performed first with the PE_PGRS16-F and PE_PGRS16-R primers that were used for the PCR amplification and then with the primers PE_PGRS16-F2 and PE_PGRS16-R2, located within the PE_PGRS16 gene. DNA sequencing of the PE_PGRS26 gene was first performed with PE_PGRS26-F and PE_PGRS26-R primers that were used for the PCR amplification. All SNPs were confirmed by double strand sequencing. Primers used for the PE_PGRS16 SNP confirmation were PE_PGRS16-SeqR, PE_PGRS16-SeqR2, and PE_PGRS-SeqF. Primers used for the PE_PGRS26 SNP confirmation were PE_PGRS26-SeqR and PE_PGRS26-SeqF (Table 1). Sequencing was performed in Applied Biosystems DNA Sequencers (Models 3700 and 3730 sequencers) at the Sequencing Core of the University of Michigan.
Sequence data analysis
The PE_PGRS16 and PE_PGRS26 gene sequences were compared to that of the M. tuberculosis reference strain H37Rv using the BLAST program of the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/BLAST). The effect of each identified insertion, deletion, and SNP on the predicted amino acid sequence of the PE_PGRS16 and the PE_PGRS26 proteins was then determined. The locations of the sequence variations of each PE_PGRS gene were mapped on to the respective gene sequence map of strain H37Rv to illustrate patterns in the distribution of the sequence variations. The profile of PE_PGRS16, PE_PGRS26, and the previously investigated PE_PGRS33 9 amino acid sequences for each of the 200 clinical M. tuberculosis strains was then examined to investigate the potential antigenic diversity generated by sequence variations in these three PE_PGRS genes.
The relationships among the sequence variations within the PE_PGRS16 and PE_PGRS26 genes were examined by constructing a branched diagram of the PE_PGRS16 alleles and the PE_PGRS26 alleles, respectively. Each branched diagram was constructed by connecting each observed PE_PGRS allele with the most similar observed allele or alleles, based on the presence or absence of sequence variations. In-frame deletions and insertions, frameshifts, and SNPs were included in the analysis and each was scored as one sequence variation. The length of the lines connecting the observed PE_PGRS alleles represents the number of differing sequence variations between the two alleles.
Results
PE_PGRS16 gene sequence variations
The genetic diversity of the PE_PGRS16 gene, the expression of which was up-regulated during the course of persistent infection 7, was investigated. Among the 200 strains, 98 (49.0%) had PE_PGRS16 gene sequences identical to that of strain H37Rv and 102 (51.0%) had one or more sequence variations within the PE_PGRS16 gene sequence. Thirty-three different sequence variations were observed including two in-frame insertions, 11 in-frame deletions, 17 SNPs, and three frameshift mutations resulting from a single nucleotide insertion or deletion (Table 2).
Table 2.
Frequencies and positions of 33 PE_PGRS16 gene sequence variations observed among 200 clinical M. tuberculosis strains.
| Sequence variation | Position | Genomic change | Amino acid change | No. (%) of strains n=200 | Principal Genetic Group | |
|---|---|---|---|---|---|---|
| H37Rv-typea | 98 (49.0) | 1, 2, & 3 | ||||
| Deletions | D1 | 1017 – 1082 | 66 bp deletion | 5 ( 2.5) | 1, 2, & 3 | |
| D2 | 1809 – 1814 | 6 bp deletion | 4 ( 2.0) | 1 | ||
| D3 | 1579 – 1830 | 252 bp deletion | 3 ( 1.5) | 2 | ||
| D4 | 1036–1083 | 48 bp deletion | 1 ( 0.5) | 2 | ||
| D5 | 1401 – 1550 | 150 bp deletion | 1 ( 0.5) | 1 | ||
| D6 | 1501 – 1641 | 141 bp deletion | 1 ( 0.5) | 2 | ||
| D7 | 1063 – 1086 | 24 bp deletion | 1 ( 0.5) | 2 | ||
| D8 | 1067 – 1069 | 3 bp deletion | 1 ( 0.5) | 3 | ||
| D9 | 1219 – 1221 | 3 bp deletion | 1 ( 0.5) | 2 | ||
| D10 | 1091 – 1162 | 72 bp deletion | 1 ( 0.5) | 2 | ||
| D11 | 1480 – 1497 | 18 bp deletion | 1 ( 0.5) | 3 | ||
| Insertions | I1 | 1797 | 171 bp insertion | 2 ( 1.0) | 3 | |
| I2 | 1376–1393 | 18 bp insertion | 1 ( 0.5) | 3 | ||
| Frameshifts | FS1b | 1969 | 1 bp insertion | Premature stop | 72 (36.0) | 2 |
| FS2 | 1609 | 1 bp deletion | Premature stop | 7 ( 3.5) | 3 | |
| FS3 | 1608 | 1 bp insertion | Premature stop | 1 ( 0.5) | 2 | |
| nsSNPs | S1 | 881 | a → g | Asn → Ser | 23 (11.5) | 2 |
| S2 | 605 | g → c | Gly → Ala | 2 ( 1.0) | 2 | |
| S3 | 2671 | a → c | Thr → Pro | 2 ( 1.0) | 1 | |
| S4 | 1262 | g → a | Gly → Glu | 1 ( 0.5) | 2 | |
| S5 | 22 | c → t | Pro → Ser | 1 ( 0.5) | 1 | |
| S6 | 1990 | g → a | Val → Met | 1 ( 0.5) | 2 | |
| S7 | 1099 | g → c | Ala → Pro | 1 ( 0.5) | 3 | |
| S8 | 1100 | c → a | Ala → Glu | 1 ( 0.5) | 3 | |
| S9 | 1136 | t → c | Ile → Thr | 1 ( 0.5) | 3 | |
| S10 | 722 | c → t | Ala → Val | 1 ( 0.5) | 2 | |
| S11 | 2557 | a → c | Ser → Arg | 1 ( 0.5) | 2 | |
| S12 | 2122 | g → t | Gly → Trp | 1 ( 0.5) | 1 | |
| sSNPs | S13 | 1944 | c → t | Gly → Gly | 2 ( 1.0) | 1 |
| S14 | 429 | c → a | Gly → Gly | 1 ( 0.5) | 2 | |
| S15 | 1368 | c → t | Thr → Thr | 1 ( 0.5) | 1 | |
| S16 | 2676 | c → t | Ser → Ser | 1 ( 0.5) | 1 | |
| S17 | 2736 | c → t | Tyr → Tyr | 1 ( 0.5) | 2 |
The PE_PGRS16 gene of M. tuberculosis strain 210 is identical to that of strain H37Rv
Sequence variation present in M. tuberculosis strain CDC1551 in addition to a one bp insertion at position 2075
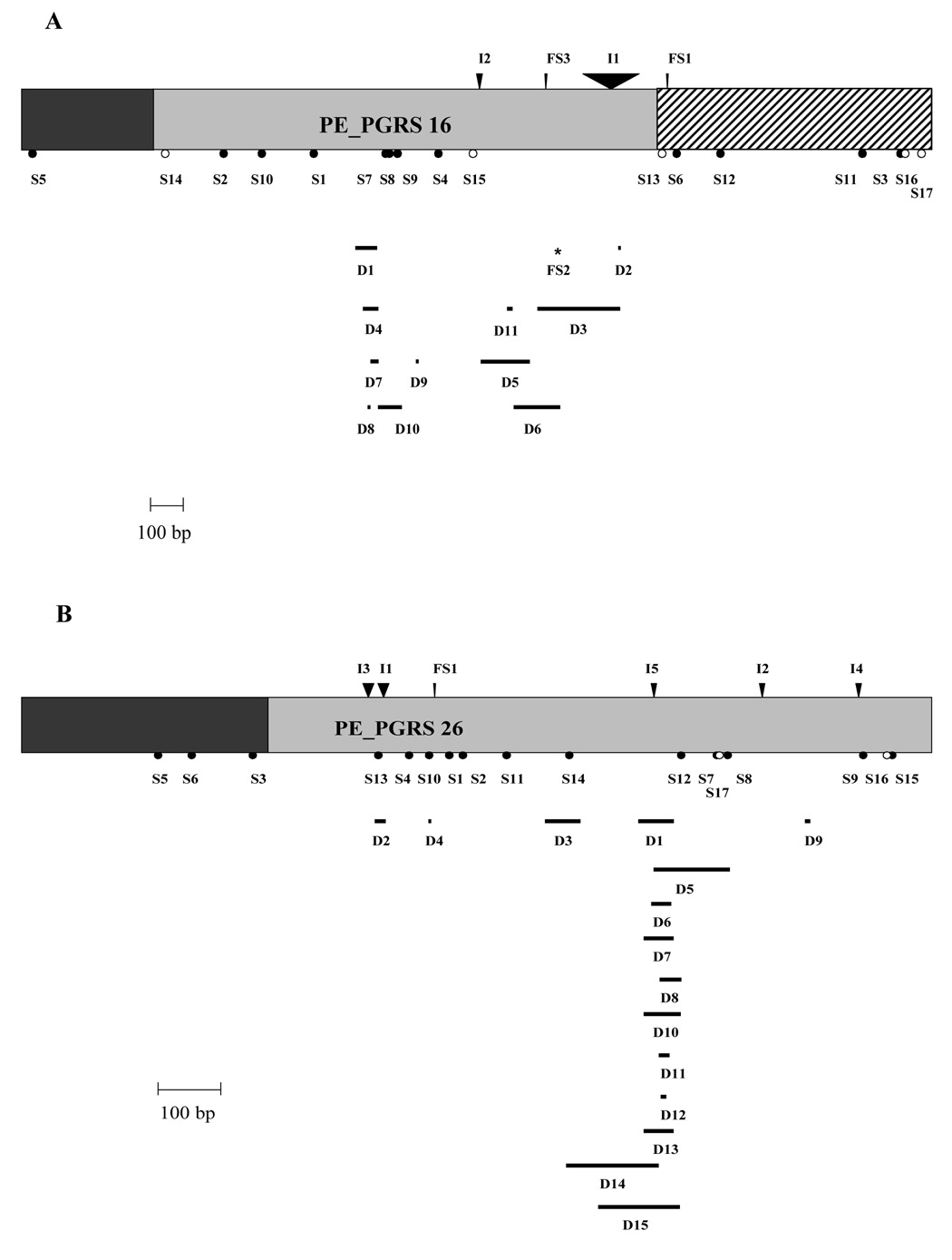
The two in-frame insertions, one 18 bp and one 171 bp, were repeats of the region of PE_PGRS16 gene sequence adjacent to the insertion site. The 11 in-frame deletions ranged in size from three bp to 252 bp. The 17 SNPs included 12 non-synonymous SNPs and five synonymous SNPs (Table 2). All the in-frame insertions and in-frame deletions and two of the three frameshift mutations were located within the PGRS domain of the PE_PGRS16 gene. The remaining frameshift mutation, FS1, was located where the PGRS domain joins with an atypical sequence at the carboxy-terminus. Of the 17 SNPs, one was in the PE domain, 11 were in the PGRS domain, and seven were in the atypical domain of the PE_PGRS16 gene (Figure 1A).
Figure 1.
Map of the positions of different sequence variations found in the M. tuberculosis PE_PGRS16 (Panel A) and PE_PGRS26 (Panel B) genes. The PE domain is shaded in dark grey and the PGRS domain is shaded in light grey. The striped region in Panel A represents an additional atypical sequence at the carboxy-terminus of PE_PGRS16. Triangles represent insertions, bold lines represent deletions, open circles represent synonymous SNPs, and solid circles represent non-synonymous SNPs. The asterisk in Panel A represents a one bp deletion.
Seventy-nine (39.5%) of the 200 strains (77.5% of the 102 strains having PE_PGRS16 gene sequence variations) had a frameshift mutation in the PE_PGRS16 gene that would result in a premature stop codon. The FS1 frameshift would result in a change in amino acid sequence for 86 amino acids and then a loss of the last 181 amino acids due to the premature stop codon, affecting a total of 29% of the M. tuberculosis H37Rv PE_PGRS16 amino acid sequence that is 923 amino acid long in the atypical domain at the carboxy-terminus. The FS2 frameshift and the FS3 frameshift would both affect 42% of the M. tuberculosis H37Rv PE_PGRS16 amino acid sequence. The FS2 frameshift would result in a change in amino acid sequence for 115 amino acids and then a loss of the last 270 amino acids due to the premature stop codon while the FS3 frameshift would result in a change in amino acid sequence for 27 amino acids and then a loss of the last 358 amino acids due to the premature stop codon.
PE_PGRS16 alleles
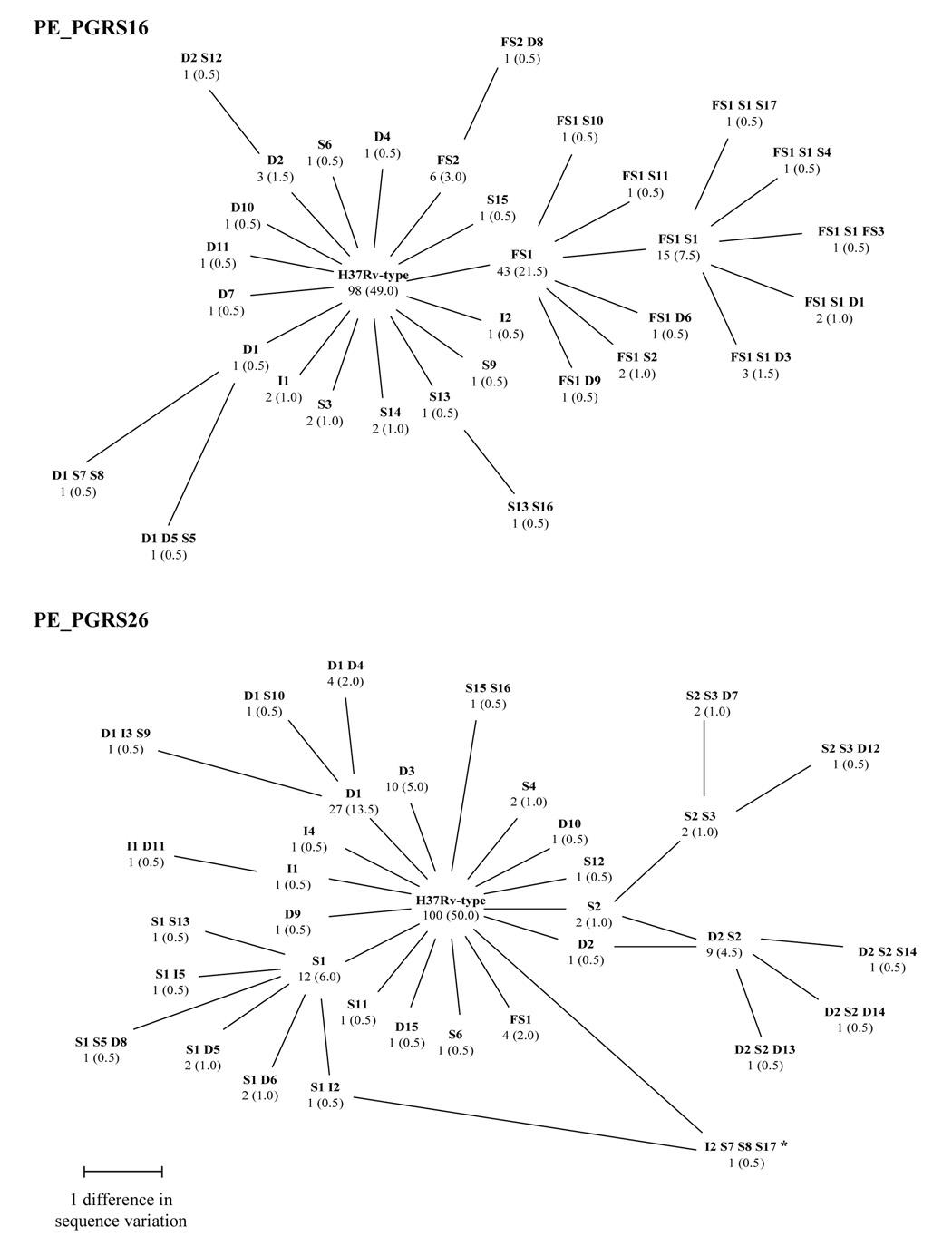
The PE_PGRS16 allele observed with the highest frequency in the sample was the H37Rv-type allele, with 98 (49.0%) of the 200 strains having this allele. Among the 102 (51.0%) of the 200 strains having at least one sequence variation, 32 different PE_PGRS16 alleles were observed, resulting from different combinations of sequence variations. Six of the 102 strains having a non-H37Rv-type PE_PGRS16 allele had only synonymous SNPs and therefore would have an H37Rv-type amino acid sequence. The most frequently observed non-H37Rv-type allele was the FS1 allele, having a frameshift mutation that would result in a premature stop codon, and this allele was observed in 43 (21.5%) of the 200 strains. The other 31 PE_PGRS16 alleles were each observed in less than 10% of the strains (Figure 2).
Figure 2.
Branched diagrams showing the relationships among the different alleles of the PE_PGRS16 and PE_PGRS26 genes based on the in-frame deletions, in-frame insertions, frameshifts, and SNPs identified within the two genes' sequences. For each of the observed PE_PGRS16 and PE_PGRS26 alleles, the number and percentage of the 200 clinical M. tuberculosis strains having a given allele are indicated. The length of the lines connecting the observed PE_PGRS alleles represents the number of differing sequence variations between the two alleles. The PE_PGRS16 gene sequence variations are described in Table 2 and the PE_PGRS26 gene sequence variations are described in Table 3.
* This PE_PGRS26 allele shares only the I2 insertion with the S1 I2 allele.
A branched diagram of the 32 non-H37Rv-type PE_PGRS16 alleles and the H37Rv-type allele illustrates the relationships among the PE_PGRS16 sequence variations (Figure 2). Twelve PE_PGRS16 alleles had the FS1 frameshift and six of these 12 alleles had the FS1 frameshift and the S1 non-synonymous SNP together. The S13 synonymous SNP, the D2 deletion, and the FS2 frameshift were each present in two PE_PGRS16 alleles. Four different PE_PGRS16 alleles had the D1 deletion. However, the D1 FS1 S1 allele was more similar to the FS1 S1 allele than to the other three alleles having the D1 deletion and therefore these four alleles are not all connected in the branched diagram.
PE_PGRS26 gene sequence variations
The genetic diversity of the PE_PGRS26 gene, the expression of which was down-regulated during the course of persistent infection 7, was then examined. One hundred (50.0%) of the 200 strains had PE_PGRS26 gene sequences identical to that of H37Rv. Among the remaining 100 strains, which had one or more sequence variations within the PE_PGRS26 gene sequence, 38 different sequence variations were observed. These 38 sequence variations included five in-frame insertions, 15 in-frame deletions, 17 SNPs, and one frameshift mutation resulting from a one bp insertion. All five in-frame insertions, either nine or 18 bp, were repeats of the region of PE_PGRS26 gene sequence adjacent to the insertion site. The 15 in-frame deletions ranged in size from three bp to 150 bp. The 17 SNPs included 15 non-synonymous SNPs and two synonymous SNPs. The frameshift mutation, observed in only 4 (2%) of the 200 strains, would result in a premature stop codon (Table 3).
Table 3.
Frequencies and positions of 38 PE_PGRS26 gene sequence variations observed among 200 clinical M. tuberculosis strains.
| Sequence variation | Position | Genomic change | Amino acid change | No. (%) of strains n=200 | Principal Genetic Group | |
|---|---|---|---|---|---|---|
| H37Rv-type | 100 (50.0) | 2 & 3 | ||||
| Deletions | D1 | 1000 – 1056 | 57 bp deletion | 33 (16.5) | 2 | |
| D2a | 567 – 584 | 18 bp deletion | 13 ( 6.5) | 1 | ||
| D3 | 842 – 898 | 57 bp deletion | 10 ( 5.0) | 3 | ||
| D4 | 652 – 654 | 3 bp deletion | 4 ( 2.0) | 2 | ||
| D5 | 1020 – 1142 | 123 bp deletion | 2 ( 1.0) | 2 | ||
| D6 | 1016 – 1048 | 33 bp deletion | 2 ( 1.0) | 2 | ||
| D7 | 1034 – 1069 | 36 bp deletion | 2 ( 1.0) | 1 | ||
| D8 | 1004 – 1051 | 48 bp deletion | 1 ( 0.5) | 2 | ||
| D9 | 1267 – 1275 | 9 bp deletion | 1 ( 0.5) | 2 | ||
| D10 | 1005 – 1064 | 60 bp deletion | 1 ( 0.5) | 2 | ||
| D11 | 1030 – 1047 | 18 bp deletion | 1 ( 0.5) | 3 | ||
| D12 | 1034 – 1042 | 9 bp deletion | 1 ( 0.5) | 1 | ||
| D13 | 1005 – 1052 | 48 bp deletion | 1 ( 0.5) | 1 | ||
| D14 | 877 – 1026 | 150 bp deletion | 1 ( 0.5) | 1 | ||
| D15 | 930 – 1061 | 132 bp deletion | 1 ( 0.5) | 3 | ||
| Insertions | I1 | 584 | 18 bp insertion | 2 ( 1.0) | 3 | |
| I2 | 1205 | 9 bp insertion | 2 ( 1.0) | 2 | ||
| I3 | 562 | 18 bp insertion | 1 ( 0.5) | 2 | ||
| I4 | 1353 | 9 bp insertion | 1 ( 0.5) | 2 | ||
| I5 | 1066 | 9 bp insertion | 1 ( 0.5) | 2 | ||
| Frameshifts | FS1 | 666 | 1 bp insertion | Premature stop | 4 ( 2.0) | 2 & 3 |
| nsSNPs | S1b | 686 | g → a | Gly → Asp | 20 (10.0) | 2 |
| S2a | 707 | a → g | Asp → Gly | 19 ( 9.5) | 1 | |
| S3 | 368 | g → c | Gly → Ala | 5 ( 2.5) | 1 | |
| S4 | 620 | a → c | Asn → Thr | 2 ( 1.0) | 2 | |
| S5 | 214 | t → c | Phe → Leu | 1 ( 0.5) | 2 | |
| S6 | 269 | c → a | Ala → Asp | 1 ( 0.5) | 3 | |
| S7 | 1118 | t → c | Val → Ala | 1 ( 0.5) | 2 | |
| S8 | 1136 | c → g | Ala → Gly | 1 ( 0.5) | 2 | |
| S9 | 1355 | g → c | Gly → Ala | 1 ( 0.5) | 2 | |
| S10 | 653 | g → c | Gly → Ala | 1 ( 0.5) | 2 | |
| S11 | 778 | g → a | Gly → Met | 1 ( 0.5) | 2 | |
| S12 | 1061 | c → a | Thr → Asn | 1 ( 0.5) | 3 | |
| S13 | 571 | g → t | Ala → Ser | 1 ( 0.5) | 2 | |
| S14a | 880 | g → a | Gly → Ser | 1 ( 0.5) | 1 | |
| S15 | 1402 | a → g | Thr → Ala | 1 ( 0.5) | 3 | |
| sSNPs | S16 | 1392 | c → g | Gly → Gly | 1 ( 0.5) | 3 |
| S17 | 1122 | t → c | Gly → Gly | 1 ( 0.5) | 2 |
Sequence variation also present in M. tuberculosis strain 210
Sequence variation also present in M. tuberculosis strain CDC1551 in addition to a one bp deletion at position 937
The five in-frame insertions, 15 in-frame deletions, and one frameshift mutation were all located within the PGRS domain of the PE_PGRS26 gene. While the insertions were distributed throughout the PGRS domain, the deletions were mostly in one region of the PGRS domain. Twelve of the 15 deletions were contained within a 300 bp region of the PGRS domain and 56 (28.0%) of the 200 strains (56% of the 100 strains having PE_PGRS26 gene sequence variations) had a deletion in this region. Of the 17 SNPs, 14 were distributed throughout the PGRS domain and three, all non-synonymous, were in the PE domain of the PE_PGRS26 gene (Figure 1B).
PE_PGRS26 alleles
In addition to the H37Rv-type allele, 34 non-H37Rv-type PE_PGRS26 alleles, resulting from a combination of different sequence variations, were observed among the 200 strains. All of the non-H37Rv-type alleles would result in a change to the PE_PGRS26 amino acid sequence. The H37Rv-type PE_PGRS26 allele, present in 100 (50.0%) of the 200 strains, was the PE_PGRS26 allele observed with the highest frequency. The non-H37Rv-type PE_PGRS26 allele observed with the highest frequency was the D1 allele, having a 57 bp deletion, and this allele was observed in 27 (13.5%) of the 200 strains. The other PE_PGRS26 alleles were observed with a frequency of less than 10% (Figure 2).
A branched diagram of the 34 non-H37Rv-type PE_PGRS26 alleles and the 37Rv-type allele illustrates the relationships among the PE_PGRS26 sequence variations (Figure 2). Seven PE_PGRS26 alleles had the S1 non-synonymous SNP, while four had the D1 deletion, and two had the I1 insertion. The I2 insertion was present in two of the PE_PGRS26 alleles. However, the I2 S7 S8 S17 allele was as similar to the I2 S1 allele as it was to the H37Rv-type allele. Five PE_PGRS26 alleles had the D2 deletion and eight PE_PGRS26 alleles had the S2 non-synonymous SNP. Four of these alleles had both the D2 deletion and the S2 non-synonymous SNP. In addition, three of the eight PE_PGRS26 alleles having the S2 non-synonymous SNP also had the S3 non-synonymous SNP.
Genetic diversity among the Principal Genetic Groups
Twenty (10%), 130 (65%), and 50 (25%) of the 200 strains belonged to PGG1, PGG2, and PGG3, respectively. Of the 20 strains belonging to PGG1, 10 (50%) had a PE_PGRS16 gene sequence that was H37Rv-type and 10 (50%) had PE_PGRS16 gene sequence variations. All 20 of the PGG1 strains had non-H37Rv-type PE_PGRS26 alleles and all had either the S2 non-synonymous SNP or the D2 18 bp deletion. Nineteen (95%) of the 20 strains had the S2 non-synonymous SNP and 13 (65%) of the 20 strains had the D2 18 bp deletion in the PE_PGRS26 gene.
Of the 130 strains belonging to PGG2, 51 (39.2%) had a PE_PGRS16 gene sequence that was H37Rv-type and 79 (60.8%) had non-H37Rv-type alleles. Of these 79 PE_PGRS16 variants, 72 (91.1%) had a frameshift. Sixty-seven (51.5%) of the 130 PGG2 strains had an H37Rv-type PE_PGRS26 allele and 63 (48.5%) had a sequence variation in the PE_PGRS26 gene. Fifty-three (84.1%) of these 63 variants had either the D1 57 bp deletion or the S1 non-synonymous SNP.
Of the 50 PGG3 strains, 37 (74%) had H37Rv-type PE_PGRS16 alleles and 13 (26%) had non-H37Rv-type alleles. Seven (53.8%) of the 13 PGG3 strains with non-H37Rv-type PE_PGRS16 alleles had a frameshift. Thirty-three (66%) of the 50 PGG3 strains had PE_PGRS26 gene sequences that were H37Rv-type and 17 (34%) had non-H37Rv-type PE_PGRS26 alleles.
Comparison of genetic diversity between the PE_PGRS16 and PE_PGRS26 genes
Of the 200 strains, 40 (20%) had H37Rv-type alleles for both the PE_PGRS16 and PE_PGRS26 genes, 60 (30%) had sequence variations exclusively in the PE_PGRS16 gene, 58 (29%) had sequence variations exclusively in the PE_PGRS26 gene, and 42 (21%) had sequence variations in both the PE_PGRS16 and PE_PGRS26 genes. In-frame insertions and SNPs were observed with a similar frequency in the PE_PGRS16 and PE_PGRS26 genes among the 200 strains. Three (1.5%) and 7 (3.5%) of the 200 strains had in-frame insertions in the PE_PGRS16 and PE_PGRS26 genes, respectively (p = 0.20), and 39 (19.5%) and 48 (24.0%) of the 200 strains had SNPs in the PE_PGRS16 and PE_PGRS26 genes, respectively (p = 0.28). However, a difference in the frequency of frameshifts and in-frame deletions between these two PE_PGRS genes was observed. Seventy-nine (39.5%) of the 200 strains had a frameshift mutation in the PE_PGRS16 gene, while only 4 (2.0%) strains had a frameshift within the PE_PGRS26 gene (p < 0.0001). On the other hand, 68 (34%) of the 200 strains had in-frame deletions within the PE_PGRS26 gene, while only 19 (9.5%) strains had in-frame deletions within the PE_PGRS16 gene (p < 0.0001).
Profile of three PE_PGRS amino acid sequences
The profile of the PE_PGRS16, PE_PGRS26, and PE_PGRS33 amino acid sequences for each of the 200 clinical M. tuberculosis strains was examined to investigate the potential antigenic diversity generated by DNA sequence variations in these three PE_PGRS genes. The H37Rv-type amino acid sequence was the most frequently observed amino acid sequence for each of the three PE_PGRS. Among the 200 strains, 103 (51.5%), 100 (50.0%), and 71 (35.5%) had H37Rv-type amino acid sequences for PE_PGRS16, PE_PGRS26, and PE_PGRS33, respectively. However, only 26 (13%) of the 200 strains had an H37Rv-type amino acid sequence for all of the three PE_PGRS proteins. Among the 200 strains, 72 different profiles were observed, based on a combination of amino acid sequences of the PE_PGRS16, PE_PGRS26, and PE_PGRS33 proteins. The frequencies of the different profiles ranged from 1 (0.5%) to 26 (13%) of the 200 strains, with the H37Rv-type amino acid sequence profile being the most frequently observed. Forty-nine (68.1%) of the 72 different amino acid sequence profiles were unique among the 200 sample strains.
Discussion
To better understand the M. tuberculosis PE_PGRS gene polymorphisms and the potential antigenic diversity generated by sequence variations in members of this gene family, we investigated the genetic diversity of the PE_PGRS16 and PE_PGRS26 genes, which are hypothesized to have a role in mediating the latent state through differential regulation based on a previous study 7. A high frequency of genetic variation was observed for both the PE_PGRS16 and the PE_PGRS26 genes, although there were differences in the frequency and location of different types of sequence variations between the two genes. In addition, 72 different amino acid sequence profiles of PE_PGRS16, 26, and 33 were observed among the 200 strains, demonstrating the potential for antigenic variability generated from the genetic diversity of just three of the ~60 PE_PGRS genes.
The genetic diversity of the PE_PGRS33 gene among 123 clinical M. tuberculosis strains, which are included within the sample of 200 strains in the present study, was previously reported 9. All three PE_PGRS genes have a high frequency of genetic variations and a higher frequency of non-synonymous changes than synonymous changes, a finding that supports the role of these genes as variable surface antigens that interact with the host immune system. In addition, all three genes had a higher number of different in-frame deletions than in-frame insertions, in relation to the sequence of M. tuberculosis H37Rv, and the in-frame insertions/deletions and frameshifts occurred within the PGRS domain whereas the SNPs were distributed throughout the gene. However, while frameshifts are the predominant type of sequence variation found within the PE_PGRS16 gene and in-frame deletions are the predominant type of sequence variation found within the PE_PGRS26 gene, the predominant types of sequence variations within the PE_PGRS33 gene were in-frame insertions, observed in 68 (55.3%) of the 123 strains, and SNPs, observed in 72 (58.5%) of the 123 strains.
There is evidence that naturally-occurring PE_PGRS gene sequence variations can have an effect on the function of the protein. A recent study of sequence variations observed in the PE_PGRS33 gene found that large insertions and deletions in the PE_PGRS33 gene significantly decreased the stimulation of TNF-α production but small insertions and deletions as well as SNPs did not show a significant difference in stimulation of TNF-α production compared to the H37Rv-type PE_PGRS33 15. While PE_PGRS16 and PE_PGRS26 may have a role in mediating the latent state by differential regulation, the specific function of either of these genes is not known. The potential implications of the sequence variations observed in the PE_PGRS16 and PE_PGRS26 genes remains to be understood in the context of the specific role that these genes play in M. tuberculosis infection. Although as a typical comparative genomics study, the present study did not provide a functional analysis of the PE_PGRS16 and 26 genes, it generated data useful to prioritizing the selection of alleles for studies of the naturally-occurring sequence variations’ impact on the protein function and clinical relevance using either functional assays or population-based association studies. In particular, studies of sequence variations in the PE_PGRS16 and PE_PGRS26 genes between the isolates obtained from patients' primary disease and isolates obtained from their reactivated diseases will aid in the understanding of the role of these two genes in latency.
The finding that 40% of the 200 strains had a frameshift that would result in a premature stop codon in the PE_PGRS16 gene was surprising because a five-fold increase in expression of this gene was observed over the course of persistent M. tuberculosis infection of mice, suggesting an important role in immune evasion or latency 7. It is not known if a PE_PGRS16 gene having one of the naturally-occurring frameshifts is expressed by M. tuberculosis or if the significant change to the protein has an effect on the course of M. tuberculosis infection. It is interesting to note that 71 (89.9%) of the 79 isolates having a frameshift in the PE_PGRS16 gene would only have loss of the atypical sequence at the carboxy-terminus of the PE_PGRS16 protein. While it is not clear what the role of this atypical sequence is, it is possible that the FS1 frameshift could be an adaptive change to the PE_PGRS16 protein. However, it is also possible that the PE_PGRS16 protein does not have a specific, non-redundant function or, i it does, that changes in other PE_PGRS genes in the strains having the PE_PGRS16 frameshift are able to compensate for the loss of function of the PE_PGRS16 protein.
There are also potential biological implications of the in-frame deletion hotspot in the PE_PGRS26 gene. Twelve of the 15 deletions observed in the PE_PGRS26 gene were within a 300 bp region of the PGRS domain and 56 (28.0%) of the 200 strains had a deletion in this region. The high number of in-frame deletions in this region could be due to a greater number of repeated nucleotide sequences in that region compared to the rest of the gene, resulting in a higher frequency of replication slippage events. Examination of the deletions revealed that many different repeated nucleotide sequences of varying lengths were responsible for the slippage events and repeated sequences are found throughout the PE_PGRS26 gene. Another possible explanation is that there may be selective forces acting on the PE_PGRS26 gene that are either favoring changes in the hotspot region or conservation of the rest of the gene or both. If the hotspot region is a part of the protein that interacts with the host immune system, then selective pressure may favor deletions in this region. On the other hand, if regions of the PE_PGRS26 protein other than the hotspot region are critical to the function of this protein, then there may be selection against deletions in the functionally important regions. However, this is unlikely to be the case because of the number of insertions and non-synonymous SNPs that are distributed throughout the length of the gene.
Along with the variable expression of the PE_PGRS genes, the genetic diversity of the members of this gene family among clinical strains also contributes to the antigenic diversity of M. tuberculosis. Examining the genetic diversity of just three of the ~60 PE_PGRS genes demonstrates the potential for generating differences in available antigenic repertoire among different M. tuberculosis strains. It is possible that each M. tuberculosis strain has a unique profile of PE_PGRS antigens. Investigation of the genetic diversity of all the members of the PE_PGRS gene family among clinical M. tuberculosis strains will help to answer this question. While we have not seen evidence that PE_PGRS genes change rapidly during the course of an infection 10, genetic variability of the profile of PE_PGRS genes among M. tuberculosis strains may contribute to the ability of M. tuberculosis to cause exogenous re-infection.
Future studies of the expression patterns of the entire family of PE_PGRS genes among different M. tuberculosis strains will need to also take into account the genetic variability of the PE_PGRS genes. If variability in PE_PGRS gene expression is seen among different strains, knowledge of the PE_PGRS genetic variations of the strains will be necessary to determine if the differences are attributable to significant genetic disruption of the PE_PGRS gene (i.e. as the result of a frameshift) or differences in regulation among the strains. The genetic variability of a PE_PGRS gene also needs to be evaluated if expression of the gene is being considered for use as a diagnostic marker. A high frequency of genetic variations that would change the amino acid sequence was observed in both the PE_PGRS16 gene and the PE_PGRS26 gene and 79 (39.5%) of the 200 strains in the study sample had a frameshift in the PE_PGRS16 gene that would result in a premature stop codon. Sequence variations resulting in significant changes to the PE_PGRS protein could affect the detection of expression and therefore the ability to use inverse expression of PE_PGRS16 and PE_PGRS26 as a diagnostic marker of latent M. tuberculosis infection.
In summary, a high frequency of PE_PGRS16 and PE_PGRS26 genetic variation was observed among a sample of 200 clinical M. tuberculosis strains. The observed sequence variations could impact the function of these two PE_PGRS proteins and be associated with different M. tuberculosis clinical manifestations. Additionally, investigation of the genetic diversity of three members of this gene family demonstrates the variability in the PE_PGRS protein profile among M. tuberculosis isolates. Understanding of the antigenic variation generated by genetic changes in the PE_PGRS genes coupled with the variability in PE_PGRS expression will further our knowledge of how M. tuberculosis is able to evade the immune system and remain persistent in the host. The genetic diversity described by this study can inform the rational design of studies of association that investigate the clinical relevance of the PE_PGRS16 and PE_PGRS26 gene polymorphisms and also provide a basis for functional studies of the naturally-occurring PE_PGRS16 and PE_PGRS26 alleles. Future studies of the association between these gene polymorphisms and clinical characteristics will generate hypotheses for functional studies and, in turn, future studies of the impact of the gene polymorphisms on the function the PE_PGRS16 and PE_PGRS26 proteins will aid in the design and the interpretation of association studies.
Acknowledgements
This study was supported by a grant (NIH-R01-AI151975) from the National Institutes of Health. The authors would like to thank Dong Yang for her assistance in the culture of M. tuberculosis isolates and the preparation of the genomic DNA used for this study. We thank Sara Sandstedt for helpful discussion of the DNA sequence analysis. We also want to acknowledge the contributions of many colleagues at the Mycobacteriology Research Laboratory at the Central Arkansas Veterans Healthcare Center to the collection of M. tuberculosis isolates and genotyping data that were used in this study.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cole S, Brosch R, Parkhill J, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 2.Singh KK, Zhang X, Patibandla AS, Chien P, Jr, Laal S. Antigens of Mycobacterium tuberculosis expressed during preclinical tuberculosis: serological immunodominance of proteins with repetitive amino acid sequences. Infect Immun. 2001;69:4185–4191. doi: 10.1128/IAI.69.6.4185-4191.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Delogu G, Brennan MJ. Comparative immune response to PE and PE_PGRS antigens of Mycobacterium tuberculosis. Infect Immun. 2001;69:5606–5611. doi: 10.1128/IAI.69.9.5606-5611.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brennan MJ, Delogu G, Chen Y, et al. Evidence that mycobacterial PE_PGRS proteins are cell surface constituents that influence interactions with other cells. Infect Immun. 2001;69:7326–7333. doi: 10.1128/IAI.69.12.7326-7333.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banu S, Honore N, Saint-Joanis B, et al. Are the PE-PGRS proteins of Mycobacterium tuberculosis variable surface antigens? Mol Microbiol. 2002;44:9–19. doi: 10.1046/j.1365-2958.2002.02813.x. [DOI] [PubMed] [Google Scholar]
- 6.Voskuil MI, Schnappinger D, Rutherford R, Liu Y, Schoolnik GK. Regulation of the Mycobacterium tuberculosis PE/PPE genes. Tuberculosis (Edinb) 2004;84:256–262. doi: 10.1016/j.tube.2003.12.014. [DOI] [PubMed] [Google Scholar]
- 7.Dheenadhayalan V, Delogu G, Sanguinetti M, Fadda G, Brennan MJ. Variable expression patterns of Mycobacterium tuberculosis PE_PGRS genes: evidence that PE_PGRS16 and PE_PGRS26 are inversely regulated in vivo. J Bacteriol. 2006;188:3721–3725. doi: 10.1128/JB.188.10.3721-3725.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delogu G, Sanguinetti M, Pusceddu C, et al. PE_PGRS proteins are differentially expressed by Mycobacterium tuberculosis in host tissues. Microbes Infect. 2006;8:2061–2067. doi: 10.1016/j.micinf.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 9.Talarico S, Cave MD, Marrs CF, et al. Variation of the Mycobacterium tuberculosis PE_PGRS 33 gene among clinical isolates. J Clin Microbiol. 2005;43:4954–4960. doi: 10.1128/JCM.43.10.4954-4960.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Talarico S, Cave MD, Foxman B, et al. Association of Mycobacterium tuberculosis PE_PGRS33 polymorphism with clinical and epidemiological characteristics. Tuberculosis (Edinb) 2007;87:338–346. doi: 10.1016/j.tube.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barnes PF, Yang Z, Preston-Martin S, et al. Patterns of tuberculosis transmission in Central Los Angeles. JAMA. 1997;278:1159–1163. [PubMed] [Google Scholar]
- 12.Chaves F, Yang Z, el Hajj H, et al. Usefulness of the secondary probe pTBN12 in DNA fingerprinting of Mycobacterium tuberculosis. J Clin Microbiol. 1996;34:1118–1123. doi: 10.1128/jcm.34.5.1118-1123.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Embden J, Cave MD, Crawford J, et al. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendation for a standardized methodology. J Clin Microbiol. 1993;31:406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sreevatsan S, Pan X, Stockbauer KE, et al. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc Natl Acad Sci U S A. 1997;94:9869–9874. doi: 10.1073/pnas.94.18.9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Basu S, Pathak SK, Banerjee A, et al. Execution of macrophage apoptosis by PE_PGRS33 of Mycobacterium tuberculosis is mediated by Toll-like receptor 2-dependent release of tumor necrosis factor-alpha. J Biol Chem. 2007;282:1039–1050. doi: 10.1074/jbc.M604379200. [DOI] [PubMed] [Google Scholar]