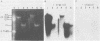Abstract
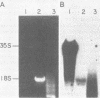
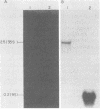
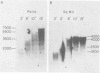
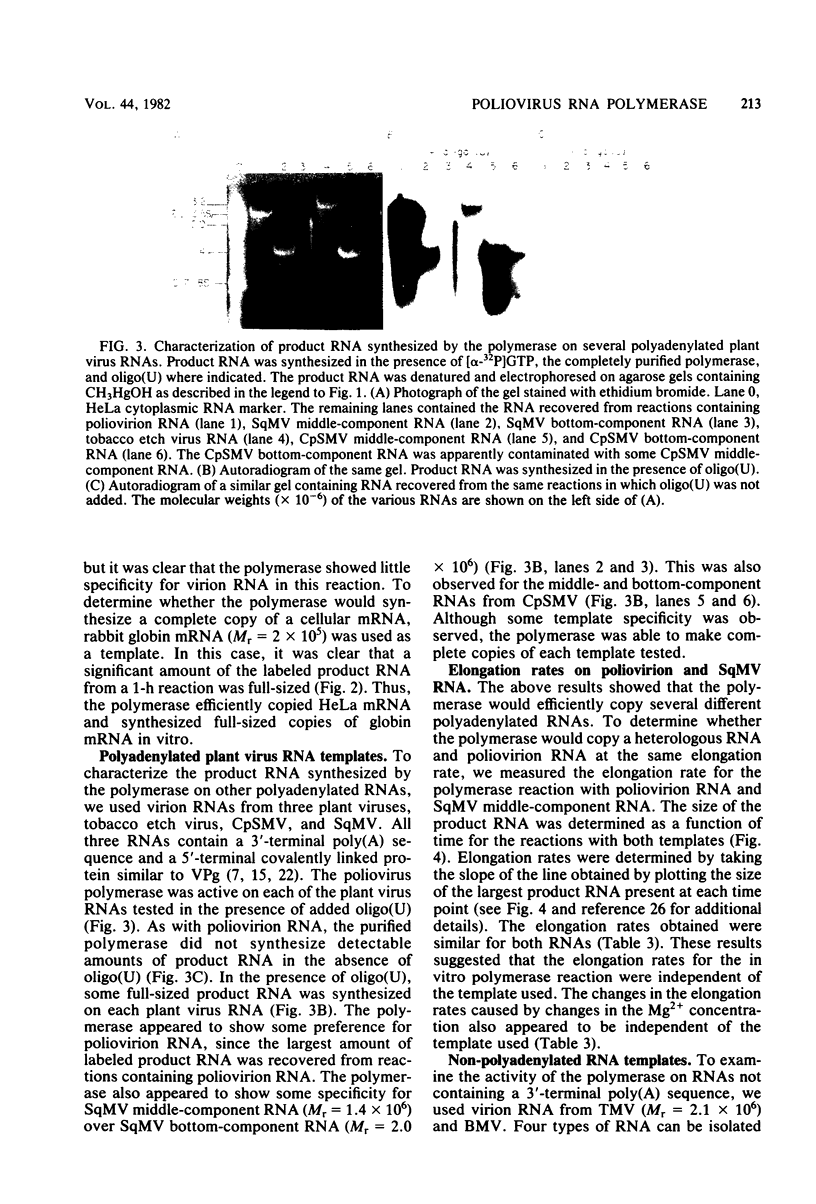
The poliovirus RNA-dependent RNA polymerase was active on synthetic homopolymeric RNA templates as well as on every natural RNA tested. The polymerase copied polyadenylate· oligouridylate [oligo(U)], polycytidylate · oligoinosinate, and polyinosinate· oligocytidylate templates to about the same extent. The observed activity on polyuridylate· oligoadenylate was about fourfold less. Full-length copies of both poliovirion RNA and a wide variety of other polyadenylated RNAs were synthesized by the polymerase in the presence of oligo(U). Polymerase elongation rates on poliovirion RNA and a heterologous RNA (squash mosaic virus RNA) were about the same. Changes in the Mg2+ concentration affected the elongation rates on both RNAs to the same extent. With two non-polyadenylated RNAs (tobacco mosaic virus RNA and brome mosaic virus RNA3), the results were different. The purified polymerase synthesized a subgenomic-sized product RNA on brome mosaic virus RNA3 in the presence of oligo(U). This product RNA appeared to initiate on oligo(U) hybridized to an internal oligoadenylate sequence in brome mosaic virus RNA3. No oligo(U)-primed product was synthesized on tobacco mosaic virus RNA. When partially purified polymerase was used in place of the completely purified enzyme, some oligo(U)-independent activity was observed on the brome mosaic virus and tobacco mosaic virus RNAs. The size of the product RNA from these reactions suggested that at least some of the product RNA was full-sized and covalently linked to the template RNA. Thus, the polymerase was found to copy many different types of RNA and to make full-length copies of the RNAs tested.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahlquist P., Dasgupta R., Kaesberg P. Near identity of 3- RNA secondary structure in bromoviruses and cucumber mosaic virus. Cell. 1981 Jan;23(1):183–189. doi: 10.1016/0092-8674(81)90283-x. [DOI] [PubMed] [Google Scholar]
- Bailey J. M., Davidson N. Methylmercury as a reversible denaturing agent for agarose gel electrophoresis. Anal Biochem. 1976 Jan;70(1):75–85. doi: 10.1016/s0003-2697(76)80049-8. [DOI] [PubMed] [Google Scholar]
- Brakke M. K., Van Pelt N. Linear-log sucrose gradients for estimating sedimentation coefficients of plant viruses and nucleic acids. Anal Biochem. 1970 Nov;38(1):56–64. doi: 10.1016/0003-2697(70)90155-7. [DOI] [PubMed] [Google Scholar]
- Cole C. N., Smoler D., Wimmer E., Baltimore D. Defective interfering particles of poliovirus. I. Isolation and physical properties. J Virol. 1971 Apr;7(4):478–485. doi: 10.1128/jvi.7.4.478-485.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta A., Baron M. H., Baltimore D. Poliovirus replicase: a soluble enzyme able to initiate copying of poliovirus RNA. Proc Natl Acad Sci U S A. 1979 Jun;76(6):2679–2683. doi: 10.1073/pnas.76.6.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta A., Zabel P., Baltimore D. Dependence of the activity of the poliovirus replicase on the host cell protein. Cell. 1980 Feb;19(2):423–429. doi: 10.1016/0092-8674(80)90516-4. [DOI] [PubMed] [Google Scholar]
- Flanegan J. B., Baltimore D. Poliovirus polyuridylic acid polymerase and RNA replicase have the same viral polypeptide. J Virol. 1979 Jan;29(1):352–360. doi: 10.1128/jvi.29.1.352-360.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanegan J. B., Baltimore D. Poliovirus-specific primer-dependent RNA polymerase able to copy poly(A). Proc Natl Acad Sci U S A. 1977 Sep;74(9):3677–3680. doi: 10.1073/pnas.74.9.3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanegan J. B., Petterson R. F., Ambros V., Hewlett N. J., Baltimore D. Covalent linkage of a protein to a defined nucleotide sequence at the 5'-terminus of virion and replicative intermediate RNAs of poliovirus. Proc Natl Acad Sci U S A. 1977 Mar;74(3):961–965. doi: 10.1073/pnas.74.3.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanegan J. B., Van Dyke T. A. Isolation of a soluble and template-dependent poliovirus RNA polymerase that copies virion RNA in vitro. J Virol. 1979 Oct;32(1):155–161. doi: 10.1128/jvi.32.1.155-161.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooding G. V., Jr, Hebert T. T. A simple technique for purification of tobacco mosaic virus in large quantities. Phytopathology. 1967 Nov;57(11):1285–1285. [PubMed] [Google Scholar]
- Hall T. C. Transfer RNA-like structures in viral genomes. Int Rev Cytol. 1979;60:1–26. doi: 10.1016/s0074-7696(08)61257-7. [DOI] [PubMed] [Google Scholar]
- Hewlett M. J., Rose J. K., Baltimore D. 5'-terminal structure of poliovirus polyribosomal RNA is pUp. Proc Natl Acad Sci U S A. 1976 Feb;73(2):327–330. doi: 10.1073/pnas.73.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lowe P. A., Brown F. Isolation of a soluble and template-dependent foot-and-mouth disease virus RNA polymerase. Virology. 1981 May;111(1):23–32. doi: 10.1016/0042-6822(81)90650-4. [DOI] [PubMed] [Google Scholar]
- Lowe P. A., King A. M., McCahon D., Brown F., Newman J. W. Temperature-sensitive RNA polymerase mutants of a picornavirus. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4448–4452. doi: 10.1073/pnas.78.7.4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundquist R. E., Ehrenfeld E., Maizel J. V., Jr Isolation of a viral polypeptide associated with poliovirus RNA polymerase. Proc Natl Acad Sci U S A. 1974 Dec;71(12):4773–4777. doi: 10.1073/pnas.71.12.4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merril C. R., Goldman D., Sedman S. A., Ebert M. H. Ultrasensitive stain for proteins in polyacrylamide gels shows regional variation in cerebrospinal fluid proteins. Science. 1981 Mar 27;211(4489):1437–1438. doi: 10.1126/science.6162199. [DOI] [PubMed] [Google Scholar]
- Semler B. L., Anderson C. W., Kitamura N., Rothberg P. G., Wishart W. L., Wimmer E. Poliovirus replication proteins: RNA sequence encoding P3-1b and the sites of proteolytic processing. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3464–3468. doi: 10.1073/pnas.78.6.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector D. H., Baltimore D. Polyadenylic acid on poliovirus RNA. II. poly(A) on intracellular RNAs. J Virol. 1975 Jun;15(6):1418–1431. doi: 10.1128/jvi.15.6.1418-1431.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyke T. A., Flanegan J. B. Identification of poliovirus polypeptide P63 as a soluble RNA-dependent RNA polymerase. J Virol. 1980 Sep;35(3):732–740. doi: 10.1128/jvi.35.3.732-740.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyke T. A., Rickles R. J., Flanegan J. B. Genome-length copies of poliovirion RNA are synthesized in vitro by the poliovirus RNA-dependent RNA polymerase. J Biol Chem. 1982 Apr 25;257(8):4610–4617. [PubMed] [Google Scholar]
- Villa-Komaroff L., McDowell M., Baltimore D., Lodish H. F. Translation of reovirus mRNA, poliovirus RNA and bacteriophage Qbeta RNA in cell-free extracts of mammalian cells. Methods Enzymol. 1974;30:709–723. doi: 10.1016/0076-6879(74)30068-7. [DOI] [PubMed] [Google Scholar]