Abstract
The 37-residue amylin peptide, also known as islet amyloid polypeptide, forms fibrils that are the main peptide or protein component of amyloid that develops in the pancreas of type 2 diabetes patients. Amylin also readily forms amyloid fibrils in vitro that are highly polymorphic under typical experimental conditions. We describe a protocol for the preparation of synthetic amylin fibrils that exhibit a single predominant morphology, which we call a striated ribbon, in electron microscope and atomic force microscope images. Solid state nuclear magnetic resonance (NMR) measurements on a series of isotopically labeled samples indicate a single molecular structure within the striated ribbons. We use scanning transmission electron microscopy and several types of one-dimensional and two-dimensional solid state NMR techniques to obtain constraints on the peptide conformation and supramolecular structure in these amylin fibrils, and derive molecular structural models that are consistent with the experimental data. The basic structural unit in amylin striated ribbons, which we call the protofilament, contains four-layers of parallel β-sheets, formed by two symmetric layers of amylin molecules. The molecular structure of amylin protofilaments in striated ribbons closely resembles the protofilament in amyloid fibrils with similar morphology formed by the 40-residue β-amyloid peptide that is associated with Alzheimer's disease.
Keywords: islet amyloid polypeptide, type 2 diabetes, amyloid fibril structure, electron microscopy, atomic force microscopy
Amylin, also called islet amyloid polypeptide or IAPP, is a 37-residue peptide that is cosecreted with insulin by the β-cells of pancreas. The normal biological effects of amylin secretion include inhibition of glucagon secretion and reduction of the rate of gastric emptying, effects that contribute to the maintenance of postprandial glucose homeostasis (1). Amylin is the major peptide or protein component of the islet amyloid found in the pancreas of approximately 90% of type 2 (or non-insulin-dependent) diabetes patients (2, 3). Fibrillar amylin has been shown to be toxic to cultured β-cells (4), and has been proposed to play a significant role in the pathogenesis of type 2 diabetes by contributing to the destruction of β-cells in the later stages of the disease (5-7). Studies with transgenic mice that express human amylin suggest that amyloid could develop uniformly throughout the islets of the pancreas from an early stage of the disease (8). In a mouse model for type 2 diabetes, development of persistent hyperglycemia and insulin deficiency was found to require expression of human amylin and to correlate positively with islet amyloid formation (9). Knowledge of the detailed molecular structure of amylin fibrils may contribute to the development of drugs to prevent the formation of islet amyloid and to an understanding of their cytotoxic properties.
Amyloid fibrils are self-assembled, β-sheet-rich aggregates that are formed by a large class of peptides and proteins (10-12). Amyloid fibrils typically exhibit diameters in the 5-15 nm range and lengths in the 0.1-10 μm range. From the standpoint of molecular structure, the defining feature of an amyloid fibril is the presence of cross-β supramolecular structure, meaning that the β-sheets within the fibril are formed by β-strand segments that run approximately perpendicular to the long axis of the fibril and are linked by hydrogen bonds that run approximately parallel to this axis (11-13). Although determination of the molecular structures of amyloid fibrils is made difficult by their inherent noncrystallinity and insolubility, techniques such as solid state nuclear magnetic resonance (NMR) (12, 14-33), electron paramagnetic resonance (EPR) (34-36), electron microscopy (37-43), hydrogen/deuterium exchange (29, 44-47), scanning mutagenesis (48), chemical crosslinking (27, 49, 50), and x-ray diffraction of amyloid-like crystals (51, 52) have recently shed substantial light on these structures.
In vitro, amylin readily forms amyloid fibrils with a variety of morphologies as seen in transmission electron microscope (TEM) and atomic force microscope (AFM) images (40, 41), and with typical cross-β diffraction patterns (53). Early solid state NMR studies by Griffiths et al. focused on fibrils formed by a peptide representing residues 20-29 of amylin. These studies provided evidence for a distorted β-strand backbone conformation in residues 24-26 and an antiparallel β-sheet structure (25). More recent EPR studies of spin-labeled, full-length amylin fibrils by Jayasinghe and Langen indicate a parallel arrangement of the peptides within the β-sheets and demonstrate that amino acid sidechains throughout the peptide are immobilized in the fibrils, but with less rigidity at the N-terminus (36).
Structural studies of peptides and proteins generally require homogeneous sample preparations. Studies by Petkova et al. of fibrils formed by the 40-residue β-amyloid peptide associated with Alzheimer's disease (Aβ1-40) have clearly shown that amyloid fibrils with distinct morphologies possess distinct molecular structures (21, 22). Compared to other amyloid-forming peptides (such as Aβ1-40 or fragments thereof), full-length amylin is difficult to synthesize and purify, has poor initial solubility in aqueous solution, and forms fibrils relatively rapidly in aqueous solution, factors that complicate the preparation of structurally homogeneous amylin fibrils. In this paper, we report a protocol that generates amylin fibrils with uniform morphology and molecular structure, in quantities that are sufficient for solid state NMR or other structural measurements. We use solid state NMR techniques to obtain constraints on the peptide conformation and supramolecular organization within these amylin fibrils, and combine these constraints with information from TEM, AFM, and scanning transmission electron microscopy (STEM) in order to develop preliminary models for the molecular structure of amylin fibrils. Finally, we compare the solid state NMR data and preliminary structural models with reported results from earlier investigations of amylin fibril structure and related issues.
Materials and Methods
Synthesis and purification of amylin
Rink amide MBHA Resin (0.69 mmol/g), Fmoc-Ser(tBu)-Ser(ψMe,Mepro)-OH, Fmoc-(FmocHmb)Ala-OH and Fmoc-(FmocHmb)Gly-OH were purchased from Novabiochem (San Diego, CA). Unlabeled Fmoc amino acids were from Anaspec (San Jose, CA); isotopically labeled Fmoc amino acids were from Cambridge Isotopes Laboratories (Andover, MA) and from Spectra Stable Isotopes (Columbia, MD). All other reagents were obtained from Applied Biosystems (Foster City, CA), Sigma-Aldrich (St. Louis, MO) and EMD Chemicals (Gibbstown, NJ). Amylin was synthesized on a 0.1 mmol scale by solid-phase peptide synthesis on an Applied Biosystems 433A synthesizer. For most residues, coupling reactions were performed for 20 min using Fmoc amino acids (10 eq) activated with 2-(1H-benzotriazol-l-yl)−1,1,3,3-tetramethyluronium hexafluorophosphate (HBTU, 10 eq) and N,N-diisopropylethylamine (DIEA, 10 eq). Coupling times of 6 hr were used for Thr9 and Gln10. Isotopically labeled residues (3 eq) were activated with HBTU (3 eq) and DIEA (3 eq) and coupled for 45 min. Capping of free amino groups with acetic anhydride was performed after each coupling step.
One amylin sample, called AMY04, was synthesized with 13C labels at the methyl carbon of Ala13 and the carbonyl carbon of Phe23 to permit measurements of intermolecular 13C-13C dipole-dipole couplings using the constant-time finite-pulse radiofrequency-driven recoupling (fpRFDR-CT) technique (54). Seven samples were prepared with uniform 15N and 13C labeling of selected residues, as summarized in Table 1. Labeled residues in each sample were chosen to minimize overlap in 13C NMR spectra, permitting measurements of 13C NMR chemical shifts, signal intensities, and linewidths throughout the amylin sequence. In certain cases, uniform labeling of consecutive pairs of residues also permitted measurements of intramolecular 15N-15N dipole couplings that serve as constraints on peptide backbone conformation. Duplication of certain labeled residues in different samples (e.g., Ile26 in AMY05 and AMY06) permitted confirmation of the reproducibility of amylin fibril formation with the protocol described below.
Table 1.
Isotopic labeling patterns and synthesis strategies for amylin samples
| sample | isotopically labeled residues (uniform 15N and 13C labeling except in AMY04) | Pseudoproline dipeptide positions | N-FmocHmb-protected position |
|---|---|---|---|
| AMY04 | 13CH3-Ala13, 13CO-Phe23 | Ser19/Ser20, Ser28/Ser29 | none |
| AMY05 | Ala5, Leu16, Phe23, Ile26, Val32, Gly33 | Ser19/Ser20 | none |
| AMY06 | Arg11, Ala13, Phe15, Val17, Ile26, Ser28 | Ser19/Ser20 | none |
| AMY07 | Thr9, Gln10, Leu12, Asn21, Gly24, Ala25, Ser34 | Ser19/Ser20 | none |
| AMY08 | Ala8, Ser20, Phe23, Gly24, Leu27, Asn35, Thr36 | none | Ala13 |
| AMY09 | Leu12, Ala13, His18, Ser19, Thr30, Asn31 | Ser28/Ser29 | Gly24 |
| AMY10 | Ala13, Asn14, Phe15, Ile26, Leu27, Ser29, Thr30 | Ser19/Ser20 | none |
| AMY11 | Asn22, Phe23, Ala25, Ile26, Gly33, Ser34, Tyr37 | Ser19/Ser20, Ser28/Ser29 | none |
Synthesis of full-length amylin from standard Fmoc-protected amino acids was found to be inefficient due to poor couplings after Arg11 and Gln10. Yields of full-length amylin were improved significantly by incorporation of either the pseudoproline dipeptide derivative Fmoc-Ser(tBu)-Ser(ψMe,Mepro)-OH, as described by Abedini and Raleigh (55), or N-FmocHmb-protected residues at certain sites. Locations of these amino acid derivatives (which were converted to standard amino acids after cleavage from the synthesis resin) were varied according to the isotopic labeling patterns in each amylin sample, as summarized in Table 1. Coupling times of 3 hr for the pseudoproline dipeptide derivative and the subsequent residue (His18 or Leu27) were used. N-FmocHmb-protected residues were coupled for 3 hr, and the subsequent residue (Phe23 or Leu12) was activated with O-(7-azabenzotriazol-l-yl)−1,1,3,3-tetramethyluronium hexafluorophosphate and coupled for 12 hr.
Crude peptide obtained by standard trifluoracetic acid (TFA) cleavage and ether precipitation was purified by high-performance liquid chromatography (HPLC) using a preparative C4 reverse-phase column (Grace Vydac, Hesperia, CA) and a water/acetonitrile gradient containing 0.1% TFA and running from 25% to 45% acetonitrile over 30 min. Purified amylin was then lyophilized and stored at −20°C. Yields of purified amylin were approximately 10-30% of the theoretical maximum, depending on the isotopic labeling pattern and the locations of pseudoproline and N-FmocHmb derivatives. Before preparation of fibrils, the peptide was solubilized at 1.3 mM in 30% dimethyl sulfoxide, 3% acetic acid and allowed to form intramolecular disulfide bonds (between Cys2 and Cys7) overnight. A second HPLC purification using a C18 column was performed after disulfide formation. Oxidized amylin was collected, frozen in liquid nitrogen, lyophilized, and used immediately. Mass spectrometry was utilized after each HPLC purification to confirm the correct assembly of synthetic amylin and the disulfide bond formation. HPLC chromatograms for reduced and oxidized amylin are shown in Figure S1 of the supporting information.
Preparation of amylin fibrils
Immediately prior to fibril formation, monomeric amylin was prepared by gel filtration as follows. Lyophilized, oxidized amylin was dissolved in 7 M guanidine hydrochloride at a concentration of 400 μM (verified by absorption at 280 nm using an extinction coefficient of 1400 M−1 cm−1). A 500 μl aliquot was injected at 0.25 ml/min on a Superdex Peptide Column 10/300 (GE Healthcare, Piscataway, NJ 08855) which had been equilibrated in 10 mM sodium phosphate, 15 mM sodium chloride, pH 7.4. Monomeric amylin was collected with a dilution factor between 8 to 10 (final concentration of 40-50 μM). About 30% of the amylin was lost in this gel filtration purification, probably due to aggregation. The column was therefore cleaned with 70% formic acid and re-equilibrated with buffer before the next injection. Fast protein liquid chromatography (FPLC) traces for human amylin and rodent amylin (which does not aggregate (56, 57)), verifying the monomeric state prepared, are shown in Figure S2 of the supporting information.
Amylin fibrils with uniform morphology were prepared initially by collecting monomeric amylin in a polypropylene tube and allowing fibrils to form overnight in a sonicator bath which was automatically turned on for 10 seconds every 30 min. This intermittent sonication ensured the formation of short fibrils and may favor the propagation of fibril morphologies that are more susceptible to fragmentation. On the second day, a small aliquot (2.5% v/v) of these fibrils was placed in a new polypropylene tube (to act as seeds) and another batch of monomeric amylin was collected directly in this tube, while gently swirling it by hand. Fibrillization was then allowed to proceed overnight under the same intermittent sonication. On the third day, a third generation of amylin fibrils was prepared by repeating this seeding procedure. The third generation fibrils were investigated by TEM and AFM and found to be highly uniform in morphology. Subsequent samples of isotopically labeled amylin fibrils were prepared by seeding with the same third generation fibrils, with addition of fresh monomeric amylin to the polypropylene tube over the course of several days. After incubation for one week at room temperature, amylin fibrils for solid state NMR measurements were collected by centrifugation, washed once with deionized water, and lyophilized. Final yields of amylin fibrils were 40% relative to the peptide obtained from the initial HPLC purification.
For comparison, polymorphic amylin fibrils were prepared by dissolving oxidized amylin (purified by HPLC and lyophilized as described above) twice in neat hexafluoroisopropanol (HFIP) and drying under nitrogen gas. HFIP-treated amylin was then dissolved at 50 μM in the same phosphate buffer described above and allowed to fibrillize for one week.
Other methods for preparing structurally homogeneous amylin fibrils were unsuccessful. For example, we tried to dissolve the peptide at approximately 500 μM concentration in dimethyl sulfoxide, HFIP, or 7 M guanidine hydrochloride prior to dilution into aqueous buffer that contained seeds. AFM images of the resulting fibrils indicated a high degree of polymorphism. In addition, FPLC traces of amylin dissolved in dimethyl sulfoxide indicated a non-monomeric state.
Electron and Atomic Force Microscopy
After one week of incubation, amylin fibrils were diluted by a factor of 10 with deionized water for TEM and STEM measurements. For TEM, diluted solutions were applied to glow-discharged carbon films on 300 mesh copper grids with lacey carbon supports. After adsorption for 2 min and blotting, grids were washed with deionized water, blotted, negatively stained with 3% uranyl acetate for 1 min, blotted, and dried in air. TEM images were recorded with FEI CM120 microscope (Amsterdam, NL) operating at 120 kV. Grids for STEM measurements were prepared similarly, but tobacco mosaic virus (TMV) was first deposited on grids and the staining step was omitted. STEM images were recorded with a VG Microscopes HB501 field-emission STEM as previously described (15) and were analyzed with ImageJ software (available at http://rsb.info.nih.gov/ij/). For AFM, fibril solutions were diluted by a factor of 10 with 1% acetic acid and immediately applied to freshly cleaved mica. The mica was washed twice with deionized water and air-dried after blotting. Images were acquired in tapping mode using a MultiMode AFM and Nanoscope IV controller (Veeco, Santa Barbara, CA), with either standard silicon nitride probes (5-6 nm tip radius, 40 N/m force constant) or micro-actuated silicon probes for faster scanning (nominal 10 nm tip radius, 3 N/m force constant). Similar images were obtained in both cases.
Solid state NMR
Measurements were performed using 9.4 T and 14.1 T magnets (100.4 MHz and 150.7 MHz 13C NMR frequencies), InfinityPlus NMR spectrometers (Varian, Inc., Palo Alto, CA 94304), and triple-channel magic-angle spinning (MAS) probes from Varian or the National Institute of Chemical Physics and Biophysics (Tallinn, Estonia 12618). Lyophilized fibrils in quantities up to 3.5 mg were packed in 3.2 mm or 2.1 mm MAS rotors with teflon spacers. After studying the dry samples, the rotors were opened and the fibrils were hydrated with deionized water (about 1 μl per mg of dry peptide, added directly into the rotor). AMY07 and AMY10 fibrils were studied only in the hydrated state. In all solid state NMR experiments, cross-polarization was used to enhance 13C and 15N NMR sensitivity (58), with a linearly ramped radiofrequency (rf) field amplitude of 50 ± 8 kHz on the 13C or 15N channel and a corresponding rf field matching the n = +1 Hartmann-Hahn condition on the 1H channel. Continuous-wave or two-pulse phase-modulated proton decoupling (59) was employed in all experiments, with 1H rf field amplitudes of approximately 110 kHz.
Two types of two-dimensional (2D) 13C NMR measurements were performed on samples with uniformly labeled residues. In 2D finite-pulse radiofrequency-driven recoupling (fpRFDR) measurements, the fpRFDR sequence (16, 60) was applied during exchange periods of 1.5 or 2.2 ms between the t1 and t2 dimensions, with 13C π pulse lengths set to 30% of the MAS rotation period. Strong crosspeaks in the 2D fpRFDR spectra connect 13C NMR lines of directly-bonded carbon sites. In 2D radiofrequency-assisted diffusion (RAD) measurements, exchange periods ranging from 500 ms to 1000 ms were used, during which an rf field amplitude equal to the MAS frequency was applied to protons (61, 62). With long exchange periods, strong crosspeaks in 2D RAD spectra connect all 13C NMR lines arising from a single labeled residue or from pairs of labeled residues that occur sequentially (i.e., consecutively in the amylin sequence). Weaker inter-residue crosspeaks arising from contacts between nonsequential residue pairs provide constraints on peptide conformation. One-dimensional (1D) RAD measurements were also performed on AMY10 fibrils to confirm the 2D results. In the 1D RAD measurements, a frequency-selective 13C π/2 pulse following cross-polarization was used to prepare longitudinal spin polarization on the Phe15 aromatic carbons, which could then be transferred to other 13C-labeled sites during the exchange period prior to signal detection after a hard π/2 pulse.
Measurements of 13C-13C and 15N-15N nuclear magnetic dipole-dipole couplings (which are proportional to the inverse cube of internuclear distances) were carried out with the constant-time fpRFDR (fpRFDR-CT) technique, as previously described (54) and applied to other amyloid-forming peptides and proteins (17, 18, 21, 63), using 20 kHz MAS frequencies and 15 μs fpRFDR π pulses. In 15N-15N distance measurements, designed to probe the peptide backbone conformation at sites where uniformly labeled residues occurred sequentially, spin polarization was transferred from backbone 15N nuclei to directly bonded 13Cα nuclei for signal detection using a transferred echo double resonance (TEDOR) sequence (64, 65) after the 15N fpRFDR-CT period, as previously described (66). In 13C-13C distance measurements on AMY04, designed to probe the organization of β-sheets in the fibrils, pulsed spin-locked detection was used to improve sensitivity (67).
13C chemical shifts were referenced to tetramethylsilane, using the carboxylate signal of 1-13C-L-alanine powder as an external standard at 177.95 ppm. 2D data were processed with nmrPipe and plotted with nmrDraw (68). Numerical simulations of fpRFDR-CT measurements were performed using the SIMPSON program (69) or specific Fortran programs, using the following spin systems: (i) linear chains of five equally spaced 13C nuclei, with spin polarization initially on the central nucleus to minimize end effects and thereby approximate the behavior of an infinite chain; (ii) two 15N nuclei with the same separation as backbone nitrogen atoms in sequential residues for a given ψ torsion angle, with spin polarization on both nuclei. In the latter case, the 15N-13C TEDOR transfer was omitted from the simulations, as it has a negligible effect on the results (66).
Construction of Molecular Models
Models for amylin fibrils were constructed within the Amber 9 suite (70) in two steps. First, a starting structure was generated using the ff99SB force field (71) using the LEaP program (70), consisting of five copies of amylin in an extended conformation (including the amidated C-terminus and the disulfide bond between Cys2 and Cys7), placed 20 Å apart in a direction perpendicular to the peptide backbones. Four types of experimentally-based restraints were used. (i) Residues 8-18 and residues 28-37 were forced to form parallel, in-register β-sheets by imposing harmonic potentials between the N and O backbone atoms of all even-number residues (2j) of a given peptide and the O and N backbone atoms of residues 2j−1 and 2j+1 of an adjacent peptide. The well of these harmonic potentials was set between 2.75 and 3.15 Å with a force constant of 20 kcal/mol/Å2. These restraints represent backbone hydrogen bonds, and are supported by the 13C fpRFDR-CT and chemical shift data presented below. (ii) ϕ and ψ backbone torsion angles for residues 8-18 and 28-37 were constrained to −130 ± 20° and 130 ± 20°, respectively, based on the observation of β-strand-like chemical shifts for most residues in these segments. Several ψ angles were constrained to the values shown in Table S1 (see Supporting Information), based on 15N fpRFDR-CT data. A force constant of 100 kcal/mol/rad2 was used for these restraints. (iii) Sidechains of either Asn14 and Ile26 or Phe15 and Leu27 were kept in proximity by imposing harmonic potentials between Cγ and Cδ1atoms, with wells between 3 and 7 Å and force constants of 10 kcal/mol/Å2. These restraints were based on the observation of nonsequential spin polarization transfers in 2D and 1D RAD measurements. (iv) Cα atoms from residue pairs Ala8-Val32, Gln10-Thr30, and Leu12-Ser28 or residue pairs Ala8-Ser34, Gln10-Val32, Leu12-Thr30, and Asn14-Ser28 in each molecule were restrained to be in proximity by a special potential with a well between 9 and 11 Å and a force constant of 10 kcal/mol/Å2 which flattens out for large deviations (see Amber 9 documentation at http://amber.scripps.edu/doc9/index.html). These restraints serve to bring the β-sheets formed by residues 8-18 and 28-37 in contact with one another, as required by the experimental fibril dimensions observed in TEM and AFM images. In addition, the trans conformation of peptide bonds and the chirality of sidechains were preserved by torsional potentials with 100 kcal/mol/rad2 force constants.
After unrestrained energy minimization of the starting structure to relieve strain (especially in the disulfide bond), restrained Langevin dynamics simulations were performed using a generalized Born continuum solvent (72, 73). The initial temperature was set to 100 K and random velocities were generated for this temperature. The system was then heated to 1000 K over 20 ps, while the restraints were linearly scaled from 0 to 10%. At 1000 K, the constraints were further scaled to 100% over 20 ps and Langevin dynamics were continued for an additional 20 ps. The system was then cooled to 100 K over 20 ps and to 0 K over 20 ps. A total of 100 trajectories were calculated under this protocol, and pentameric structures with the lowest potential energies were selected.
In the second step, two copies of a pentamer (i.e., a final structure from the first step) were manually rotated and displaced such that the β-sheet segments comprised of residues 28-37 were antiparallel and about 20 Å apart, producing a two-fold symmetric structure consistent with experimental STEM data and solid state 13C NMR spectra (see below). In addition to the restraints within each pentamer, intermolecular restraints between Cα atoms of Leu27-Val32 pairs were added to attract the two pentamers to one another. Langevin dynamics were then simulated for the decamer, with heating from 0 K to 1000 K and scaling of all constraints from 0% to 100% over 5 ps, equilibration for 25 ps at 1000 K, and final cooling to 0 K over 5 ps. The resulting structural models were visualized and images were produced using the VMD software (74).
Results
Characterization of amylin fibril morphology and mass-per-length
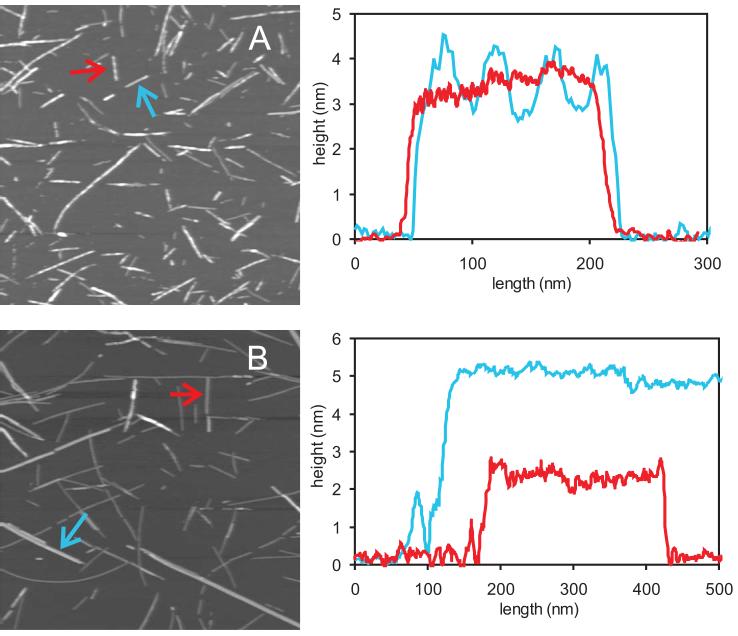
Representative TEM images of amylin fibrils are shown in Figure 1. A polymorphic fibril sample (Figure 1A), prepared with HFIP pretreatment as described above, contains fibrils with an apparent axial twist and fibrils that appear to be comprised of untwisted, rod-like protofilaments that associate laterally into ribbons. We refer to fibrils with the latter morphology as “striated ribbons”. Twisted fibrils have apparent twist periods (i.e., distances between the apparent maxima in fibril width) between 50 and 250 nm, and width maxima of 9 ± 1 nm. Protofilaments in striated ribbons have widths of 4.5-5.0 nm and commonly associate in groups of 2-8. In contrast, amylin fibrils prepared with the gel filtration protocol described above are predominantly striated ribbons (Figure 1B). Striated ribbons can be as long as 50 μm. The very high level of morphological homogeneity in amylin fibrils prepared with the gel filtration protocol leads to high-quality solid state NMR spectra and permits the development of molecular structural models, as described below.
Figure 1.
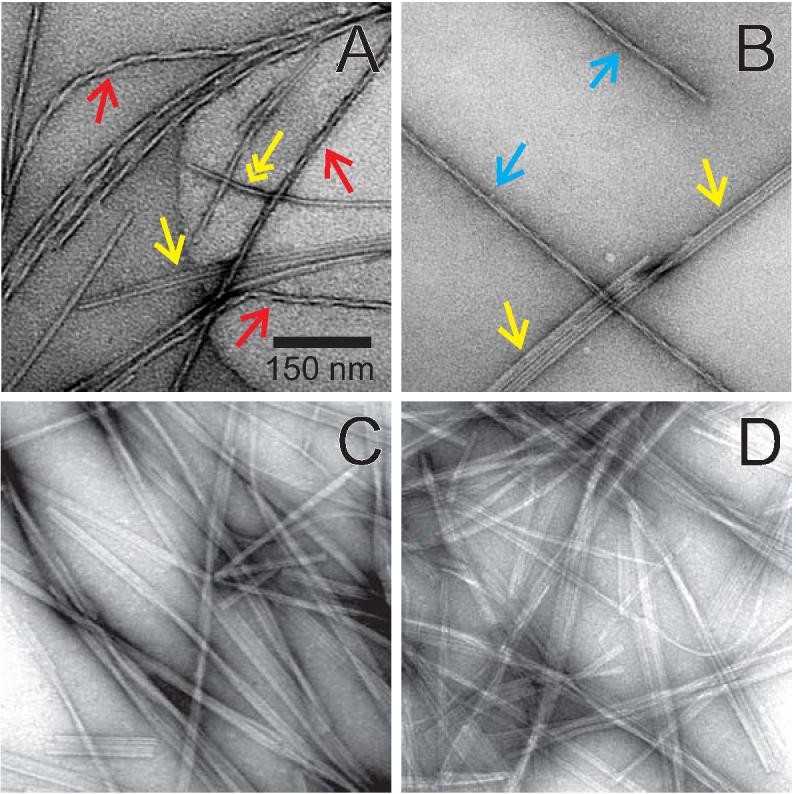
TEM images of negatively stained amylin fibrils from polymorphic (A,B) and morphologically homogeneous (C,D) preparations. Red and blue arrows indicate fibrils with apparent periodic twists. Yellow arrows indicate striated ribbons. Double-headed yellow arrow indicates a single protofilament that appears to be the building block of striated ribbons. Homogeneous samples prepared by the gel filtration protocol described in the text are primarily striated ribbons.
Figure 2 shows topographic AFM images of polymorphic and morphologically homogeneous amylin fibril samples. Twisted and striated ribbon fibrils can be distinguished in the AFM image of the polymorphic sample (Figure 2A). Twisted fibrils vary between 4.0 nm and 10.0 nm in height, with a modulation that reveals twist periods of 25 nm and 50 nm in most cases. Striated ribbons have heights between 2.5 nm and 5.0 nm and do not exhibit periodic height modulation. In AFM images of morphologically homogeneous fibrils (Figure 2B), only striated ribbons are observed. The two classes of apparent heights for striated ribbons (roughly 2.5 nm and 5.0 nm) are identical to heights observed by Goldsbury et al. in earlier AFM studies of polymorphic amylin fibrils (75).
Figure 2.
Topographic AFM images of amylin fibrils absorbed to mica from (A) polymorphic and (B) morphologically homogeneous preparations. Image areas are 2 µm X 2 µm. Height profiles for selected fibrils (indicated by arrows of corresponding colors) are shown to the right.
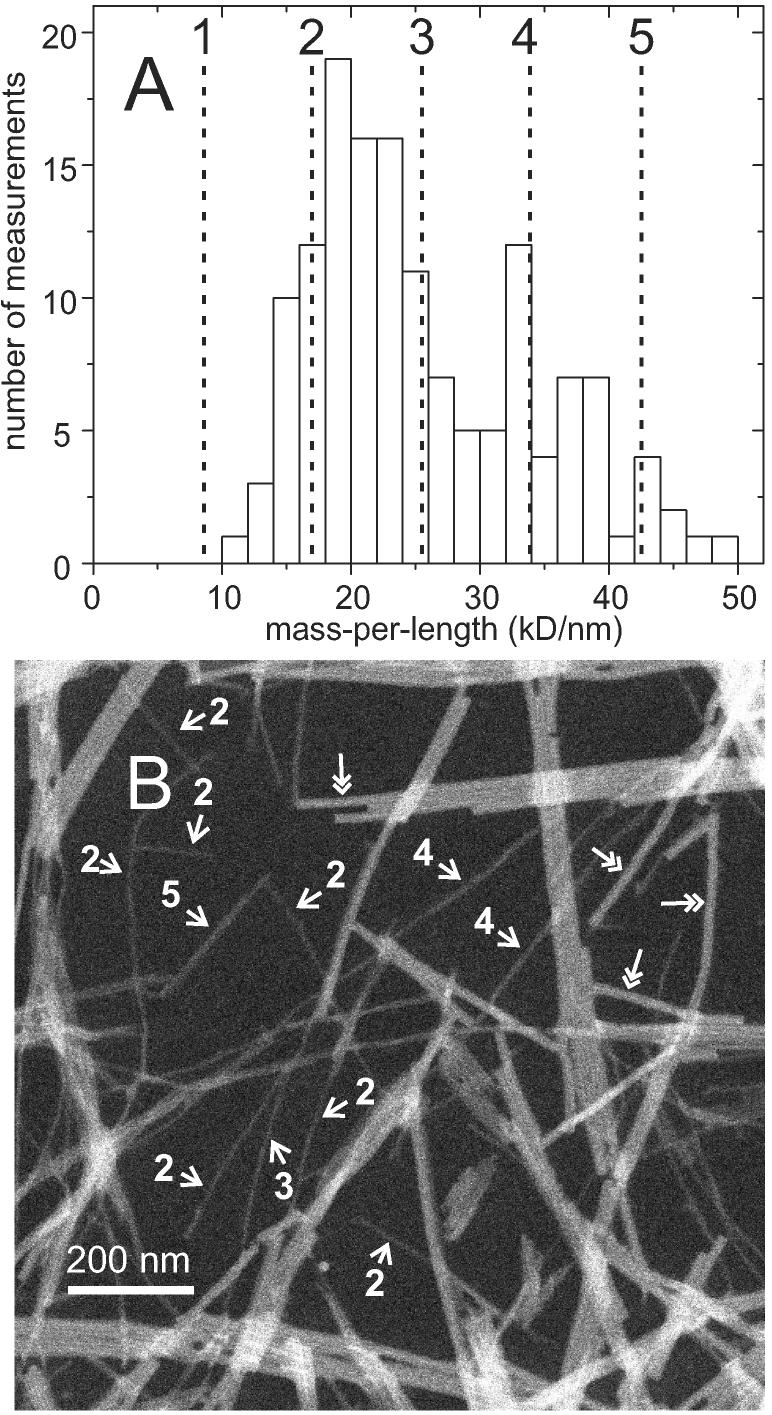
STEM measurements were performed on a morphologically homogeneous amylin fibril sample in order to determine the protofilament MPL, based on the principle that intensities in STEM images of unstained samples are directly proportional to projected mass densities (76). A representative STEM image is shown in Figure 3B. As previously described (15), MPL values were determined from integrated image intensities within rectangular regions that enclose fibril segments, after subtraction of background intensities determined by integration over empty rectangular regions. The constant of proportionality between mass density and image intensity was determined for each STEM image from similar measurements on TMV particles. For fibrils with striated ribbon morphologies, determination of the MPL value of protofilaments is complicated by their tendency to associate laterally into ribbons and bundles of ribbons. Measurements of MPL values were therefore deliberately restricted to fibrils that appeared as thin, single lines in the STEM images. Due to the low resolution and contrast in the STEM images, single protofilaments could not always be distinguished from laterally associated pairs of protofilaments or from fibrils with other morphologies, which represent a small fraction of the total material but are less prone to lateral association.
Figure 3.
(A) Histogram of MPL values extracted from STEM images of amylin fibrils. Vertical dashed lines indicate the ideal MPL values of integral numbers of molecular layers with cross-β structures. (B) Representative STEM image (50,000 X magnification). Approximate MPL values of individual fibrils are indicated in units of the MPL of one molecular layer. Double-headed arrows indicate tobacco mosaic virus particles used to determine the ratio of MPL to image intensity.
Figure 3A shows a histogram of MPL values extracted from 16 STEM images. Assuming an ideal cross-β structure for amylin fibrils, the MPL value for a single layer of molecules would be 8.1 kD/nm, corresponding to the quotient of the 3.91 kD molecular weight and the 0.48 nm spacing between β-strands in ideal, planar β-sheets. MPL values below 10 kD/nm are not observed, but values in the 13-19 kD/nm range represent 24% of the total number of MPL values. Uncertainty in each value is expected to be approximately ±3 kD/nm, due to background noise and carbon film thickness variations (15). We therefore conclude that the protofilament consists of two cross-β molecular layers, with an ideal MPL value of 16.2 kD/nm. We attribute higher MPL values in Figure 3B to lateral association of protofilaments and to a minority fraction of other fibril morphologies. Examples of MPL values for individual amylin fibrils are given in Figure 3B.
Goldsbury et al. have reported STEM data for amylin fibrils with a variety of morphologies, including morphologies similar to our striated ribbons (41). A minimum MPL value of approximately 15 kD/nm was reported. The MPL data were fit to three Gaussian distributions, centered at 19.9, 29.0, and 40.3 kD/nm. Goldsbury et al. interpreted their data as evidence for a “protofibril” with MPL of approximately 10 kD/nm. In contrast, we interpret the approximate 10 kD/nm MPL increment as the MPL of one molecular layer and conclude that the protofilament contains two molecular layers. We attribute the observation of MPL values around 30 kD/nm to polymorphism, as supported by our studies of Aβ1-40 fibrils in which distinct polymorphs (with distinct molecular structures) were found to have minimum MPL values corresponding to either two molecular layers or three molecular layers (21).
Characterization of secondary structure by 13C NMR
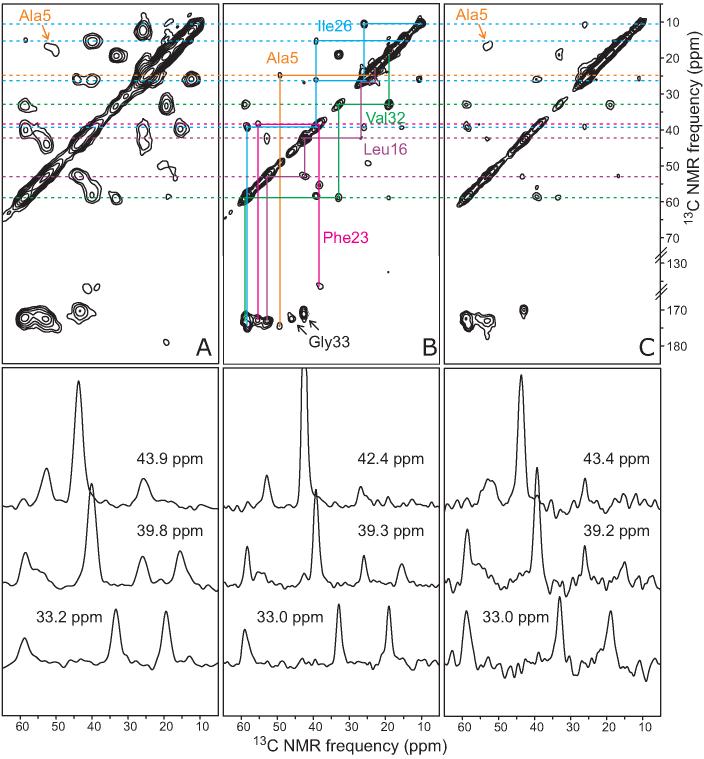
Figures 4A and 4B show 2D 13C NMR spectra of polymorphic amylin fibrils and morphologically homogeneous fibrils, respectively. Both samples were prepared from AMY05 and were hydrated in these measurements. 13C chemical shifts at the positions of maximum crosspeak intensity are similar in the two spectra, with the exception of chemical shifts for Ala5, for which the Cβ chemical shifts differ by 8 ppm, and Ile26, for which the Cδ chemical shifts differ by 2 ppm. However, linewidths in the spectrum of polymorphic fibrils are greater than 2.5 ppm (full width at half maximum, determined from crosspeaks in the 2D spectra), while linewidths in the spectrum of morphologically homogeneous fibrils range from 1.0 ppm to 2.0 ppm and are typically less than 1.5 ppm. The greater linewidths in the polymorphic case reflect the presence of a distribution of structures, which produces a range of chemical shifts for individual 13C-labeled sites. In morphologically homogeneous samples (Figures 4, 5, and 6), nearly all residues exhibit a single set of 13C chemical shifts, indicating a single molecular structure. Only Gly33 exhibits resolved major and minor 13C NMR signals, with intensity ratios of approximately 4:1, which we attribute to the existence of a minor population of an alternative peptide backbone conformation at Gly33.
Figure 4.
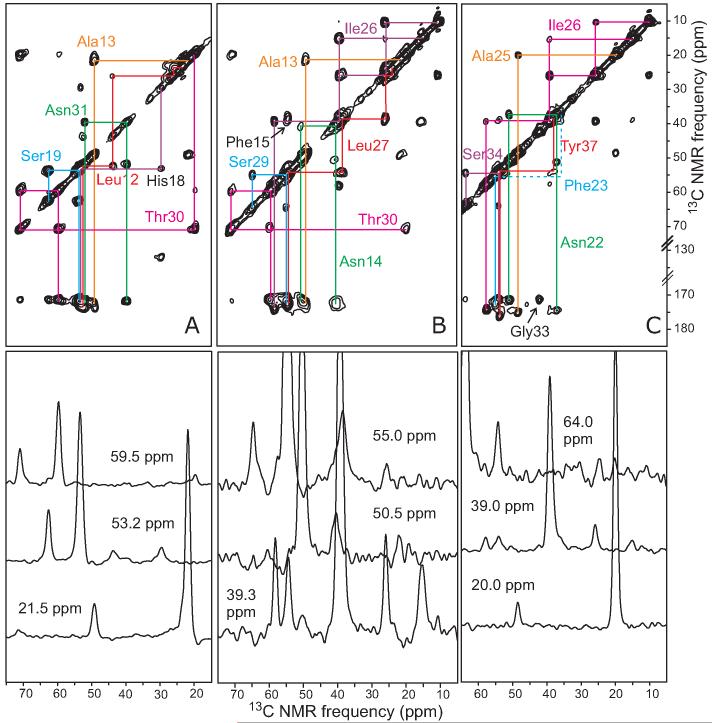
Selected regions from 2D 13C fpRFDR spectra of polymorphic (A) and morphologically homogenous (B,C) amylin fibrils with the AMY05 labeling pattern. Fibrils are rehydrated after lyophilization in A and B, but lyophilized without rehydration in C. Chemical shift assignment paths for uniformly 15N,13C-labeled residues are shown in B, with dashed lines extending to A and C to permit comparisons of 13C chemical shifts in the three samples. 1D slices from each spectrum at the indicated chemical shift positions are shown directly below. Measurements were performed at 150.7 MHz 13C NMR frequency, with a 2.2 ms fpRFDR exchange period and a 25.00 kHz MAS frequency. Gaussian line broadening of 150 Hz was applied in both dimensions. In the 2D plots, the lowest contour levels are slightly above the noise level and increase by successive factors of 1.7.
Figure 5.
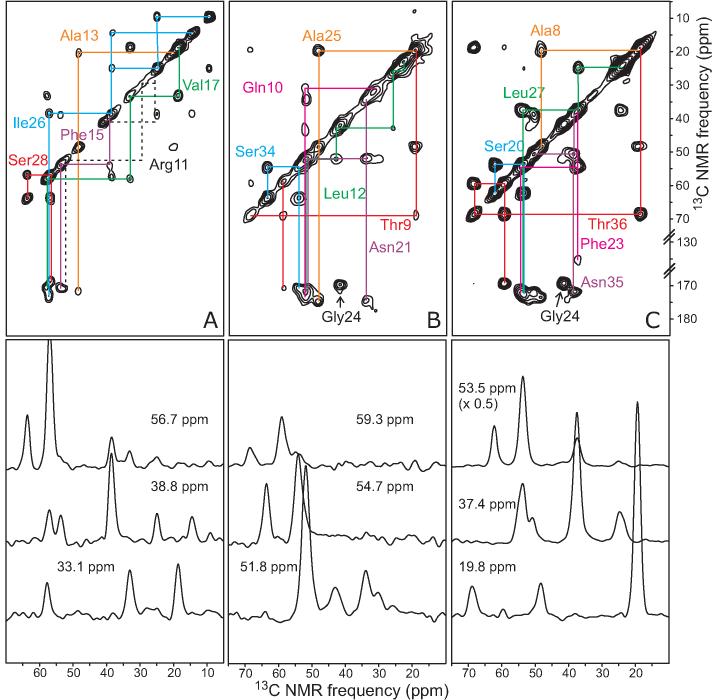
Same as in Figure 3B, for morphologically homogeneous amylin fibrils with the AMY06, AMY07, AMY08 labeling patterns (A, B, and C, respectively). Measurements were performed at 100.4 MHz 13C NMR frequency, with a 1.6 ms fpRFDR exchange period and a 20.00 kHz MAS frequency.
Figure 6.
Same as in Figure 3B, for morphologically homogeneous amylin fibrils with the AMY09, AMY10, and AMY11 labeling patterns (A, B, and C, respectively). Measurements were performed at 150.7 MHz 13C NMR frequency, with a 2.2 ms fpRFDR exchange period and MAS frequencies of 25.00 kHz, 30.00 kHz, and 15.00 kHz (A, B, and C, respectively).
Figure 4C shows the 2D 13C NMR spectrum of morphologically homogeneous AMY05 fibrils in the dry, lyophilized state. Crosspeaks in Figure 4C are broader than in Figure 4B, as observed in previous solid state NMR studies of other amyloid fibrils when comparing lyophilized and hydrated states of the same sample (18, 19, 22, 77, 78). Line-narrowing in the hydrated state is attributed to increased molecular motions, which partially average out the inhomogeneous broadening of NMR chemical shifts that is present in all noncrystalline solids. Surprisingly, the Cβ chemical shift of Ala5 changes significantly in the lyophilized state. All other chemical shifts are unchanged, indicating that the fibril structure is not dependent on hydration. The anomalous behavior of Ala5 is consistent with conformational flexibility in the disulfide-linked loop between Cys2 and Cys7. EPR studies of spin-labeled amylin fibrils also indicate mobility in the N-terminal segment, although the native disulfide linkage was not present in the EPR studies (36).
13C NMR signals of all residues from Ala8 to Tyr37 were investigated in morphologically homogeneous samples of AMY05-AMY11 fibrils. 1D 13C NMR spectra of all samples in hydrated and lyophilized states are given in Figures S3-S5 of the supporting information. 2D 13C NMR spectra are shown in Figures 5 and 6. Assignments of 13C NMR signals were based on the characteristic chemical shifts ranges for each amino acid residue type and on the patterns of crosspeak connectivities in the 2D spectra. Residues labeled in multiple samples displayed identical chemical shifts within experimental errors in all cases. 13C chemical shifts are summarized in Table S1 of the supporting information.
13C NMR linewidths in the 1.0-2.0 ppm range indicate that the entire segment from Ala8 to Tyr37 is structured and part of the fibril core. Crosspeak signals from Arg11 were missing in spectra of AMY06 (Figure 5C), and crosspeaks from neighboring residues (i.e., Ala8, Thr9, Gln10, Leu12, and Ala13) were found to be weak when compared to crosspeaks from other residues within the same sample. These observations suggest that residues 8-13 have somewhat greater amplitudes of motion or different time scales for motion than residues 14-37.
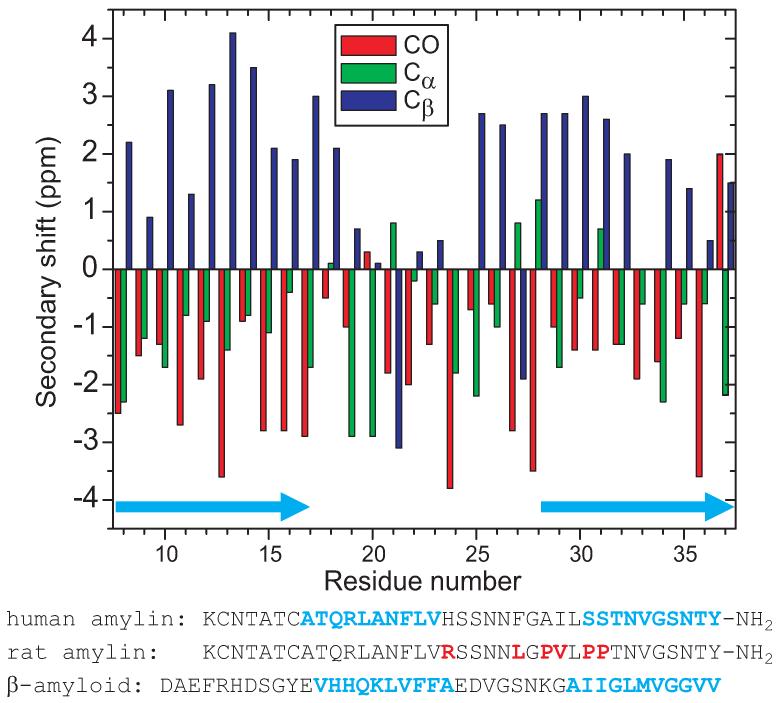
Secondary 13C chemical shifts, defined as δsecondary = δexp − δcoil, where δexp is the chemical shift for a given site in amylin fibrils and δcoil is the chemical shift for the same site in a “random coil” peptide, are plotted in Figure 7, using random coil shifts determined by Wishart et al. (79). The majority of residues exhibit secondary shifts consistent with β-strand conformations, i.e., positive secondary shifts for Cβ sites and negative secondary shifts for CO and Cα sites (16, 80). Deviations from this pattern are observed at His18 (for Cα), Ser20 (for CO), Asn21 (for Cα and Cβ), Leu27 (for Cα and Cβ), Ser28 (for Cα), and Asn31 (for Cα). The large positive CO secondary shift for Tyr37 is a consequence of Tyr37 being the C-terminal residue. Qualitatively, the secondary shifts support the existence of two β-strand segments, comprised approximately of residues 8-17 and 28-37 and separated by a “bend” or “loop” segment comprised of residues 18-27.
Figure 7.
13C NMR secondary chemical shifts for carbonyl, Cα, and Cβ sites in amylin fibrils with the morphology in Figs. 1C and 1D, determined from spectra in Figs. 4, 5, and 6. Light blue arrows indicate likely β-strand segments. Amino acid sequences of human amylin (with likely β-strands in light blue), rodent amylin (with amino acid differences in red), and β-amyloid (with previously determined β-strands (20-22) in light blue) are shown.
Experimentally determined 13C NMR chemical shifts were used as input to the TALOS program, which provides predictions of backbone torsion angles based on comparisons with a chemical shift database for proteins of known structure (81). TALOS predictions (i.e., average backbone torsion angles for the ten best matches to the TALOS database, containing 186 proteins with known structures and chemical shifts; see Table S1 of supporting information) were within common β-strand ranges (i.e., ϕ = −140°±30° and ψ = 140°±30°) for all residues except Leu16, His18, Ser20, Asn21, Asn22, Phe23, Leu27, and Asn35. Of the ten best matches to the TALOS database, at least nine were in the β-strand region for all residues except Ser20, Asn21, Asn22, Gly24, Leu27, Ser28, and Gly33.
Quantitative constraints on backbone conformation
When two consecutive residues k and k+1 are isotopically labeled, a quantitative constraint on the backbone torsion angle ψ of residue k can be obtained by measuring the magnetic dipole-dipole coupling between consecutive backbone amide 15N nuclei. The strength of this coupling is proportional to R−3, where R is the internuclear distance, which is in turn a function of ψ. Assuming standard chemical bonding geometry, R ranges between 2.70 Å at ψ = ±30° to 3.65 Å at ψ = ±180°, corresponding to coupling constants (defined as γ2h/2πR3, where h is Planck's constant and γ is the nuclear magnetogyric ratio) between 25 and 63 Hz. The 15N-15N dipole-dipole coupling can be measured with the fpRFDR-CT recoupling technique (54), which has been shown to give accurate data for this range of coupling strengths (17-19). Note that the 15N-15N distance, and hence the fpRFDR-CT data, is independent of the sign of ψ, assuming ideal bonding geometry. In samples where more than two residues are isotopically labeled, it is advantageous to detect the 15N fpRFDR-CT curves through 13C NMR signals of directly-bonded Cα sites, because the spectral resolution in the 13C NMR spectrum is generally higher than in the 15N NMR spectrum. This is accomplished by transferring spin polarization from 15N nuclei to directly bonded 13Cα sites after the fpRFDR-CT period, using a frequency-selective TEDOR pulse sequence (64-66). As long as the 13Cα NMR line of either residue k or residue k+1 is at least partially resolved in the 1D 13C NMR spectrum, a constraint on ψ of residue k can be obtained.
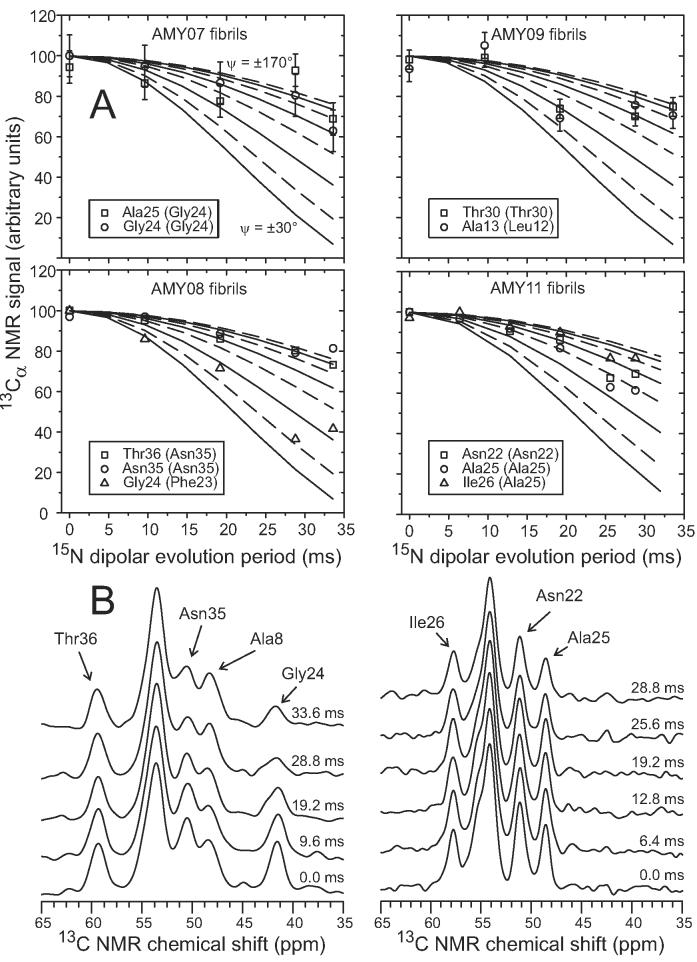
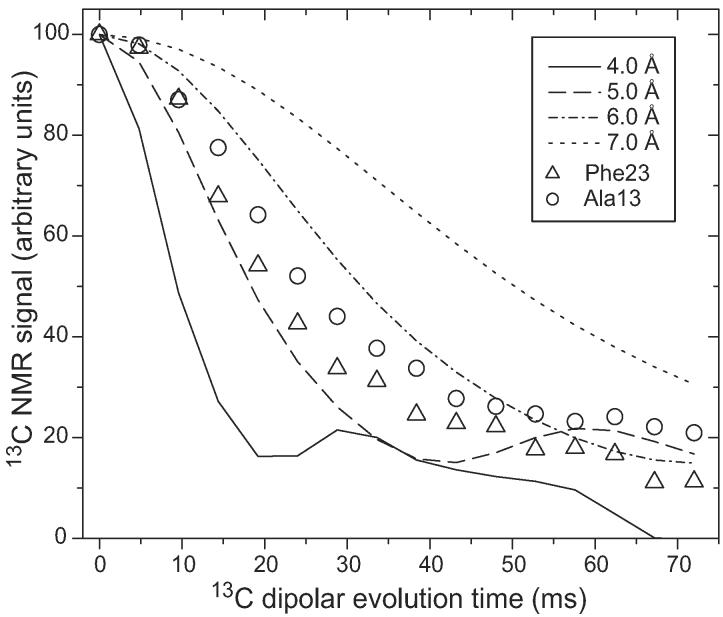
Figure 8A compares experimental 13Cα-detected 15N fpRFDR-CT data for amylin fibrils with simulated curves for various values of ψ. Figure 8B shows examples of the 13C NMR spectra from which the data in Figure 8A were obtained by measurement of peak areas after least-squares fitting to a sum of Gaussian lineshapes with fixed positions and widths, but variable amplitudes. Of the ten residues (and seven ψ angles) for which data could be obtained, only the labeled Phe23/Gly24 pair in AMY08 yielded data that are clearly inconsistent with a β-strand conformation. Data for AMY08 fibrils indicate |ψ| ≈ 60° for Phe23, but for other residues |ψ| > 100°.
Figure 8.
(A) Experimental (symbols) and simulated (solid and dashed lines) 15N fpRFDR-CT data for amylin fibrils with AMY07, AMY08, AMY09, and AMY11 isotopic labeling patterns, detected through 13C NMR signals of Cα sites in the indicated residues. The experimental data serve as constraints on the backbone ψ torsion angles of residues in parentheses. Simulations are for ψ values ranging from ±30° (most rapidly decreasing solid line) to ±170° (least rapidly decreasing dashed line), in ±20° steps. Data for AMY07, AMY08, and AMY09 were obtained at 100.8 MHz 13C NMR frequency and 20.00 kHz MAS frequency. Data for AMY11 were obtained at 150.7 MHz 13C NMR frequency and 15.00 kHz MAS frequency. (B) 13C NMR spectra from which AMY08 (left) and AMY11 (right) data were obtained, with 15N dipolar evolution periods and assignments of 13Cα lines indicated.
A non-β-strand conformation at Phe23 is consistent with the existence of a bend in residues 18-27. Although the TALOS analysis suggests that Asn22 has a non-β-strand conformation, |ψ| ≈ 100° for Asn22 according to the data in Figure 8. Of course, these 15N fpRFDR-CT data do not place constraints on backbone ϕ torsion angles, and the value of ψ alone does not fully determine the backbone conformation.
Identification of sidechain contacts
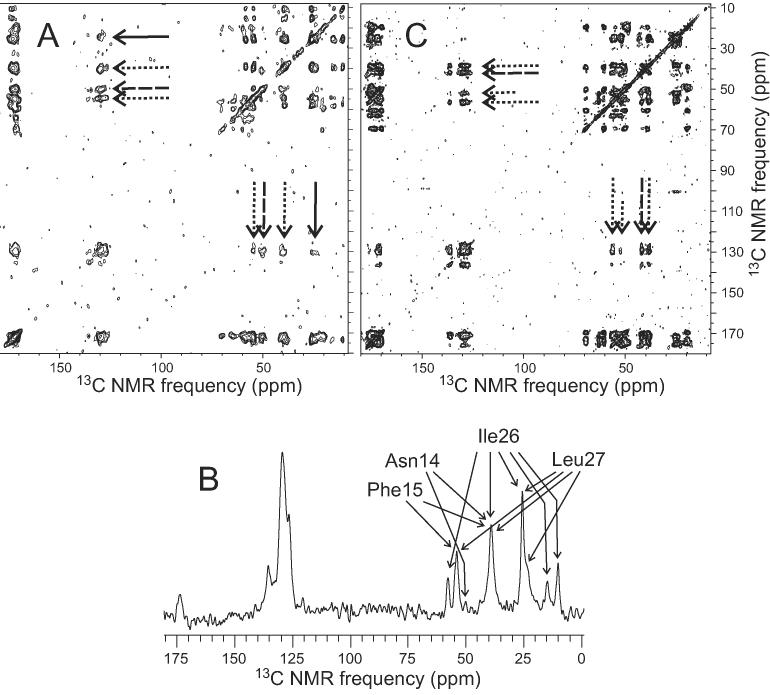
Figure 9A shows a 2D RAD spectrum of AMY10 fibrils, in which relatively weak nonsequential, inter-residue crosspeaks are observed between aromatic 13C NMR lines of Phe15 and aliphatic 13C NMR lines of Ile26 and Leu27. Spin polarization transfer from Phe15 to Ile26 and Leu27 is confirmed by the 1D RAD spectrum in Figure 9B. The simplest interpretation of these data, as discussed in similar studies of Aβ1-40 fibrils (22), is that the phenyl ring of Phe15 is in close proximity (approximately 6 Å or less) to sidechain methyl carbons of either Ile26 or Leu27. However, because Asn14 is also uniformly 15N,13C-labeled in AMY10, the observed crosspeaks and polarization transfer may also result from proximity between the Asn14 sidechain and the Ile26 sidechain or the Asn14 sidechain and the Leu27 sidechain. This uncertainty is due to the long exchange periods employed in the RAD measurements, which produce near equilibration of spin polarizations for sequential labeled residues. Although the precise sidechain contact indicated by the RAD data can not be determined, these data do support the existence of non-β-strand conformations at certain sites in residues 18-27, leading to an overall bend that can bring residues 14-15 close to residues 26-27.
Figure 9.
(A) 2D RAD spectrum of AMY10 fibrils, obtained at 150.7 MHz 13C NMR frequency with MAS at 21.00 kHz and a 1000 ms exchange period. Arrows indicate crosspeaks between 13C NMR signals of Phe15 aromatic carbons and Ile26/Leu27 methyl carbons (solid lines), Asn14 Cα carbons (dashed lines), and Phe15 Cα and Cβ carbons (dotted lines). (B) 1D RAD spectrum of AMY10 fibrils, obtained at 150.7 MHz 13C NMR frequency with MAS at 25.00 kHz and a 1000 ms exchange period after selective preparation of longitudinal spin polarization on Phe15 aromatic carbons. Assignments of aliphatic 13C NMR signals that arise from polarization transfer from the Phe15 aromatic carbons are shown. (C) 2D RAD spectrum of AMY08 fibrils, obtained at 150.7 MHz 13C NMR frequency with MAS at 18.00 kHz and a 500 ms exchange period. Arrows indicate crosspeaks between 13C NMR signals of Phe23 aromatic carbons and Gly24 Cα carbons (dashed lines) and Phe23 Cα and Cβ carbons (dotted lines). Short dotted arrows indicate crosspeaks that arise from MAS sidebands of Phe23 carbonyl signals. Contour levels in 2D plots increase by successive factors of 1.5.
Figure 9C shows a 2D RAD spectrum of AMY08 fibrils, in which crosspeaks between Phe23 and Gly24, but no nonsequential crosspeaks, are observed. The absence of nonsequential crosspeaks in this spectrum demonstrates the structural specificity of 2D RAD measurements. 2D RAD measurements were also performed on AMY05, AMY06, AMY07, and AMY09, and AMY11 fibrils, without detection of crosspeaks that place constraints on the amylin conformation.
Evidence for in-register parallel β-sheets
As originally demonstrated by Lynn, Meredith, Botto, and coworkers (26, 27), the β-sheet structures in amyloid fibrils can be investigated by applying dipolar recoupling techniques to samples that are prepared from peptides in which a single site is 13C-labeled. With single-site labeling, the dipolar recoupling data represent measurements of intermolecular distances, which can readily distinguish parallel from antiparallel β-sheets and establish the precise registry of backbone hydrogen bonds within the β-sheets. We have previously used 13C fpRFDR-CT measurements to investigate β-sheet structures in fibrils formed by β-amyloid peptides (17, 19, 21) and by yeast prion proteins (18, 63).
Figure 10 compares experimental 13C fpRFDR-CT data for AMY04 fibrils with simulations for ideal linear chains of 13C nuclei with various internuclear spacings. The large chemical shift difference between the 13C-labeled methyl site of Ala13 and the carbonyl site of Phe23 allowed intermolecular distances for Ala13 methyls and Phe23 carbonyls to be measured separately with a single AMY04 fibril sample. Assuming a β-strand conformation for residues 8-17, the intramolecular distance between the labeled methyl and carbonyl sites must exceed 10 Å (even if residues 18-27 are in a loop that folds back on residues 8-17, as can be seen by examination of molecular models), making intramolecular dipole-dipole couplings insignificant. For both labeled sites, the fpRFDR-CT data indicate nearest-neighbor intermolecular distances between 5.0 Å and 5.5 Å. In an ideal in-register parallel β-sheet, these distances would be 4.8Å. A one-residue shift from in-register alignment would produce nearest-neighbor distances of approximately 5.3 Å for a backbone carbonyl label, but nearest-neighbor distances of approximately 6.7 Å for a Cβ label (18). Thus, although the Phe23 carbonyl data are consistent with either in-register or shifted parallel β-sheet structures, the Ala13 methyl data rule out a shifted β-sheet structure. The fact that the experimental fpRFDR-CT data decay more slowly than simulated data for a 4.8 Å distance is attributable to effects of transverse nuclear spin relaxation on the experimental data (17).
Figure 10.
Experimental and simulated 13C fpRFDR-CT data for amylin fibrils with the AMY04 labeling pattern. Separate measurements were performed on the labeled Phe23 carbonyl (triangles) and Ala13 methyl (circles) sites. Experimental data are corrected for signal contributions from natural-abundance 13C nuclei by subtraction of a constant baseline equal to 16.5% and 13.2% of the initial signal (based on the numbers of carbonyl and methyl sites). Simulations are for linear chains of 13C nuclei with the indicated nearest-neighbor spacings.
EPR data for spin-labeled amylin fibrils also indicate a parallel β-sheet structure (36). The EPR measurements were carried out on samples that were polymorphic, based on the reported TEM images (36), and were prepared from peptides that lacked the native disulfide bond. The solid state NMR data in Figure 10 therefore strengthen the existing evidence for inregister parallel β-sheets in amylin fibrils, particularly for fibrils with the striated ribbon morphology and the native disulfide bond.
Discussion
Development of molecular models for amylin fibrils
Although the solid state NMR and electron microscopy data presented above are not sufficient to determine the molecular structure of amylin fibrils uniquely, it is nonetheless useful to develop tentative structural models based on these data, both as a means of summarizing the information content of the experimental data and as a guide for future experiments. Models for amylin decamers in configurations that represent a segment of the amylin protofilament were developed as described above (see Materials and Methods), using restrained Langevin dynamics to identify low-energy structures that are consistent with the experimental data. Restraints included: (i) backbone torsion angle restraints for β-strand segments identified from 13C chemical shifts (Figure 7) and supported by 15N fpRFDR-CT data (Figure 10), and for the non-β-strand ψ angle of Phe23; (ii) intermolecular distance restraints between backbone amide nitrogen and carbonyl oxygen sites in β-strand segments, in order to produce an in-register parallel alignment of neighboring peptide chains that is consistent with 13C fpRFDR-CT data (Figure 10) and the earlier EPR data; (iii) intramolecular distance restraints between sidechains of Asn14 and Ile26 (model in Figure 11C) or Phe15 and Leu27 (model in Figure 11D), supported by nonsequential polarization transfers in 1D and 2D RAD spectra (Figure 9); (iv) intramolecular distance restraints between certain pairs of Cα sites in the β-strand segments, in order to produce a reversal of the peptide chain direction by the bend in residues 18-27, as supported by the amylin protofilament diameter observed in TEM images (Figure 1). In addition, the models were constructed as two layers of amylin pentamers with C2 rotational symmetry about the fibril axis (i.e., about the direction approximately perpendicular to the β-strands and approximately parallel to the interstrand hydrogen bonds). The double-layered structure is consistent with MPL measurements (Figure 3). The C2 symmetry is consistent with the observation of relatively narrow 13C NMR lines with a single set of chemical shifts, which indicates that all amylin molecules have nearly identical conformations and are in nearly identical structural environments (although the conformations and environments are not strictly identical, due to the noncrystallinity of amyloid fibrils).
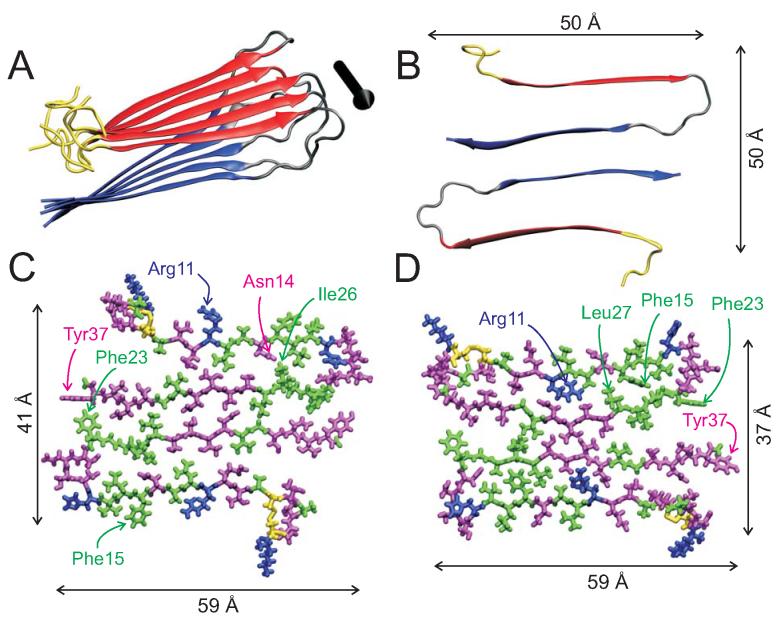
Figure 11.
Molecular structural models for the protofilament in amylin fibrils with striated ribbon morphologies, generated using restrained Langevin dynamics simulations as described in the text. (A) Ribbon representation of one cross-β molecular layer, with N-terminal and C-terminal β-strand segments colored red and blue, respectively. The black arrow indicates the fibril axis. (B) Cross-sectional view of two amylin molecules in the protofilament. This configuration, with approximate C2 symmetry about the fibril axis, is supported by the STEM data in Figure 3 and the observation of a single set of chemical shifts in 13C NMR spectra. The overall dimensions are consistent with TEM and AFM images. (C, D) All-atom representations of two possible models, with hydrophobic residues in green, polar residues in magenta, positively charged residues in blue and disulfide-linked cysteine residues in yellow.
Figure 11A shows the ribbon representation of a typical structure for one pentameric layer of amylin molecules after simulated annealing with experimentally-based restraints, starting from well-separated molecules with extended conformations. The N-terminal segment includes the Cys2-Cys7 disulfide bond, but is otherwise unrestrained, leading to substantial disorder consistent with the NMR observations for Ala5. Figure 11B shows a cross-sectional view of a pair of amylin molecules with the C2 symmetry discussed above. Protofilament models with C2 symmetry and with two β-strands per molecule, separated by a chain-reversing bend in residues 18-27, have overall widths of approximately 5 nm, consistent with the apparent protofilament widths in TEM images of striated ribbons. All-atom representations are shown in Figures 11C and 11D for the final structural models from two independent modeling runs. These two models are shown to illustrate the range of amylin protofilament structures that are consistent with existing data. The main differences in peptide conformation in the two models are in residues 19-21, 24, and 25, resulting in different sets of sidechain contacts between β-sheet layers. In Figure 11C, sidechains of Gln10, Leu12, Asn14, and Leu16 make contacts to the β-sheet formed by residues 28-37, while sidechains of Arg11, Ala13, and Phe15 are on the exterior of the protofilament. In Figure 11D, sidechains of Arg11, Ala13, and Phe15 make contacts to the β-sheet formed by residues 28-37, while sidechains of Gln10, Leu12, Asn14, and Leu16 are on the exterior of the protofilament. In Figure 11C, sidechains of Leu27, Ser29, and Asn31 are located in the interface between the two layers of amylin molecules, while sidechains of Leu26, Ser 28, Thr30, and Val32 are in the interior of each layer. In Figure 11D, sidechains of Leu26, Ser 28, Thr30, and Val32 are located in the interface between the two layers of amylin molecules, while sidechains of Leu27, Ser29, and Asn31 are in the interior of each layer. We interpret the qualitative observation that crosspeaks for residues 8-13 in 2D fpRFDR spectra of hydrated amylin fibrils are relatively weak as support for structural models in which β-sheets formed by residues 8-17 are located on the exterior of the protofilament, rather than participating in the interface between molecular layers. Sidechains on the exterior are likely to have greater amplitudes of motion, leading to a reduction of motionally-averaged 13C-13C dipole-dipole couplings and possibly a reduction in 13C T2 relaxation times for these residues. Both effects would reduce the crosspeak intensities in 2D fpRFDR spectra.
We attribute the observation of 2.5 nm fibril heights in AFM images of striated ribbons (Figure 2) to disassembly of the two molecular layers within the amylin protofilament under AFM scanning conditions. Fibrils with 5.0 nm heights represent intact protofilaments.
Comparison with Aβ1-40 fibril structure
Models in Figure 11 are remarkably similar to models developed by Petkova et al. for Aβ1-40 fibrils (22), especially for the protofilaments in Aβ1-40 fibrils that are grown with gentle agitation, which also have morphologies that closely resemble striated ribbons (21). In Aβ1-40fibrils, residues 1-9 are most strongly disordered (although not disulfide-linked), residues 12-21 and 30-40 form β-strands, and residues 22-29 form a chain-reversing bend. The protofilament is comprised of two layers of Aβ1-40 molecules, with C2 rotational symmetry about the fibril axis and with the C-terminal β-strands forming the interface between molecular layers. Figure 7 shows an alignment of the amylin and Aβ1-40 sequences based on structural similarity. This alignment differs slightly from a previously proposed alignment that was based on sequence similarity (82), in that the FGAIL segment of amylin (residues 23-27) aligns with residues 26-30 of Aβ1-40 in Figure 7, rather than with residues 28-32 of Aβ1-40. Nonetheless, our evidence that the peptide conformations in amylin and Aβ1-40 fibrils are similar supports the earlier proposal that the two peptides have similar secondary structure patterns in the fibrillar state (36).
Although the protofilament structures in amylin striated ribbons and in “agitated” Aβ1-40fibrils are apparently very similar, differences in amino acid sequence imply that the interactions that stabilize the two structures are different. The hydrophobic nature of the β-strand segments in Aβ1-40 fibrils implies that sidechain interactions within and between β-sheet layers are predominantly hydrophobic. In the models of Petkova et al., the only non-hydrophobic sidechains in the Aβ1-40 protofilament core are oppositely charged Asp and Lys sidechains, which form salt bridges (22). In contrast, the core of the amylin protofilament (Figures 11C and 11D) contains both hydrophobic and polar sidechains. Hydrophobic sidechains of residues 23-27 interact with hydrophobic sidechains of residues 15-17 and 32. Simultaneously, polar sidechains of residues 28-31 in one molecular layer may interact favorably with sidechains of the same residues in the other molecular layer. Additional experimental constraints on sidechain contacts are required before these interactions can be determined definitively. However, it appears that amylin fibrils are stabilized by a mixture of hydrophobic and polar interactions.
Gazit and coworkers have proposed that sidechain interactions among aromatic residues have important stabilizing effects on amyloid structures (83, 84). The role of interactions among aromatic sidechains in amylin fibril formation has also been addressed by Marek et al. (85). In the absence of higher-resolution restraints on sidechain conformations, the models in Figure 11 do not permit an assessment of the significance of these interactions.
Solid state NMR studies of Aβ1-40 fibrils have shown that fibrils with different morphologies in TEM images have different underlying molecular structures. Data acquired to date indicate that Aβ1-40 polymorphs have similar secondary structures (i.e., similar locations of β-strand, non-β-strand, and conformationally disordered segments) and tertiary structures (i.e., the same in-register parallel β-sheet organization), but different quaternary structures (i.e., different protofilament MPL values, symmetry, and contacts between β-sheets). Similar structural differences among amylin polymorphs may be uncovered in future experiments.
Comparison with earlier studies of amylin fibrils
Figure 7 also compares the human and rodent amylin sequences (86, 87). Rodent amylin does not form amyloid fibrils under typical in vitro conditions (57) or in vivo (56), nor do diabetic mice that express rodent (rather than human) amylin develop pancreatic amyloid (9). In light of the models in Figure 11, the critical difference between the two sequences seems to be the proline substitutions at Ser28 and Ser29, which would disrupt the C-terminal β-sheets. Biophysical studies by Abedini and Raleigh (88) have shown that amyloid formation by human amylin can also be inhibited by simultaneous proline substitutions at residues 17, 19, and 30, without changing the sequence in residues 20-29, a segment that has been suggested (89) and subsequently discussed (56, 83, 90, 91) as the “core” for amylin fibril formation. According to our data, residues 17 and 30 are in the C-terminal and N-terminal β-strands in amylin fibrils, respectively.
Identification of residues 20-29 as the core amyloid-forming segment of amylin seems erroneous, particularly if one expects the core segment to form β-sheets in the mature fibrils. In light of current knowledge that the ability to form amyloid fibrils is not an unusual property for a peptide (10), the fact that short peptides representing residues 20-29 form fibrils (56, 89) does not imply that this segment constitutes the core of full-length amylin fibrils. In fact, the molecular structures of fibrils formed by short peptides can be qualitatively different from the structures of the corresponding segments in the context of full-length peptide fibrils, as demonstrated by findings that certain β-amyloid fragments form antiparallel β-sheets in amyloid fibrils (16, 19, 24, 78), while other fragments (15, 26, 27) and full-length β-amyloid (15, 17, 21) form parallel β-sheets. Evidence from Griffiths et al. for antiparallel β-sheets in fibrils formed by residues 20-29 of human amylin (25) indicates that, as in the case of β-amyloid fibrils, the molecular structures in fibrils formed by amylin fragments need not reflect the molecular structure of full-length amylin fibrils.
Scrocchi et al. have reported that peptides representing residues 20-25 and 24-29 can act as inhibitors of fibril formation by full-length amylin, while a peptide representing residues 22-27 accelerates fibril formation (91). Similarly, Porat et al. have shown that a peptide representing residues 22-28, with a F23Y substitution, inhibits fibril formation (83). Assuming that these amylin fragments interact with the corresponding segments of the full-length sequence and that residues 8-17 and 28-37 comprise the β-sheet core of full-length amylin fibrils as indicated by our data, it appears that interactions outside the β-sheet core can affect fibril formation. Efforts to design fibrillization inhibitors need not focus exclusively on the β-sheetforming segments of amylin.
Green et al. have shown that rodent amylin with R18H, L23F, and V26I substitutions (producing an amylin variant that differs from the human form only by proline substitutions at residues 25, 28, and 29) is capable of forming amyloid fibrils, but with much lower yield than human amylin under the same experimental conditions (92). The structural models in Figure 11 do not provide a simple rationalization for the effect of these substitutions. One possibility, which would require additional solid state NMR data to verify, is that the fibrils observed by Green et al. have somewhat different β-strand locations than fibrils formed by human amylin.
An alternative, “β-serpentine” model for amylin fibrils has been proposed by Kajava et al. (90), based on analysis of the amylin sequence, experimental data on fibril formation by amylin fragments, evidence for parallel β-sheets from EPR (36), STEM data from Goldsbury et al. (41), and analogies to known structures of β-helical proteins. The model of Kajava et al. contains three β-strands, formed by residues 12-17, 22-27, and 31-37 and separated by short chain-reversing bends in which His18, Asn21, and Thr30 have non-β-strand backbone conformations. The details of this model are inconsistent with our 15N fpRFDR-CT data for Phe23 and Thr30 and with our interpretation of the STEM data, as discussed above. Nonetheless, the possibility exists that the model of Kajava et al. may apply to amylin fibrils with morphologies that differ from the striated ribbons examined in our experiments.
Padrick and Miranker have used fluorescence spectroscopy as a probe of amylin fibril structure and structural changes that accompany fibril formation (93). Their data indicate reduced solvent accessibility and mobility for the Tyr37 sidechain upon fibril formation, and proximity of the Tyr37 sidechain to sidechains of Phe15 and/or Phe23 (estimated distances less than 11 Å). Although Tyr37 is not buried in the protofilament core in the models in Figure 11, solvent accessibility and sidechain mobility are restricted by interactions within parallel β-sheets and between β-sheet layers, and possibly by lateral association of protofilaments. Intermolecular distances from the Tyr37 sidechain to the Phe23 sidechain in the models in Figure 11 are roughly 5-12 Å, consistent with the fluorescence data. Tyr37-Phe15 distances are greater than 20 Å in Figure 11, although Padrick and Miranker's analysis of their fluorescence data suggests that sidechains of both Phe15 and Phe23 are close to Tyr37. The source of this disagreement with the earlier analysis of fluorescence data remains to be determined.
In conclusion, we have developed a protocol for the preparation of morphologically homogeneous amylin fibrils that leads to high-quality solid-state NMR data. We have obtained structural constraints from solid state NMR, electron microscopy, and AFM that support the low-resolution structural models shown in Figure 11. Additional constraints that permit refinement of these models may be obtained in future experiments. The current structural models apply to amylin fibrils with striated ribbon morphologies. Variations in molecular structure that underlie variations in fibril morphology may also be identified in future experiments, provided that protocols for the preparation of alternative morphologies can be developed.
It has been suggested that the toxicity of human amylin aggregates may be attributed to soluble oligomeric species and not to fibrils, based in part on the findings that certain transgenic mice (homozygous males that overexpress human amylin) develop hyperglycemia and insulin deficits prior to the appearance of detectable islet amyloid (94), and that rifampicin (a drug that may inhibit fibril formation but not oligomer formation) does not protect cultured β-cells from amylin-induced apoptosis (95). The interpretation of the rifampicin studies is unclear, given that Tomiyama et al. show that rifampicin binds to, but does not inhibit or dissociate, amylin fibrils(96). In future work, it will be interesting to compare the molecular structures of amylin oligomers with the structures of amylin fibrils, as has been done recently in the case of Aβ1-40(97).
Acknowledgements
We thank Dr. Aneta Petkova and Erin Reese for important contributions to our initial efforts to synthesize, purify, and characterize the fibrillar structure of human amylin and Dr. Anant Paravastu for assistance with electron microscopy and numerous scientific discussions.
Supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, and by an Otto Hahn Award to SL from the Max Planck Society.
Abbreviations
- IAPP
islet amyloid polypeptide
- EPR
electron paramagnetic resonance
- TEM
transmission electron microscope
- AFM
atomic force microscope
- NMR
nuclear magnetic resonance
- HBTU
2-(1H-benzotriazol-l-yl)−1,1,3,3-tetramethyluronium hexafluorophosphate
- DIEA
N,N- diisopropylethylamine
- STEM
scanning transmission electron microscope
- TFA
trifluoroacetic acid
- HPLC
high-performance liquid chromatography
- FPLC
fast protein liquid chromatography
- fpRFDR
finite-pulse radiofrequency-driven recoupling
- fpRFDR-CT
constant-time finite-pulse radiofrequency-driven recoupling
- MAS
magic-angle spinning
- RAD
radiofrequency-assisted diffusion
- Fmoc
fluorenylmethoxycarbonyl
- Hmb
2-hydroxy-4-methoxybenzyl
- HFIP
hexafluoroisopropanol
- rf
radiofrequency
- TEDOR
transferred echo double resonance
- MPL
mass per length
Footnotes
Supporting Information Available HPLC chromatograms from the purification of both reduced and oxidized amylin by reverse-phase chromatography (Figure S1); FPLC chromatograms from the purification of both human and rodent amylin by size-exclusion chromatography (Figure S2); 1D 13C spectra of amylin fibrils with various isotopic labeling patterns, in lyophilized and rehydrated conditions (Figures S3-S5); Table of 13C NMR chemical shifts in amylin fibrils, TALOS predictions of backbone torsion angles based on these shifts, and torsion angles determined from 15N fpRFDR-CT measurements (Table S1). This material is freely available on the World Wide Web at http://www.acs.org.
Supplementary Material
References
- 1.Gedulin BR, Jodka CM, Herrmann K, Young AA. Role of endogenous amylin in glucagon secretion and gastric emptying in rats demonstrated with the selective antagonist, AC187. Regul. Pept. 2006;137:121–127. doi: 10.1016/j.regpep.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 2.Cooper GJS, Willis AC, Clark A, Turner RC, Sim RB, Reid KBM. Purification and characterization of a peptide from amyloid rich pancreases of type 2 diabetic patients. Proc. Natl. Acad. Sci. U. S. A. 1987;84:8628–8632. doi: 10.1073/pnas.84.23.8628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Westermark P, Wernstedt C, Wilander E, Hayden DW, Obrien TD, Johnson KH. Amyloid fibrils in human insulinoma and islets of Langerhans of the diabetic cat are derived from a neuropeptide like protein also present in normal islet cells. Proc. Natl. Acad. Sci. U. S. A. 1987;84:3881–3885. doi: 10.1073/pnas.84.11.3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lorenzo A, Razzaboni B, Weir GC, Yankner BA. Pancreatic islet cell toxicity of amylin associated with type 2 diabetes mellitus. Nature. 1994;368:756–760. doi: 10.1038/368756a0. [DOI] [PubMed] [Google Scholar]
- 5.Kahn SE. The relative contributions of insulin resistance and beta-cell dysfunction to the pathophysiology of type 2 diabetes. Diabetologia. 2003;46:3–19. doi: 10.1007/s00125-002-1009-0. [DOI] [PubMed] [Google Scholar]
- 6.Kahn SE, Andrikopoulos S, Verchere CB. Islet amyloid: A long-recognized but underappreciated pathological feature of type 2 diabetes. Diabetes. 1999;48:241–253. doi: 10.2337/diabetes.48.2.241. [DOI] [PubMed] [Google Scholar]
- 7.Höppener JWM, Lips CJM. Role of islet amyloid in type 2 diabetes mellitus. Int. J. Biochem. Cell Biol. 2006;38:726–736. doi: 10.1016/j.biocel.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 8.Wang F, Hull RL, Vidal J, Cnop M, Kahn SE. Islet amyloid develops diffusely throughout the pancreas before becoming severe and replacing endocrine cells. Diabetes. 2001;50:2514–2520. doi: 10.2337/diabetes.50.11.2514. [DOI] [PubMed] [Google Scholar]
- 9.Höppener JWM, Oosterwijk C, Nieuwenhuis MG, Posthuma G, Thijssen JHH, Vroom TM, Ahrén B, Lips CJM. Extensive islet amyloid formation is induced by development of type 2 diabetes mellitus and contributes to its progression: Pathogenesis of diabetes in a mouse model. Diabetologia. 1999;42:427–434. doi: 10.1007/s001250051175. [DOI] [PubMed] [Google Scholar]
- 10.Dobson CM. Protein folding and misfolding. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 11.Sunde M, Blake CCF. From the globular to the fibrous state: protein structure and structural conversion in amyloid formation. Q. Rev. Biophys. 1998;31:1–39. doi: 10.1017/s0033583598003400. [DOI] [PubMed] [Google Scholar]
- 12.Tycko R. Molecular structure of amyloid fibrils: insights from solid state NMR. Q. Rev. Biophys. 2006;39:1–55. doi: 10.1017/S0033583506004173. [DOI] [PubMed] [Google Scholar]
- 13.Eanes ED, Glenner GG. X-ray diffraction studies on amyloid filaments. J. Histochem. Cytochem. 1968;16:673–677. doi: 10.1177/16.11.673. [DOI] [PubMed] [Google Scholar]
- 14.Antzutkin ON, Balbach JJ, Leapman RD, Rizzo NW, Reed J, Tycko R. Multiple quantum solid-state NMR indicates a parallel, not antiparallel, organization of β-sheets in Alzheimer's β-amyloid fibrils. Proc. Natl. Acad. Sci. U. S. A. 2000;97:13045–13050. doi: 10.1073/pnas.230315097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Antzutkin ON, Leapman RD, Balbach JJ, Tycko R. Supramolecular structural constraints on Alzheimer's β-amyloid fibrils from electron microscopy and solid-state nuclear magnetic resonance. Biochemistry. 2002;41:15436–15450. doi: 10.1021/bi0204185. [DOI] [PubMed] [Google Scholar]
- 16.Balbach JJ, Ishii Y, Antzutkin ON, Leapman RD, Rizzo NW, Dyda F, Reed J, Tycko R. Amyloid fibril formation by Aβ(16-22), a seven-residue fragment of the Alzheimer's β-amyloid peptide, and structural characterization by solid state NMR. Biochemistry. 2000;39:13748–13759. doi: 10.1021/bi0011330. [DOI] [PubMed] [Google Scholar]
- 17.Balbach JJ, Petkova AT, Oyler NA, Antzutkin ON, Gordon DJ, Meredith SC, Tycko R. Supramolecular structure in full-length Alzheimer's β-amyloid fibrils: Evidence for a parallel β-sheet organization from solid-state nuclear magnetic resonance. Biophys. J. 2002;83:1205–1216. doi: 10.1016/S0006-3495(02)75244-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chan JCC, Oyler NA, Yau WM, Tycko R. Parallel β-sheets and polar zippers in amyloid fibrils formed by residues 10-39 of the yeast prion protein Ure2p. Biochemistry. 2005;44:10669–10680. doi: 10.1021/bi050724t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petkova AT, Buntkowsky G, Dyda F, Leapman RD, Yau WM, Tycko R. Solid state NMR reveals a pH-dependent antiparallel β-sheet registry in fibrils formed by a β-amyloid peptide. J. Mol. Biol. 2004;335:247–260. doi: 10.1016/j.jmb.2003.10.044. [DOI] [PubMed] [Google Scholar]
- 20.Petkova AT, Ishii Y, Balbach JJ, Antzutkin ON, Leapman RD, Delaglio F, Tycko R. A structural model for Alzheimer's β-amyloid fibrils based on experimental constraints from solid state NMR. Proc. Natl. Acad. Sci. U. S. A. 2002;99:16742–16747. doi: 10.1073/pnas.262663499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petkova AT, Leapman RD, Guo ZH, Yau WM, Mattson MP, Tycko R. Self-propagating, molecular-level polymorphism in Alzheimer's β-amyloid fibrils. Science. 2005;307:262–265. doi: 10.1126/science.1105850. [DOI] [PubMed] [Google Scholar]
- 22.Petkova AT, Yau WM, Tycko R. Experimental constraints on quaternary structure in Alzheimer's β-amyloid fibrils. Biochemistry. 2006;45:498–512. doi: 10.1021/bi051952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jaroniec CP, MacPhee CE, Bajaj VS, McMahon MT, Dobson CM, Griffin RG. High-resolution molecular structure of a peptide in an amyloid fibril determined by magic angle spinning NMR spectroscopy. Proc. Natl. Acad. Sci. U. S. A. 2004;101:711–716. doi: 10.1073/pnas.0304849101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lansbury PT, Costa PR, Griffiths JM, Simon EJ, Auger M, Halverson KJ, Kocisko DA, Hendsch ZS, Ashburn TT, Spencer RGS, Tidor B, Griffin RG. Structural model for the b-amyloid fibril based on interstrand alignment of an antiparallel-sheet comprising a C-terminal peptide. Nat. Struct. Biol. 1995;2:990–998. doi: 10.1038/nsb1195-990. [DOI] [PubMed] [Google Scholar]
- 25.Griffiths JM, Ashburn TT, Auger M, Costa PR, Griffin RG, Lansbury PT. Rotational resonance solid state NMR elucidates a structural model of pancreatic amyloid. J. Am. Chem. Soc. 1995;117:3539–3546. [Google Scholar]
- 26.Gregory DM, Benzinger TLS, Burkoth TS, Miller-Auer H, Lynn DG, Meredith SC, Botto RE. Dipolar recoupling NMR of biomolecular self-assemblies: determining inter- and intrastrand distances in fibrilized Alzheimer's β-amyloid peptide. Solid State Nucl. Magn. Reson. 1998;13:149–166. doi: 10.1016/s0926-2040(98)00086-1. [DOI] [PubMed] [Google Scholar]
- 27.Benzinger TLS, Gregory DM, Burkoth TS, Miller-Auer H, Lynn DG, Botto RE, Meredith SC. Propagating structure of Alzheimer's β-amyloid(10-35) is parallel β-sheet with residues in exact register. Proc. Natl. Acad. Sci. U. S. A. 1998;95:13407–13412. doi: 10.1073/pnas.95.23.13407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kammerer RA, Kostrewa D, Zurdo J, Detken A, García-Echeverría C, Green JD, Müller SA, Meier BH, Winkler FK, Dobson CM, Steinmetz MO. Exploring amyloid formation by a de novo design. Proc. Natl. Acad. Sci. U. S. A. 2004;101:4435–4440. doi: 10.1073/pnas.0306786101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ritter C, Maddelein ML, Siemer AB, Lührs T, Ernst M, Meier BH, Saupe SJ, Riek R. Correlation of structural elements and infectivity of the HET-s prion. Nature. 2005;435:844–848. doi: 10.1038/nature03793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heise H, Hoyer W, Becker S, Andronesi OC, Riedel D, Baldus M. Molecular-level secondary structure, polymorphism, and dynamics of full-length alpha-synuclein fibrils studied by solid-state NMR. Proc. Natl. Acad. Sci. USA. 2005;102:15871–15876. doi: 10.1073/pnas.0506109102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferguson N, Becker J, Tidow H, Tremmel S, Sharpe TD, Krause G, Flinders J, Petrovich M, Berriman J, Oschkinat H, Fersht AR. General structural motifs of amyloid protofilaments. Proc. Natl. Acad. Sci. U. S. A. 2006;103:16248–16253. doi: 10.1073/pnas.0607815103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iwata K, Fujiwara T, Matsuki Y, Akutsu H, Takahashi S, Naiki H, Goto Y. 3D structure of amyloid protofilaments of β2-microglobulin fragment probed by solid state NMR. Proc. Natl. Acad. Sci. U. S. A. 2006;103:18119–18124. doi: 10.1073/pnas.0607180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naito A, Kamihira M, Inoue R, Saito H. Structural diversity of amyloid fibril formed in human calcitonin as revealed by site-directed 13C solid state NMR spectroscopy. Magn. Reson. Chem. 2004;42:247–257. doi: 10.1002/mrc.1323. [DOI] [PubMed] [Google Scholar]
- 34.Der-Sarkissian A, Jao CC, Chen J, Langen R. Structural organization of alpha-synuclein fibrils studied by site-directed spin labeling. J. Biol. Chem. 2003;278:37530–37535. doi: 10.1074/jbc.M305266200. [DOI] [PubMed] [Google Scholar]
- 35.Török M, Milton S, Kayed R, Wu P, McIntire T, Glabe CG, Langen R. Structural and dynamic features of Alzheimer's Aβ peptide in amyloid fibrils studied by site-directed spin labeling. J. Biol. Chem. 2002;277:40810–40815. doi: 10.1074/jbc.M205659200. [DOI] [PubMed] [Google Scholar]
- 36.Jayasinghe SA, Langen R. Identifying structural features of fibrillar islet amyloid polypeptide using site-directed spin labeling. J. Biol. Chem. 2004;279:48420–48425. doi: 10.1074/jbc.M406853200. [DOI] [PubMed] [Google Scholar]
- 37.Jiménez JL, Nettleton EJ, Bouchard M, Robinson CV, Dobson CM, Saibil HR. The protofilament structure of insulin amyloid fibrils. Proc. Natl. Acad. Sci. U. S. A. 2002;99:9196–9201. doi: 10.1073/pnas.142459399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiménez JL, Guijarro JL, Orlova E, Zurdo J, Dobson CM, Sunde M, Saibil HR. Cryo-electron microscopy structure of an SH3 amyloid fibril and model of the molecular packing. EMBO J. 1999;18:815–821. doi: 10.1093/emboj/18.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Serpell LC, Smith JM. Direct visualisation of the beta-sheet structure of synthetic Alzheimer's amyloid. J. Mol. Biol. 2000;299:225–231. doi: 10.1006/jmbi.2000.3650. [DOI] [PubMed] [Google Scholar]
- 40.Goldsbury C, Goldie K, Pellaud J, Seelig J, Frey P, Müller SA, Kistler J, Cooper GJS, Aebi U. Amyloid fibril formation from full-length and fragments of amylin. J. Struct. Biol. 2000;130:352–362. doi: 10.1006/jsbi.2000.4268. [DOI] [PubMed] [Google Scholar]
- 41.Goldsbury CS, Cooper GJS, Goldie KN, Müller SA, Saafi EL, Gruijters WTM, Misur MP, Engel A, Aebi U, Kistler J. Polymorphic fibrillar assembly of human amylin. J. Struct. Biol. 1997;119:17–27. doi: 10.1006/jsbi.1997.3858. [DOI] [PubMed] [Google Scholar]
- 42.Goldsbury CS, Wirtz S, Müller SA, Sunderji S, Wicki P, Aebi U, Frey P. Studies on the in vitro assembly of Aβ(1-40): Implications for the search for Aβ fibril formation inhibitors. J. Struct. Biol. 2000;130:217–231. doi: 10.1006/jsbi.2000.4259. [DOI] [PubMed] [Google Scholar]
- 43.Goldsbury C, Frey P, Olivieri V, Aebi U, Müller SA. Multiple assembly pathways underlie amyloid-β fibril polymorphisms. J. Mol. Biol. 2005;352:282–298. doi: 10.1016/j.jmb.2005.07.029. [DOI] [PubMed] [Google Scholar]
- 44.Ippel JH, Olofsson A, Schleucher J, Lundgren E, Wijmenga SS. Probing solvent accessibility of amyloid fibrils by solution NMR spectroscopy. Proc. Natl. Acad. Sci. U. S. A. 2002;99:8648–8653. doi: 10.1073/pnas.132098999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kheterpal I, Lashuel HA, Hartley DM, Wlaz T, Lansbury PT, Wetzel R. Aβ protofibrils possess a stable core structure resistant to hydrogen exchange. Biochemistry. 2003;42:14092–14098. doi: 10.1021/bi0357816. [DOI] [PubMed] [Google Scholar]
- 46.Whittemore NA, Mishra R, Kheterpal I, Williams AD, Wetzel R, Serpersu EH. Hydrogen-deuterium (H/D) exchange mapping of Aβ(1-40) amyloid fibril secondary structure using nuclear magnetic resonance spectroscopy. Biochemistry. 2005;44:4434–4441. doi: 10.1021/bi048292u. [DOI] [PubMed] [Google Scholar]
- 47.Yamaguchi KI, Katou H, Hoshino M, Hasegawa K, Naiki H, Goto Y. Core and heterogeneity of β2-microglobulin amyloid fibrils as revealed by H/D exchange. J. Mol. Biol. 2004;338:559–571. doi: 10.1016/j.jmb.2004.02.067. [DOI] [PubMed] [Google Scholar]
- 48.Williams AD, Portelius E, Kheterpal I, Guo JT, Cook KD, Xu Y, Wetzel R. Mapping Aβ amyloid fibril secondary structure using scanning proline mutagenesis. J. Mol. Biol. 2004;335:833–842. doi: 10.1016/j.jmb.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 49.Bitan G, Kirkitadze MD, Lomakin A, Vollers SS, Benedek GB, Teplow DB. Amyloid β-protein (Aβ) assembly: Aβ 40 and Aβ 42 oligomerize through distinct pathways. Proc. Natl. Acad. Sci. U. S. A. 2003;100:330–335. doi: 10.1073/pnas.222681699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shivaprasad S, Wetzel R. An intersheet packing interaction in Aβ fibrils mapped by disulfide cross-linking. Biochemistry. 2004;43:15310–15317. doi: 10.1021/bi048019s. [DOI] [PubMed] [Google Scholar]
- 51.Nelson R, Sawaya MR, Balbirnie M, Madsen AO, Riekel C, Grothe R, Eisenberg D. Structure of the cross-β spine of amyloid-like fibrils. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, Thompson MJ, Balbirnie M, Wiltzius JJW, McFarlane HT, Madsen AO, Riekel C, Eisenberg D. Atomic structures of amyloid cross-β spines reveal varied steric zippers. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 53.Makin OS, Serpell LC. Structural characterisation of islet amyloid polypeptide fibrils. J. Mol. Biol. 2004;335:1279–1288. doi: 10.1016/j.jmb.2003.11.048. [DOI] [PubMed] [Google Scholar]
- 54.Ishii Y, Balbach JJ, Tycko R. Measurement of dipole-coupled lineshapes in a many-spin system by constant-time two-dimensional solid state NMR with high-speed magic angle spinning. Chem. Phys. 2001;266:231–236. [Google Scholar]
- 55.Abedini A, Raleigh DP. Incorporation of pseudoproline derivatives allows the facile synthesis of human IAPP, a highly amyloidogenic and aggregation-prone polypeptide. Org. Lett. 2005;7:693–696. doi: 10.1021/ol047480+. [DOI] [PubMed] [Google Scholar]
- 56.Westermark P, Engström U, Johnson KH, Westermark GT, Betsholtz C. Islet amyloid polypeptide: Pinpointing amino acid residues linked to amyloid fibril formation. Proc. Natl. Acad. Sci. U. S. A. 1990;87:5036–5040. doi: 10.1073/pnas.87.13.5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Williamson JA, Miranker AD. Direct detection of transient alpha-helical states in islet amyloid polypeptide. Protein Sci. 2007;16:110–117. doi: 10.1110/ps.062486907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pines A, Gibby MG, Waugh JS. Proton-enhanced NMR of dilute spins in solids. J. Chem. Phys. 1973;59:569–590. [Google Scholar]
- 59.Bennett AE, Rienstra CM, Auger M, Lakshmi KV, Griffin RG. Heteronuclear decoupling in rotating solids. J. Chem. Phys. 1995;103:6951–6958. [Google Scholar]
- 60.Ishii Y. 13C-13C dipolar recoupling under very fast magic angle spinning in solid state nuclear magnetic resonance: Applications to distance measurements, spectral assignments, and high-throughput secondary-structure determination. J. Chem. Phys. 2001;114:8473–8483. [Google Scholar]
- 61.Morcombe CR, Gaponenko V, Byrd RA, Zilm KW. Diluting abundant spins by isotope edited radio frequency field assisted diffusion. J. Am. Chem. Soc. 2004;126:7196–7197. doi: 10.1021/ja047919t. [DOI] [PubMed] [Google Scholar]
- 62.Takegoshi K, Nakamura S, Terao T. 13C-1H dipolar-assisted rotational resonance in magic angle spinning NMR. Chem. Phys. Lett. 2001;344:631–637. [Google Scholar]
- 63.Shewmaker F, Wickner RB, Tycko R. Amyloid of the prion domain of Sup35p has an in-register parallel β-sheet structure. Proc. Natl. Acad. Sci. U. S. A. 2006;103:19754–19759. doi: 10.1073/pnas.0609638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jaroniec CP, Tounge BA, Herzfeld J, Griffin RG. Frequency selective heteronuclear dipolar recoupling in rotating solids: Accurate 13C-15N distance measurements in uniformly 13C,15N-labeled peptides. J. Am. Chem. Soc. 2001;123:3507–3519. doi: 10.1021/ja003266e. [DOI] [PubMed] [Google Scholar]
- 65.Hing AW, Vega S, Schaefer J. Transferred-echo double-resonance NMR. J. Magn. Reson. 1992;96:205–209. [Google Scholar]
- 66.Tycko R. Symmetry-based constant-time homonuclear dipolar recoupling in solid state NMR. J. Chem. Phys. 2007;126:064506. doi: 10.1063/1.2437194. [DOI] [PubMed] [Google Scholar]
- 67.Petkova AT, Tycko R. Sensitivity enhancement in structural measurements by solid state NMR through pulsed spin locking. J. Magn. Reson. 2002;155:293–299. doi: 10.1006/jmre.2002.2519. [DOI] [PubMed] [Google Scholar]
- 68.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 69.Bak M, Rasmussen JT, Nielsen NC. SIMPSON: A general simulation program for solid state NMR spectroscopy. J. Magn. Reson. 2000;147:296–330. doi: 10.1006/jmre.2000.2179. [DOI] [PubMed] [Google Scholar]
- 70.Case DA, Cheatham TE, Darden T, Gohlke H, Luo R, Merz KM, Onufriev A, Simmerling C, Wang B, Woods RJ. The Amber biomolecular simulation programs. J. Comput. Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of multiple amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Loncharich RJ, Brooks BR, Pastor RW. Langevin dynamics of peptides - the frictional dependence of isomerization rates of N-acetylalanyl-N'methylamide. Biopolymers. 1992;32:523–535. doi: 10.1002/bip.360320508. [DOI] [PubMed] [Google Scholar]
- 73.Tsui V, Case DA. Theory and applications of the generalized Born solvation model in macromolecular simulations. Biopolymers. 2000;56:275–291. doi: 10.1002/1097-0282(2000)56:4<275::AID-BIP10024>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 74.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996;14:33-8–27-8. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 75.Goldsbury C, Kistler J, Aebi U, Arvinte T, Cooper GJS. Watching amyloid fibrils grow by time-lapse atomic force microscopy. J. Mol. Biol. 1999;285:33–39. doi: 10.1006/jmbi.1998.2299. [DOI] [PubMed] [Google Scholar]
- 76.Wall JS, Hainfeld JF. Mass mapping with the scanning transmission electron microscope. Annu. Rev. Biophys. Biophys. Chem. 1986;15:355–376. doi: 10.1146/annurev.bb.15.060186.002035. [DOI] [PubMed] [Google Scholar]
- 77.Paravastu AK, Petkova AT, Tycko R. Polymorphic fibril formation by residues 10-40 of the Alzheimer's β-amyloid peptide. Biophys. J. 2006;90:4618–4629. doi: 10.1529/biophysj.105.076927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bu ZM, Shi Y, Callaway DJE, Tycko R. Molecular alignment within β-sheets in Aβ(14-23) fibrils: Solid state NMR experiments and theoretical predictions. Biophys. J. 2007;92:594–602. doi: 10.1529/biophysj.106.091017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wishart DS, Bigam CG, Holm A, Hodges RS, Sykes BD. 1H, 13C and 15N random coil NMR chemical shifts of the common amino acids. 1. Investigations of nearest neighbor effects. J. Biomol. NMR. 1995;5:67–81. doi: 10.1007/BF00227471. [DOI] [PubMed] [Google Scholar]
- 80.Saito H. Conformation dependent 13C chemical shifts: A new means of conformational characterization as obtained by high-resolution solid state 13C NMR. Magn. Reson. Chem. 1986;24:835–852. [Google Scholar]
- 81.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 82.O'Nuallain B, Williams AD, Westermark P, Wetzel R. Seeding specificity in amyloid growth induced by heterologous fibrils. J. Biol. Chem. 2004;279:17490–17499. doi: 10.1074/jbc.M311300200. [DOI] [PubMed] [Google Scholar]
- 83.Porat Y, Mazor Y, Efrat S, Gazit E. Inhibition of islet amyloid polypeptide fibril formation: A potential role for heteroaromatic interactions. Biochemistry. 2004;43:14454–14462. doi: 10.1021/bi048582a. [DOI] [PubMed] [Google Scholar]
- 84.Azriel R, Gazit E. Analysis of the structural and functional elements of the minimal active fragment of islet amyloid polypeptide (IAPP) - An experimental support for the key role of the phenylalanine residue in amyloid formation. J. Biol. Chem. 2001;276:34156–34161. doi: 10.1074/jbc.M102883200. [DOI] [PubMed] [Google Scholar]
- 85.Marek P, Abedini A, Song BB, Kanungo M, Johnson ME, Gupta R, Zaman W, Wong SS, Raleigh DP. Aromatic interactions are not required for amyloid fibril formation by islet amyloid polypeptide but do influence the rate of fibril formation and fibril morphology. Biochemistry. 2007;46:3255–3261. doi: 10.1021/bi0621967. [DOI] [PubMed] [Google Scholar]
- 86.Nishi M, Chan SJ, Nagamatsu S, Bell GI, Steiner DF. Conservation of the sequence of islet amyloid polypeptide in 5 mammals is consistent with its putative role as an islet hormone. Proc. Natl. Acad. Sci. U. S. A. 1989;86:5738–5742. doi: 10.1073/pnas.86.15.5738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Leffert JD, Newgard CB, Okamoto H, Milburn JL, Luskey KL. Rat amylin: Cloning and tissue specific expression in pancreatic islets. Proc. Natl. Acad. Sci. U. S. A. 1989;86:3127–3130. doi: 10.1073/pnas.86.9.3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Abedini A, Raleigh DP. Destabilization of human IAPP amyloid fibrils by proline mutations outside of the putative amyloidogenic domain: Is there a critical amyloidogenic domain in human IAPP? J. Mol. Biol. 2006;355:274–281. doi: 10.1016/j.jmb.2005.10.052. [DOI] [PubMed] [Google Scholar]
- 89.Glenner GG, Eanes ED, Wiley CA. Amyloid fibrils formed from a segment of the pancreatic islet amyloid protein. Biochem. Biophys. Res. Commun. 1988;155:608–614. doi: 10.1016/s0006-291x(88)80538-2. [DOI] [PubMed] [Google Scholar]
- 90.Kajava AV, Aebi U, Steven AC. The parallel superpleated β-structure as a model for amyloid fibrils of human amylin. J. Mol. Biol. 2005;348:247–252. doi: 10.1016/j.jmb.2005.02.029. [DOI] [PubMed] [Google Scholar]
- 91.Scrocchi LA, Chen Y, Waschuk S, Wang F, Cheung S, Darabie AA, McLaurin J, Fraser PE. Design of peptide-based inhibitors of human islet amyloid polypeptide fibrillogenesis. J. Mol. Biol. 2002;318:697–706. doi: 10.1016/S0022-2836(02)00164-X. [DOI] [PubMed] [Google Scholar]
- 92.Green J, Goldsbury C, Min T, Sunderji S, Frey P, Kistler J, Cooper G, Aebi U. Full-length rat amylin forms fibrils following substitution of single residues from human amylin. J. Mol. Biol. 2003;326:1147–1156. doi: 10.1016/s0022-2836(02)01377-3. [DOI] [PubMed] [Google Scholar]
- 93.Padrick SB, Miranker AD. Islet amyloid polypeptide: Identification of long-range contacts and local order on the fibrillogenesis pathway. J. Mol. Biol. 2001;308:783–794. doi: 10.1006/jmbi.2001.4608. [DOI] [PubMed] [Google Scholar]
- 94.Janson J, Soeller WC, Roche PC, Nelson RT, Torchia AJ, Kreutter DK, Butler PC. Spontaneous diabetes mellitus in transgenic mice expressing human islet amyloid polypeptide. Proc. Natl. Acad. Sci. U. S. A. 1996;93:7283–7288. doi: 10.1073/pnas.93.14.7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Meier JJ, Kayed R, Lin CY, Gurlo T, Haataja L, Jayasinghe S, Langen R, Glabe CG, Butler PC. Inhibition of human IAPP fibril formation does not prevent β-cell death: evidence for distinct actions of oligomers and fibrils of human IAPP. Am. J. Physiol. Endocrinol. Metab. 2006;291:E1317–E1324. doi: 10.1152/ajpendo.00082.2006. [DOI] [PubMed] [Google Scholar]
- 96.Tomiyama T, Kaneko H, Kataoka K, Asano S, Endo N. Rifampicin inhibits the toxicity of pre-aggregated amyloid peptides by binding to peptide fibrils and preventing amyloid-cell interaction. Biochem. J. 1997;322:859–865. doi: 10.1042/bj3220859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chimon S, Ishii Y. Capturing intermediate structures of Alzheimer's β-amyloid, Aβ(1-40), by solid-state NMR spectroscopy. J. Am. Chem. Soc. 2005;127:13472–13473. doi: 10.1021/ja054039l. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.