Abstract
Elastin-like polypeptides (ELPs) are recombinant peptide-based biopolymers that contain repetitive sequences enriched in glycine, valine, proline, and alanine. Because of the unusually large fraction of these amino acids in ELPs as compared to other cellular proteins, we hypothesized that intracellular pools of these amino acids can be selectively depleted and limit protein yields during expression. In this study, we examined how culture conditions and individual medium components affect protein yields by monitoring cell growth and protein expression kinetics of E. coli expressing an ELP tagged with a green fluorescent protein (GFP). By determining the underlying principles of superior fusion protein yields generated by the hyperexpression protocol, we further improved protein yields through the addition of glycerol and certain amino acids such as proline and alanine, and found that amino acid concentrations and the type of basal medium used strongly influenced this beneficial effect. Surprisingly, amino acids other than those that are abundant in ELPs, for example, asparagine, aspartic acid, glutamine, and glutamic acid, also enhanced protein yields even in a nutrient-rich medium. Compared to commonly-used Luria-Bertani medium, the protein yield was improved by 36-fold to the remarkable level of 1.6 g/L in shaker flask cultures with a modified medium and optimized culture conditions, which also led to a 8-fold reduction in the cost of the fusion protein. To our knowledge, this is the highest yield of an ELP-fusion protein purified from E. coli cultured in shaker flasks. This study also suggests a useful strategy to improve the yields of other ELP fusion proteins and repetitive polypeptides.
Keywords: Elastin-like polypeptides, non-chromatographic purification, amino acids, supplements, condition optimization
Introduction
Recombinant protein-based biopolymers have recently attracted much attention as a new class of materials, which provide precise control of chain length, stereochemistry, and monodispersity. Their biological, chemical, and mechanical properties are controlled by their amino acid (natural or unnatural) sequence and the number of repeat units. It has been previously shown that novel properties such as temperature- or pH-sensitivity (1, 2) and electric conductivity (3) can be introduced into biopolymers. Furthermore, biological and chemical functionalities can also be site-specifically incorporated into peptide-based biopolymers (1, 4).
Elastin-like polypeptides (ELPs) are biopolymers with a repeating pentapeptide sequence Val-Pro-Gly-Xaa-Gly (where the guest residue Xaa can be any amino acid, except Pro). ELPs are thermally responsive polypeptides that undergo an inverse temperature phase transition. ELPs are highly soluble in aqueous solution below their transition temperature (Tt) but aggregate rapidly at temperatures above their Tt. This process is completely reversible. This unique property of ELPs make them attractive for a wide range of applications in biotechnology and biomedicine, for example, non-chromatographic protein purification by a method we have termed inverse transition cycling (ITC) (1), hyperthermia-mediated drug carriers (5), injectable tissue engineering scaffolds (6, 7), and the design of regenerable surfaces for biosensors (8).
For most applications of recombinant ELPs and their fusion proteins, it is important to produce them in large quantities and high purity in a cost-efficient manner. This requirement is essential for the use of recombinant polypeptides such as ELPs as bulk, structural materials, most notably as biomaterials and as tissue engineering scaffolds, for which high yield and low cost are the two greatest barriers to their adoption. In an orthogonal set of applications for production of ELP fusion proteins, where the ELP is appended to a target protein to facilitate purification (1, 9, 10), the high level expression of the fusion protein is also imperative for economical production of peptide and protein drugs.
Motivated by the need for high-yield, low-cost production of ELPs for different applications, we investigated the impact of various culture parameters such as induction protocol and medium formulation on recombinant ELP-fusion protein expression and purification. Since ELPs and ELP-fusion proteins have disproportional amounts of glycine, valine, proline, and alanine compared to E. coli cellular proteins (Table 1), we also examined how individual medium components such as carbon source, nitrogen source, and amino acids affect cell growth and protein expression. As a test-case that could be easily followed through the different steps of expression and purification, an ELP was fused with green fluorescent protein (GFP) and was expressed in E. coli to monitor the expression kinetics of the fluorescent fusion protein at a single cell level using flow cytometry (11-13). This allowed us to readily examine protein expression and its dependence on culture conditions. An understanding of how these components affect cell and protein production enabled us to develop a systematic strategy that can be applied to improve yields of other ELPs and ELP fusion proteins. In addition, this study also suggests a rational approach for optimizing culture conditions of other repetitive polypeptides or proteins that preferentially consume intracellular amino acid pools.
Table 1.
Amino acid compositions of ELP[V5A2G3]-90, GFP-ELP, and E. coli cellular proteins
| % weight | |||
|---|---|---|---|
| Amino acid | ELP[V5A2G3]-90 | GFP-ELP | E. coli protein (17) |
| Glycine | 32.10 | 20.39 | 7.81 |
| Valine | 35.74 | 23.29 | 5.99 |
| Proline | 23.95 | 15.26 | 4.56 |
| Alanine | 3.39 | 3.25 | 13.02 |
| Other | 4.82 | 37.81 | 68.62 |
Methods and Materials
Plasmid construction
An ELP encoding ninety Val-Pro-Gly-Xaa-Gly repeats where Xaa is Val, Ala, and Gly in a 5:2:3 ratio, respectively, ELP[V5A2G3]-90 was used in this study. An ELP protein fused with GFP (GFP-ELP) was synthesized by inserting the GFP gene (Qbiogene, Carlsbad, CA) 5′ to the ELP[V5A2G3]-90 gene in a modified pET25b vector with a T-7 promoter (Novagen, Madison, WI) (Figure 1, see Supporting Information for the detailed schematic of vector construction). A synthetic gene with SfiI-generated, compatible sticky ends encoding for ELP[V5A2G3]-90 was synthesized by recursive directional ligation in pUC-19 (14). Details of monomer gene synthesis and gene oligomerization of ELP[V5A2G3]-90 have been described elsewhere (1). The modified pET25b vector was produced by replacement of the NotI to AvaI segment of pET25b with an oligonucleotide cassette encoding for an oligohistidine tag, thrombin cleavage site, and a SfII restriction site into which the ELP[V5A2G3]-90 gene was subsequently inserted (1). This vector was then digested with XbaI and SalI, followed by enzymatic dephosphorylation with calf intestinal phosphatase and purification from a low melting point agarose gel (National Diagnostics, Atlanta, Georgia). For the GFP gene, its first 723 bp was excised from the pQBIT7-GFP plasmid (Qbiogene) by digestion with XbaI and AlwnI, and the fragment was purified from a low melting point agarose gel. A linker cassette with AlwnI and SalI compatible sticky ends encoding for the remaining 35 bp of the GFP gene was created by oligonucleotide synthesis (Integrated DNA technologies, Coralville, IA). Assembly of the GFP-ELP fusion protein was accomplished by a ternary ligation of the 723 bp fragment of the GFP gene, the linker cassette, and the digested and gel-purified modified pET25b vector. Proper fusion protein assembly (GFP-ELP) was confirmed by screening with AlwnI digestion and DNA sequencing. The expression vector was then transformed into E. coli stain BLR(DE3) (Novagen), which is commonly used for expressing recombinant proteins with tandem repeats due to its deficiency in homologous recombination.
Figure 1.
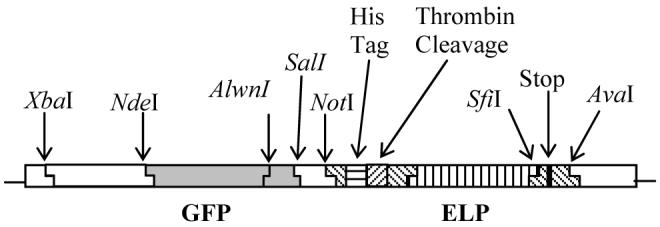
Schematic of the GFP-ELP fusion vector showing flanking restriction endonuclease sites used in construction of the fusion gene.
Protein expression
Three different media were used: Luria-Bertani (LB) medium (for 1 liter, 10 g tryptone, 5 g yeast extract, and 10 g sodium chloride, pH 7.0), Circlegrow (CG; Qbiogene), and Terrific Broth (TB) (for 1 liter, 12 g tryptone and 24 g yeast extract (TB basal, TBB). Phosphate buffer (2.31 g potassium phosphate monobasic and 12.54 g potassium phosphate dibasic) and glycerol (4 ml) (PBG) were added separately as supplements, where noted.). Stock solutions of the twenty amino acid supplements (Sigma, St. Louis, MO) were prepared in deionized water at 200 mM and were sterilized separately using 0.2 μm filters before being added to the medium. The initial pH values of cultures supplemented with various amino acids ranged from 7 to 7.2, except those with aspartic acid, glutamic acid, and histidine, which were adjusted to this range by adding an appropriate amount of 1M sodium hydroxide. A 2 ml culture of E. coli BLR(DE3) harboring a plasmid for the GFP-ELP protein was inoculated with a single colony from a freshly streaked agar plate supplemented with 100 μg/ml ampicillin. After overnight incubation at 37 °C with orbital agitation at 300 rpm, the optical density (OD600) of the culture was determined on a spectrophotometer (Shimadzu, Columbia, MD) or a microwell plate reader (Molecular Devices, Sunnyvale, CA). E. coli cells were pelleted by centrifugation (2000×g, 4 °C, 15 min), re-suspended in fresh medium, and used to inoculate 50 ml of medium in a 250 ml Erlenmeyer flask, unless otherwise stated. The inoculum volume was adjusted to obtain an initial OD600 of 0.1. The culture was incubated at 37 °C with orbital agitation at 300 rpm. For the IPTG induction protocol, isopropyl β-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM to induce protein expression when OD600 = 1. Cultures were then continued for an additional 4 h post induction, which is the typical incubation duration for induced cultures. For the hyperexpression protocol, no IPTG was added and they were allowed to grow for 24 h after inoculation in the 50 ml cultures (15). Cells were harvested from the cultures by centrifugation (2000×g, 4 °C, 15 min), re-suspended in 5 ml PBS, and stored frozen at -20 °C until further use. Cell cultures were performed in two to four separate experiments with at least two replicates total, unless otherwise stated.
Fluorescence measurements
E. coli cultures were collected during fermentation and stored at 4 °C. The E. coli cells (1 ml, OD600 of 0.1) were washed with PBS before analysis by flow cytometry (FACSVantage DiVa, Becton Dickonson, San Jose, CA). The flow cytometer was calibrated with a LinearFlow green flow cytometry intensity calibration kit (Molecular Probes, Eugene, OR). All samples were analyzed within 30 h after isolation and no significant loss in fluorescence intensities was observed during the storage procedure. E. coli expressing ELP[V5A2G3]-90 cultured in the basal medium without any supplements was used as a negative control for fluorescent intensity measurements. Cell densities were determined using a bacteria counting kit from Molecular Probes, following the manufacturer’s instructions. One OD600 unit was determined to be 8 million cells/ml. Ten thousand events were collected to ensure statistical significance and the collected data were analyzed using FlowJo software (Treestar, San Carlos, CA). Additionally, the expression of GFP-ELP protein was monitored using a microplate fluorescence reader (Cytofluor 4000, Applied Biosystems, Foster City, CA). Cells were diluted to an OD600 of less than 1 for all measurements to avoid signal saturation and non-linearity. The fluorescence intensities measured by the flow cytometer (mean fluorescence intensity per cell) and those taken by the microplate fluorometer were well correlated ((Mean FI/cell)flow cytometer = 0.106 × (Mean FI/cell)microplate fluorometer, R2 =0.97).
Protein purification and quantification
The GFP-ELP fusion protein was purified from E. coli lysate using the inverse transition cycling (ITC) protocol described elsewhere (9, 10). Briefly, one ml of cells was diluted with an equal volume of PBS and lysed by ultrasonic disruption (Fisher Scientific 550 Sonic Dismembrator using a microtip). The lysate was centrifuged (16,000×g, 4 °C, 15 min) to remove insoluble cellular debris. Next, nucleic acids were removed from the supernatant by adding polyethylenimine (0.5% w/v) and centrifugation (16,000×g, 4 °C, 15 min). The ITC protocol was performed as follows. GFP-ELP in the soluble cell lysate was precipitated by adding NaCl (0.8 - 1.3 M depending on expression level) and centrifugation (16,000×g, 45 °C, 15 min). The supernatant was discarded, and the pellet was re-suspended in PBS at 4 °C. The remaining insoluble matter was removed by centrifugation (16,000×g, 4 °C, 15 min) and the supernatant containing GFP-ELP was retained. This ITC procedure was repeated once more for a total of two cycles. SDS-PAGE with Coomassie blue (Sigma) staining was used to visualize the overexpression of recombinant ELP-fusion proteins (9). The concentrations of purified GFP-ELP was determined by its absorbance at 280 nm using a spectrophotometer (Shimadzu), with an ε280 of 25820 M-1cm-1 calculated from the primary amino acid sequence with the software program Protean (DNASTAR, Madison, WI).
Results
Effects of induction protocol on kinetics of cell growth and protein expression
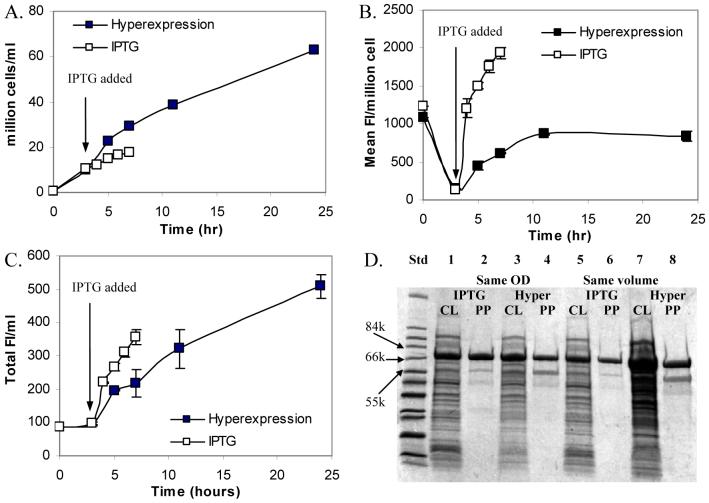
To determine the impact of culture conditions such as induction protocol on the expression of recombinant ELPs and their fusion proteins, we constructed a GFP-tagged ELP[V5A2G3]-90 to monitor protein expression kinetics by measuring intracellular fluorescence. The optical densities and fluorescence intensities were measured using a spectrophotometer and a flow cytometer to determine cell growth kinetics and protein expression kinetics, respectively. We found that the cell density of uninduced cultures using the hyperexpression protocol was 4-fold greater than induced cultures following the IPTG induction protocol at the end of the cultures (Figure 2A). Assuming that GFP fluorescence intensity reflects the amount of expressed GFP-ELP, there was a ∼10-fold decrease in the amount of protein expressed per cell in the first 3 h of both uninduced and induced cultures (Figure 2B). After induction with IPTG, the amount of protein expressed per cell dramatically increased by 20-fold until the end of the induced cultures. In contrast, the amount of protein expressed per cell in uninduced cultures slowly increased by up to 9-fold at 10 h and remained at this level until 24 h (Figure 2B). The yield of GFP-ELP per milliliter of culture (termed “protein yield” herein after) was 40% higher in uninduced cultures than in induced cultures (Figure 2C). The protein yield was higher for uninduced cultures at 24 h because the duration for protein production was longer leading to a higher cell density (Figure 2D), even though the amount of protein expressed per cell was in general lower in uninduced cultures (Figures 2B and 2D, same OD vs. same volume). This explains why the hyperexpression protocol usually results in higher yields of ELP-fusion proteins than the IPTG induction protocol (15).
Figure 2.
Effects of different induction protocols on cell density (A), the amount of protein expressed per cell as mean fluorescence intensity (mean FI) per cell determined by flow cytometry (B), and protein yield as total fluorescence intensity (total FI) per ml of culture (C) of E. coli cultures expressing GFP-ELP (two replicates total). Cells were collected from 1-liter cultures in CG medium following the hyperexpression protocol (hyperexpression) or the IPTG induction protocol (IPTG). SDS-PAGE analyses for cell lysate (lanes 1, 3, 5, 7) and purified proteins (lanes 2, 4, 6, 8) for cultured with (lanes 1, 2, 5, and 6) and without (lanes 3, 4, 7, and 8) IPTG induction (D). Samples for lanes 1 - 4 were loaded with volumes adjusted to the same OD600 and samples for lanes 5 - 8 were loaded using same volumes. The bands of 65.5 kDa on the gel represent GFP-ELP expressed by E. coli and the bands of 55 kDa are probably small amounts of co-contaminants that non-specifically bind to the GFP-ELP.
Effects of carbon/nitrogen source and amino acids on cell density and protein expression
Past work has shown that the protein yield of ELPs was higher in TB medium than in LB medium (15). To investigate whether there are limiting nutrients such as the carbon or nitrogen source that restrict cell density and protein production, protein expression was carried out in LB medium supplemented with an extra carbon source such as glucose or glycerol using the hyperexpression protocol. The concentrations of glucose (20 g/L) (16) and glycerol (4 ml/L, an identical amount as in TB medium) (15) were chosen based on reported medium compositions that led to high protein expression. Glycerol was observed to be better than glucose in increasing cell density (2.9-fold vs. 2-fold; Figure 3A) and the amount of protein expressed per cell (5.5-fold vs. 0.2-fold; Figure 3B) for E. coli expressing GFP-ELP, compared to the control (LB medium with phosphate buffer). This indicated that the enhanced protein yield using TB medium was mainly due to the presence of glycerol but not due to the higher amount of glucose in TB medium as compared to LB medium. Notably, the amount of GFP-ELP expressed per cell and protein yield decreased substantially with the addition of glucose as compared to the control medium (Figures 3B and 3C). An increase in glycerol concentration to 8 ml/L did not further increase cell density, the amount of protein expressed per cell, or protein yield (Figure 3). Similarly, the addition of both glycerol and glucose did not enhance any of these parameters to a greater extent than glycerol alone (Figure 3).
Figure 3.
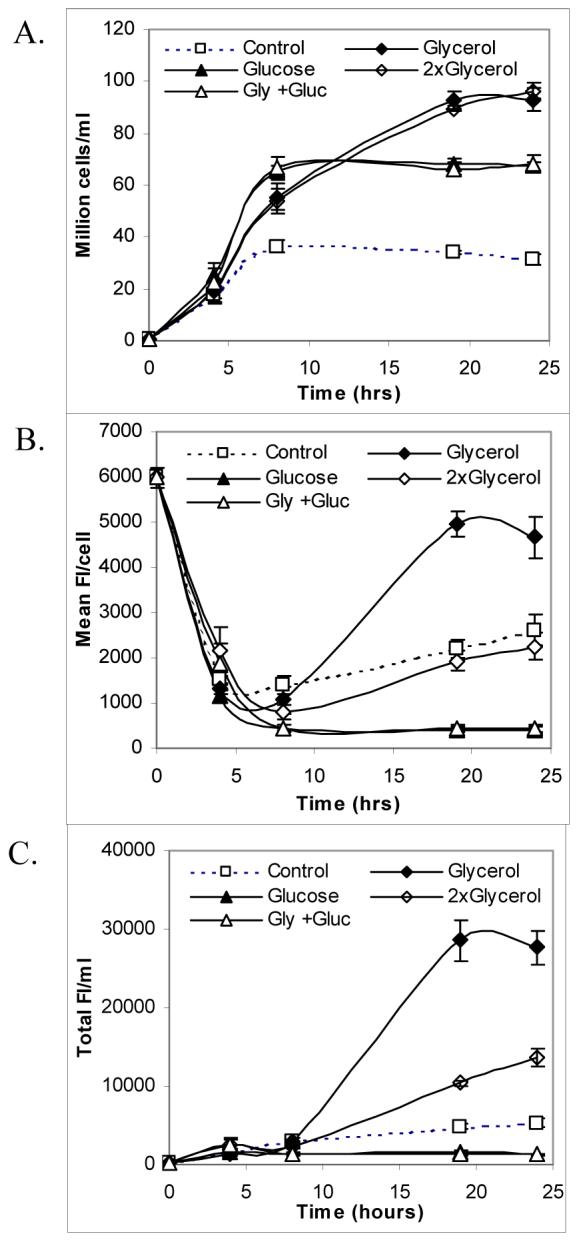
Effects of additional glucose and glycerol on cell density (A), the amount of protein expressed per cell (B), and protein yield per ml of culture (C) in 50 ml uninduced cultures of E. coli expressing GFP-ELP using LB medium with phosphate buffer (control) as a function of time (four replicates total). Supplements used are 20 g glucose/L (glucose), 4 ml glycerol/L (glyercol), 20 g glucose and 4 ml glycerol/L (Gly + Gluc), and 8 ml glycerol/L (2× glycerol).
We next examined the effect of an additional nitrogen source and amino acids that are highly abundant in ELPs on protein production. A range of concentrations was tested to determine the dose-response of each amino acid on cell density and protein production. The inclusion of ammonia as an extra nitrogen source slightly improved cell density, the amount of protein expressed per cell, and protein yield in LB medium with phosphate buffer and glycerol (PBG) (Figure 4). The addition of glycine and valine did not increase cell density, the amount of protein expressed per cell, and protein yield, regardless of their concentrations. The addition of proline and alanine at 100 mM resulted in a 3-fold increase in the amount of protein expressed per cell and protein yield using LB + PBG as a basal medium. The beneficial effects of proline and alanine were only observed at concentrations higher than 10 mM, which is higher than expected for most medium supplements (Figure 4) (17).
Figure 4.
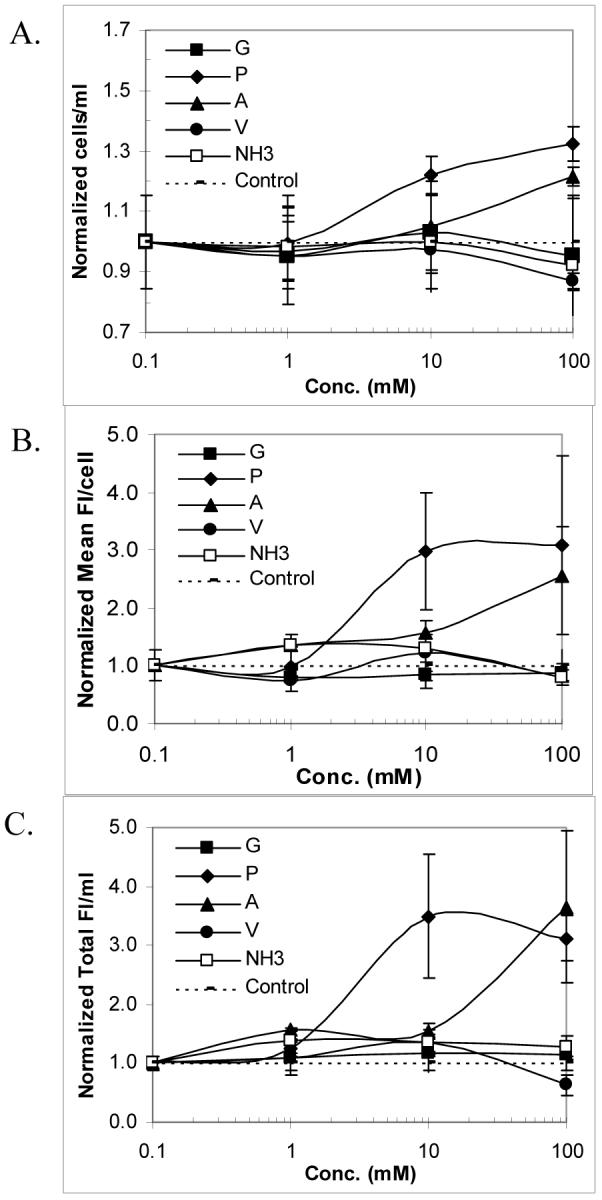
Effects of different concentrations of amino acids that are abundant in ELPs such as glycine (G), proline (P), alanine (A), and valine (V), as well as ammonia (NH3), on cell density (A), the amount of protein expressed per cell (B), and protein yield per ml of culture (C) in 50 ml uninduced cultures of E. coli expressing GFP-ELP after 24 hours (two replicates total). Data for various conditions were normalized to the control (LB + PBG). For the control, OD600, mean FI/cell, and total FI/ml are 10.66, 185.8, and 32049, respectively.
Effects of other amino acids on cell density and protein expression
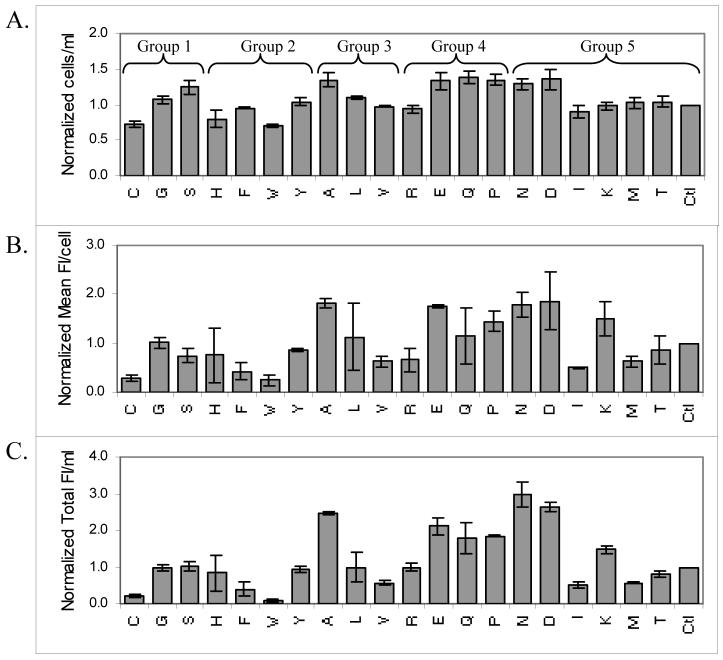
We also examined the effects of amino acids that are less abundant in the ELP sequence on the production of GFP-ELP. Amino acids are grouped into 5 families according to their initial precursors (Table 2) (18). The addition of several amino acids in groups 1, 3, 4, and 5 improved cell density to different extents, compared to LB + PBG as the basal medium (Figure 5A). The addition of serine, alanine, glutamic acid, glutamine, proline, asparagine, and aspartic acid resulted in the largest increase in cell density (Figure 5A). Asparagine, aspartic acid, alanine, and glutamic acid were better than proline, lysine, and glutamine in improving the amounts of protein expressed per cell (Figure 5B). A similar trend was observed for these amino acids in their ability to improve protein yields on a volumetric basis (Figure 5C).
Table 2.
Distributions of amino acids in different groups based on initial metabolic precursors in ELP[V5A2G3]-90, GFP-ELP, and E. coli cellular proteins
| % weight | |||
|---|---|---|---|
| Amino acid group | ELP[V5A2G3]-90 | GFP-ELP | E. coli protein (17) |
| Serine (C, G, S) (A1) | 32.79 | 22.59 | 10.7 |
| Aromatic (H, F, W, Y) (A2) | 2.69 | 9.36 | 10.3 |
| Alanine (A, L, V) (A3) | 39.44 | 30.53 | 22.3 |
| Glutamate (R, E, Q, P) (A4) | 24.39 | 21.69 | 24.7 |
| Aspartate (N, D, I, K, M, T) (A5) | 0.69 | 15.83 | 32.0 |
Figure 5.
Effect of 20 natural amino acids (denoted by single letter codes) on cell density (A), the amount of protein expressed per cell (B), and protein yield per ml of culture (C) in 50 ml uninduced cultures of E. coli expressing GFP-ELP after 24 hours (two replicates total). Various amino acids (100 mM) are divided into 5 groups according to their initial precursors. Data for various conditions were normalized to the control (Ctl, LB + PBG). For the control, OD600, mean FI/cell and total FI/ml are 11.1, 287.7, and 20932, respectively.
Additional amino acids also enhance ELP production in different media
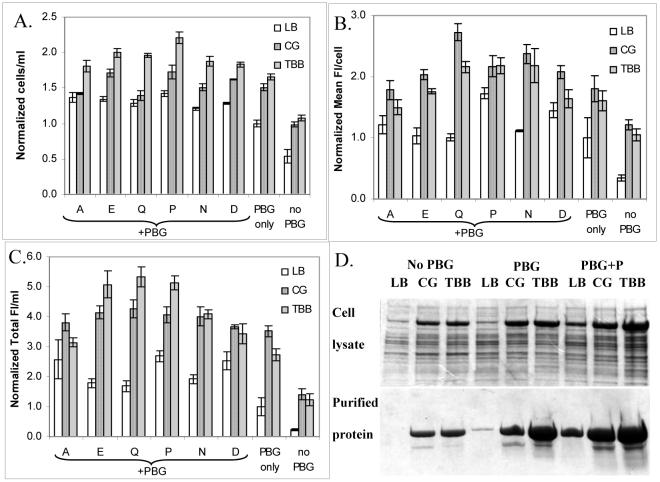
Since amino acids and glycerol enhanced cell and protein production in cultures using LB as a basal medium, we also examined how these supplements would affect protein production in other media to determine the generality of these effects. Cell growth was enhanced by at least 75% with PBG and various amino acids present in TB basal (TBB) medium, compared to the one without PBG (Figure 6A). A similar trend was also observed for CG medium, but to a lesser extent (at least 40%) (Figure 6A). Since excessive glycerol can adversely affect cell and protein production (Figure 3), TBB medium containing only tryptone and yeast extract was used for comparison. The addition of PBG to TB medium reduced cell density and protein yield (data not shown). Compared to other amino acids, the increase in cell density was the greatest (2.3-fold) when proline together with PBG was added to TBB medium (Figure 6A). The addition of PBG alone to CG or TBB medium improved the amount of protein expressed per cell (1.5-fold) (Figure 6B) and the addition of PBG together with various amino acids to these media led to a further improvement in the amount of protein expressed per cell (up to 2-fold) (Figure 6B). Protein yields on a volumetric basis in cultures using CG or TBB medium were improved with PBG alone (2- to 2.5-fold), and even more when further supplemented with glutamic acid, proline, or glutamine (3- to 4-fold) (Figure 6C). These results indicated that the extra carbon source and amino acid are important in improving cell density, the amount of protein expressed per cell, and protein yield for all media.
Figure 6.
Effect of amino acids (denoted by single letter codes) on cell density (A), the amount of protein expressed per cell (B), and protein yield per ml of culture (C) in 50 ml uninduced cultures of E. coli expressing GFP-ELP after 24 hours (four replicates total). The concentrations of amino acids used were 100 mM. Data for various conditions were normalized to the control (LB + PBG). Media without phosphate buffer and glycerol were also tested (No PBG). For the control, OD600, mean FI/cell and total FI/ml are 9.6, 7664, and 10280, respectively. SDS-PAGE analyses for cell lysate (upper panel) and purified proteins (lower panel) for various conditions in different media (LB, CG, and TBB) (D). Sample loading volumes were adjusted and normalized to the volume of cultures. Note that the sample loading volumes were different from those used in Figure 2D.
Among all conditions, the addition of proline together with PBG was the most beneficial in terms of cell density, the amount of protein expressed per cell, and protein yield, regardless of the type of medium used. SDS-PAGE visually demonstrates the remarkable enhancement in the amount of isolated, purified GFP-ELP that can be attained by this relatively simple optimization procedure. As shown in Figure 6D, the addition of PBG improved the amount of protein expressed in E. coli cells (upper panel) and the amount of purified protein isolated from 50 ml cultures (lower panel). The enhancement in GFP-ELP expression was even greater when extra proline was added, regardless of the type of medium used. Higher protein expression also facilitated the recovery of proteins through the inverse temperature cycling process because high protein concentration enhances the formation of ELP aggregates. The highest level of expression was observed upon supplementation of TBB medium with PBG and proline leading to a yield of purified GFP-ELP of 1.6 g/L.
These results are notable for two reasons: first, this simple supplementation procedure enhanced the yield by >6-fold compared to TBB medium and by an extraordinary 36-fold compared to LB medium (Table 3). Second, the maximum yield of 1.6 g/L of purified GFP-ELP fusion protein is more than an order of magnitude greater than reported for other GFP constructs. The best yields previously reported for GFP expressed in E. coli was 18 mg/L that was purified with hydrophobic interaction chromatography (19) and 26.8 mg/L with organic extraction (20), respectively. Even after accounting for the ELP tag (35.0 kDa out of 65.5 kDa total) and the efficiency of thrombin cleavage (conservatively 50%), the expression of GFP with an ELP tag and its purification by exploiting the phase transition of the fusion protein provide a huge improvement in the yield of the target protein (∼400 mg/L) over other GFP constructs expressed in E. coli and purified by chromatography.
Table 3.
Cost analysis of supplements in various media
| Yield (mg/L) | Medium cost ($/L) | Unit Price (¢/mg) | ||||
|---|---|---|---|---|---|---|
| Without | With* | Without | With* | Without | With* | |
| Luria-Bertani (LB) | 44.9 | 456.0 | 1.8 | 6.4 | 4.1 | 1.4 |
| Circlegrow (CG) | 267.7 | 1037.9 | 4.0 | 8.6 | 1.5 | 0.8 |
| Terrific Broth basal (TBB) | 263.2 | 1620.8 | 3.6 | 8.2 | 1.4 | 0.5 |
Supplements used are glycerol, phosphate buffer, and proline ($4.59/L total).
Discussion
Novel properties of recombinant protein-based biomaterials have numerous applications in biotechnology and biomedicine. In order to realize their potential, it is important to produce large quantities of these materials, which are usually limited by cell density, the amount of protein expressed per cell, and culture volume. As shown in this study, cell and protein production can be greatly enhanced through the optimization of culture conditions (IPTG induction and culture duration) and the rational design of culture medium (carbon source, nitrogen source, medium type, and amino acids).
Previously, Guda et al. have shown that a substantial increase in the expression of a synthetic gene G-(VPGVG)119-VPGV in E. coli was observed in uninduced cultures over induced cultures after 24 h, probably because of the decrease in repressor protein concentration (hyperexpression protocol) (15). A comprehensive investigation of this phenomenon provides additional insights into how the yield of recombinant protein-based biopolymers can be further improved. We found that cell growth was slower after IPTG induction, indicating that higher protein expression induces significant metabolic stress on E. coli (17). Therefore, cell density and protein yield are in general lower in induced cultures. In contrast, the amount of protein expressed per cell increased more gradually in uninduced cultures presumably because the reduced metabolic stress on cells allows the culture to reach a higher cell density. This result also suggests that the control of the promoter became less stringent at the later stage of the culture (21). It is also possible that IPTG induction altered the localization or expression pattern (for example, inclusion bodies) of recombinant proteins, which affects protein expression (recombinant protein to total protein ratio), protein recovery (protein solubility), or protein stability (protection from protease) (22). To further improve protein yields on a volumetric basis, these results indicated that one should allow a culture to reach a higher cell density by reducing metabolic stress and increasing cell viability, or increase protein expression per cell by releasing nutrient limitations and preventing product inhibition.
Higher expression of recombinant ELPs or ELP-fusion proteins obtained using the hyperexpression protocol may cause nutrient depletion, thus adversely affecting cell and protein production. Our results showed that the use of glycerol as a carbon source significantly enhanced cell and protein production because acetate, which inhibits cell growth at high concentrations, is not produced (23, 24). However, a higher concentration of glycerol (8 mg/L) did not further improve cell density and the increases in the amount of protein expressed per cell and protein yield were also more modest, probably due to a reduction in carbon flux through glycolysis (24) (Figure 3). The addition of glucose in LB medium improved cell density by 2 folds, but to a lesser extent than the 3-fold increase observed with glycerol. The presence of a high level of glucose shuts down protein expression in E. coli because catabolic products of glucose reduce the intracellular cyclic AMP level, which subsequently blocks protein expression (17).
Other than the carbon source, the nitrogen source and specific amino acids are also important in recombinant protein expression. The weight percents of glycine, valine, or proline in ELP and GFP-ELP are at least 3 times higher than E. coli cellular proteins, while the weight percent of alanine is significantly lower than E. coli cellular proteins (Table 1). We hypothesized that the disproportional amounts of these amino acids in ELPs or ELP-fusion proteins may pose a much higher demand on certain metabolic pathways than E. coli cellular proteins, and potentially lead to the depletion of intracellular amino acid pools. E. coli can synthesize all 20 amino acids required for protein synthesis from ammonia salts and the biosynthesis of amino acids is highly regulated thought end-product regulation and amino acid transporters (17). A possible method to replenish the intracellular amino acid pools is by supplementing various amino acids in the medium. It has been demonstrated that the coordinated addition of phenylalanine (a rate-limiting precursor) increases the yield of chloramphenicol-acetyl-transferase (CAT), enhances specific activity, and reduces protein degradation (18, 25). For GFP-ELP fusion protein, extra proline or alanine promoted cell and protein production, while extra glycine or valine inhibited cell and protein production. Both concentrations and types of amino acids added were found to be important. However, the addition of ammonia did not improve protein yields, suggesting that the cultures were not limited by the nitrogen source.
Our results also suggest that other regulatory mechanisms might play a role in maintaining intracellular amino acid availability. The interplay of various metabolism pathways, particularly those responsible for biosynthesis of 20 amino acids, and the availability of precursors and intermediates in such pathways, can also be an important part of this complex control mechanism. Since there are no simple principles governing how levels of intermediates affect protein synthesis, we took an empirical approach to determine whether supplementing the media with other amino acids would improve protein expression. All natural amino acids can be synthesized using five independent metabolic pathways with different initial precursors, so they can be categorized into 5 groups, namely, serine, aromatic, alanine, glutamate, and aspartate (18) (see Supporting Information for amino acid metabolic pathways of a recombinant E. coli cell).
For the ELP or GFP-ELP protein, the weight percents of amino acids in the serine and alanine groups (groups 1 and 3) are much higher than those in other groups (Table 2). However, only one amino acid (alanine) in these two groups led to an improvement in protein yields. None of the amino acids in the aromatic group (group 2) enhanced protein yields. In contrast, all amino acids in the glutamine group (group 4) except arginine improved protein yields. In the aspartate group (group 5), asparagine and aspartic acid substantially improved protein yields, while lysine also slightly improved protein yield. This pattern indicates that the consumption of certain amino acids in later steps in glycolytic pathway maybe much higher than those in earlier steps. Therefore, the supplementation of certain amino acids further downstream in the glycolytic pathway would be more beneficial in protein expression than those further upstream. The differential effects of amino acids in each group on protein expression highlight the complexity of the dynamics between amino acid availability in the medium and recombinant protein synthesis, as well as elaborate regulatory mechanisms of the intracellular amino acid pools.
The beneficial effects of these amino acids and glycerol on the production of ELP fusion proteins prompted us to examine whether the addition of these supplements can improve protein yields in other media such as CG and TBB medium. Interestingly, the addition of glycerol further enhanced protein yields, indicating that the depletion of carbon source occurs even in nutrient-rich media for the hyperexpression of ELPs. The addition of proline, glutamine, and glutamic acid was also beneficial in cell and protein production to different extents, depending on the type of medium used. However, the beneficial effects for protein yields induced by alanine, asparagine, and aspartic acid in CG and TBB medium were not as great as other amino acids. Although the amino acid composition of a recombinant protein can provide a guideline on the type of supplements that should be added, the beneficial effects on protein yields largely depend on the actual concentration of amino acids in the medium, the type of medium used, and other amino acid regulatory mechanisms (for example, amino acid transporters).
An important and practical outcome of this study is that it showed that the yield of GFP-ELP could be increased by a remarkable 36-fold, compared to a standard protocol that would typically involve the expression of a recombinant protein in LB medium without IPTG induction. When compared to a commercially available nutrient-rich medium, a 6-fold enhancement of protein yield was observed; this is notable as the initial protein yield of 270 mg/L is rather high for shaker flask E. coli cultures, so that the final 1.6 g/L protein yield for the optimized medium and culture conditions is extraordinary by any measure for protein expression in E. coli cultured in shaker flasks. In addition, our preliminary data showed that cell density and protein yield for cultures of other ELP-fusion proteins were double when TB medium with proline instead of CG medium was used, confirming that this culture condition optimization strategy is applicable to other proteins other than GFP-ELP (data not shown).
To put these expression levels in perspective, the yields of most recombinant proteins produced in shaker flask cultures and purified with chromatography columns range from 10 - 200 mg/L (10, 26-28), and the expression of highly repetitive recombinant proteins (e.g., G-protein-coupled receptors) usually suffers from even lower yields (29). Protein yields can reach 250 - 500 mg/L with specially formulated media (30), and can be further improved to above 1 g/L only by genetic engineering of metabolic pathways in E. coli cultivated in a well-controlled bench-top fermenter (31). To our knowledge, the 1.6 g/L of GFP-ELP reported here is the highest level of any ELP-fusion proteins purified from shaker flask E. coli cultures, without using laborious metabolic engineering and expensive purification techniques. These results are intriguing because they suggest that the ELP tag not only facilitates protein purification, but also appears to play a synergistic role in enhancing protein expression. Under our best culture condition, the OD600 of E. coli cultures could reach over 19, which corresponds to 4.56 g/L of total proteins (E. coli at an OD600 of 1 usually contains about 0.4 g/L dry cell weight and 0.24 g/L of total proteins (32)). Based on the facts that over 50% of the total proteins is GFP-ELP (Figure 6D) and the typical purification yield of the ITC method is about 75% (10), the estimated yield of purified GFP-ELP is about 1.7 g/L, which closely agrees with the actual protein yield.
The level of expression reported here, we believe, is hence remarkable and has practical implications at two levels. First, it enables gram-level quantities of the fusion protein be routinely produced in a laboratory setting without the necessity of large-scale fermenters, which will facilitate the use of these biopolymers for applications that require large quantities of biopolymers or fusion proteins. The second implication of these results is economical. The addition of an amino acid such as proline or alanine and an extra carbon source such as glycerol increases the cost of medium, so that a potential trade-off of the enhanced expression might be cost. We performed a cost analysis to determine whether the use of supplements is indeed cost-efficient to produce ELPs in large quantities. From our analysis, the addition of phosphate buffer, glycerol, and proline to LB medium would provide 70% cost reduction and the use of these supplements in CG and TBB medium would provide at least 50% cost reduction per mg protein (Table 3). Based on the actual amount of purified GFP-ELP and the cost of medium, the best medium design that optimizes both protein yield and production cost would be TB medium supplemented with 100 mM proline. Hence, the optimized medium that leads to high-level expression also led to at least 3-fold reduction in the cost of the fusion protein, so that high-level, gram-per-liter expression of GFP-ELP is not at the expense of greater cost of the product.
Conclusions
For polypeptides that contain highly repetitive sequences rich in specific amino acids, it is likely that the expression of such proteins in E. coli would lead to the depletion of intracellular amino acid pools and would cause tremendous metabolic stress on the expression host. In this study, we identified a rational strategy to overcome limitations on the expression of recombinant ELP-fusion proteins. By determining the underlying principles behind the superior protein yields caused by the hyperexpression protocol, we further improved protein yields through the addition of glycerol and certain amino acids such as proline and alanine. Surprisingly, amino acids other than those that are abundant in ELPs (asparagine, aspartic acid, glutamine, and glutamic acid) also enhanced protein yields, which indicated that more complicated regulatory mechanisms and the interplay between metabolite intermediates play a role in the control of intracellular amino acid pools. Further analyses revealed that such beneficial effects in protein yields induced by supplements were also observed in other nutrient-rich media, but to various extents. With the optimization of culture conditions, we showed that it is possible to improve the protein yield by 6 folds from an initial 0.27 g/L in a commercially available nutrient-rich medium to the remarkable level of 1.6 g/L using a modified medium in shaker flask cultures. To our knowledge, this is the highest yield of an ELP-fusion protein purified from E. coli in shaker flask cultures. This 6-fold increase in protein yield also led to at least 3-fold reduction in the cost of the fusion protein. Future studies on how various amino acid transporters control the intracellular amino acid pools and protein expression, as well as analyses on metabolic fluxes for biosynthesis pathways of amino acids, would be useful to develop a more robust cultivation strategy with superior protein yields for ELP-fusion proteins.
Supplementary Material
Acknowledgements
We thank Anand Patel, Bum-joon Kim, Dong Woo Lim, Li Liu, and Wenge Liu for their useful discussion and technical assistance. Technical support of the Duke University Medical Center Flow Cytometry Facility is kindly acknowledged. This research was financially supported by the National Institutes of Health (GM-061232).
References and Notes
- (1).Meyer DE, Chilkoti A. Purification of recombinant proteins by fusion with thermally-responsive polypeptides. Nat. Biotechnol. 1999;17:1112–1115. doi: 10.1038/15100. [DOI] [PubMed] [Google Scholar]
- (2).Petka WA, Harden JL, McGrath KP, Wirtz D, Tirrell DA. Reversible hydrogels from self-assembling artificial proteins. Science. 1998;281:389–392. doi: 10.1126/science.281.5375.389. [DOI] [PubMed] [Google Scholar]
- (3).Kothakota S, Mason TL, Tirrell DA, Fournier MJ. Biosynthesis of a periodic protein containing 3-thienylalanine: a step toward genetically engineered conducting polymers. J. Am. Chem. Soc. 1995;117:536–537. [Google Scholar]
- (4).Heilshorn SC, DiZio KA, Welsh ER, Tirrell DA. Endothelial cell adhesion to the fibronectin CS5 domain in artificial extracellular matrix proteins. Biomaterials. 2003;24:4245–4252. doi: 10.1016/s0142-9612(03)00294-1. [DOI] [PubMed] [Google Scholar]
- (5).Meyer DE, Kong GA, Dewhirst MW, Zalutsky MR, Chilkoti A. Targeting a genetically engineered elastin-like polypeptide to solid tumors by local hyperthermia. Cancer Res. 2001;61:1548–1554. [PubMed] [Google Scholar]
- (6).Betre H, Setton LA, Meyer DE, Chilkoti A. Characterization of a genetically engineered elastin-like polypeptide for cartilaginous tissue repair. Biomacromolecules. 2002;3:910–916. doi: 10.1021/bm0255037. [DOI] [PubMed] [Google Scholar]
- (7).McMillan RA, Conticello VP. Synthesis and characterization of elastin-mimetic protein gels derived from a well-defined polypeptide precursor. Macromolecules. 2000;33:4809–4821. [Google Scholar]
- (8).Nath N, Chilkoti A. Fabrication of a reversible protein array directly from cell lysate using a stimuli-responsive polypeptide. Anal. Chem. 2003;75:709–715. doi: 10.1021/ac0261855. [DOI] [PubMed] [Google Scholar]
- (9).Meyer DE, Trabbic-Carlson K, Chilkoti A. Protein purification by fusion with an environmentally responsive elastin-like polypeptide: effect of polypeptide length on the purification of thioredoxin. Biotechnol. Prog. 2001;17:720–728. doi: 10.1021/bp010049o. [DOI] [PubMed] [Google Scholar]
- (10).Trabbic-Carlson K, Liu L, Kim B, Chilkoti A. Expression and purification of recombinant proteins from Escherichia coli: Comparison of an elastin-like polypeptide fusion with an oligohistidine fusion. Protein Sci. 2004;13:3274–3284. doi: 10.1110/ps.04931604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Albano CR, Randers-Eichhorn L, Bentley WE, Rao G. Green fluorescent protein as a real time quantitative reporter of heterologous protein production. Biotechnol. Prog. 1998;14:351–354. doi: 10.1021/bp970121b. [DOI] [PubMed] [Google Scholar]
- (12).Cha HJ, Wu CF, Valdes JJ, Rao G, Bentley WE. Observations of green fluorescent protein as a fusion partner in genetically engineered Escherichia coli: monitoring protein expression and solubility. Biotechnol. Bioeng. 2000;67:565–574. [PubMed] [Google Scholar]
- (13).Chae HJ, DeLisa MP, Cha HJ, Weigand WA, Rao G, Bentley WE. Framework for online optimization of recombinant protein expression in high-cell-density Escherichia coli cultures using GFP-fusion monitoring. Biotechnol. Bioeng. 2000;69:275–285. doi: 10.1002/1097-0290(20000805)69:3<275::aid-bit5>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- (14).Meyer DE, Chilkoti A. Genetically encoded synthesis of protein-based polymers with precisely specific molecular weight and sequence by recursive directional ligation: examples from the elastin-like polypeptide system. Biomacromolecules. 2002;3:357–367. doi: 10.1021/bm015630n. [DOI] [PubMed] [Google Scholar]
- (15).Guda C, Zhang X, McPherson DT, Xu J, Cherry JH, Urry DW, Daniell H. Hyper expression of an environmentally friendly synthetic polymer gene. Biotechnol. Lett. 1995;17:745–750. [Google Scholar]
- (16).Wang F, Lee SY. Production of poly(3-hydroxybutyrate) by fed-batch culture of filamentation-suppressed recombinant Escherichia coli. Appl. Environ. Microbiol. 1997;63:4765–4769. doi: 10.1128/aem.63.12.4765-4769.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Bailey JE, Ollis DF. Biochemical Engineering Fundamentals. 2nd ed. Vol. 1. McGraw-Hill; New York: 1986. [Google Scholar]
- (18).Harcum SW, Ramirez DM, Bentley WE. Optimal nutrient feed policies for heterologous protein production. Appl. Biochem. Biotechnol. 1992;34/35:161–173. [Google Scholar]
- (19).McRae SR, Brown CL, Bushell GR. Rapid purification of EGFP, EYFP, and ECFP with high yield and purity. Protein Expr. Purif. 2005;41:121–127. doi: 10.1016/j.pep.2004.12.030. [DOI] [PubMed] [Google Scholar]
- (20).Yakhnin AV, Vinokurov LM, Surin AK, Alakhov YB. Green fluorescent protein purification by organic extraction. Protein Expr. Purif. 1998;14:382–386. doi: 10.1006/prep.1998.0981. [DOI] [PubMed] [Google Scholar]
- (21).Grossman TH, Kawasaki ES, Punreddy SR, Osburne MS. Spontaneous cAMP-dependent derepression of gene expression in stationary phase plays a role in recombinant expression instability. Gene. 1998;209:95–103. doi: 10.1016/s0378-1119(98)00020-1. [DOI] [PubMed] [Google Scholar]
- (22).Makrides SC. Strategies for achieving high-level expression of genes in Escherichia coli. Microbiol. Rev. 1996;60:512–538. doi: 10.1128/mr.60.3.512-538.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Holms WH. The central metabolic pathways of Escherichia coli: relationship between flux and control at a branch point, efficiency of conversion to biomass, and excretion of acetate. Curr. Top. Cell. Regul. 1986;28:69–105. doi: 10.1016/b978-0-12-152828-7.50004-4. [DOI] [PubMed] [Google Scholar]
- (24).Lee SY. High cell-density culture of Escherichia coli. Trends Biotechnol. 1996;14:98–105. doi: 10.1016/0167-7799(96)80930-9. [DOI] [PubMed] [Google Scholar]
- (25).Ramírez DM, Bentley WE. Enhancement of recombinant protein synthesis and stability via coordinated amino acid addition. Biotechnol. Bioeng. 1993;41:557–565. doi: 10.1002/bit.260410508. [DOI] [PubMed] [Google Scholar]
- (26).Scheich C, Sievert V, Büssow K. An automated method for high-throughput protein purification applied to a comparison of His-tag and GST-tag affinity chromatography. BMC Biotechnol. 2003;3 doi: 10.1186/1472-6750-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Verma R, Boleti E, George A. Antibody engineering: Comparison of bacterial, yeast, insect and mammalian expression systems. J. Immunol. Methods. 1998;216:165–181. doi: 10.1016/s0022-1759(98)00077-5. [DOI] [PubMed] [Google Scholar]
- (28).Zhang AH, Gonzalez SM, Cantor EJ, Chong SR. Construction of a mini-intein fusion system to allow both direct monitoring of soluble protein expression and rapid purification of target proteins. Gene. 2001;275:241–252. doi: 10.1016/s0378-1119(01)00663-1. [DOI] [PubMed] [Google Scholar]
- (29).Sarramegna V, Talmont R, Demange P, Milon A. Heterologous expression of G-protein-coupled receptors: comparison of expression systems from the standpoint of large-scale production and purification. Cell Mol. Life Sci. 2003;60:1529–1546. doi: 10.1007/s00018-003-3168-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Studier F. Protein production by auto-induction in high-density shaking cultures. Protein Expr. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- (31).Aristidou A, San K, Bennett G. Metabolic engineering of Escherichia coli to enhance recombinant protein production through acetate reduction. Biotechnol. Prog. 1995;11:475–478. doi: 10.1021/bp00034a019. [DOI] [PubMed] [Google Scholar]
- (32).Chen W, Graham C, Ciccarelli RB. Automated fed-batch fermentation with feedback controls based on dissolved oxygen (DO) and pH for production of DNA vaccines. J. Ind. Microbiol. Biotechnol. 1997;18:43–48. doi: 10.1038/sj.jim.2900355. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.