Abstract
Statins upregulate endothelial thrombomodulin (TM) by mechanisms that involve members of the Kruppel-like factor (KLF) family. While KLFs are unequivocally implicated in this process experimental evidence points to additional mechanisms.
Deletion/mutation analysis of reporter constructs was used to demonstrate that mutation of the SP1/KLF element in the TM promoter only partially abolishes statin-induced TM upregulation whereas simultaneous mutation of relevant heat shock elements (HSEs) and SP1/KLF element completely prevents statin-induced TM upregulation, thus demonstrating a role for heat shock factors (HSFs). We further identified the pathway by which statins increase binding of HSF1 to HSEs in the TM promoter. Specifically, statins caused NO-dependent dissociation of HSF1 from heat shock protein 90 (HSP90), nuclear translocation of HSF1, and binding to HSEs in the TM promoter. Statins also decreased nuclear content of the HSF1 chaperone 14-3-3β.
In addition to reducing TM upregulation, inhibition of HSF1 reduced statin-induced upregulation of tissue plasminogen activator (tPA), whereas, downregulation of thrombomospondin (TSP-1), plasminogen activator inhibitor 1 (PAI-1), or connective tissue growth factor (CTGF) was unaffected. Knockdown of 14-3-3β or inhibition of HSF1 phosphorylation enhanced the effect of statins on TM and tPA, but did not influence TSP-1, PAI-1, or CTGF.
These data demonstrate that HSF1 is involved in statin-induced regulation of TM. They also suggest that analogous mechanisms may apply to genes that are upregulated by statins, but not to downregulated genes. These results may have broad implications and suggest the use of heat shock protein modulators to selectively regulate pleiotropic statin effects.
Introduction
Inhibitors of 3-hydroxy-3-methylglutaryl (HMG-CoA) reductase, statins, reduce cholesterol biosynthesis and are widely used in the treatment of hyperlipidemia disorders. In addition, statins also have many so-called pleiotropic (non-lipid related) properties. Most pleiotropic effects are mediated through reduced levels of cholesterol biosynthesis pathway intermediates that serve as lipid attachments for posttranslational modification (isoprenylation) of proteins, including Ras and Ras-like proteins such as Rho and Rac 1.
Vascular endothelium is a major target for the pleiotropic effects of statins. Statins exert anti-inflammatory, anti-coagulant, and fibrinolytic properties by up-regulating and enhancing the activity of endothelial nitric oxide synthase (eNOS)2. This results in, for example, down-regulation of connective tissue growth factor (CTGF), thrombospondin (TSP-1), and plasminogen activator inhibitor-1 (PAI-1)3 and up-regulation of tissue plasminogen activator (tPA)4, and thrombomodulin (TM)5;6.
TM is expressed on the luminal surface of endothelial cells where it forms a complex with thrombin. Thrombin, when bound to TM, loses its ability to cleave fibrinogen and activate cellular thrombin receptors, but instead acquires the ability to activate the “natural anticoagulant”, protein C. Restoration of the TM-thrombin-protein C pathway has potential therapeutic benefits in many disorders associated with endothelial dysfunction, including sepsis 7, adult respiratory distress syndrome8, coagulation disorders 9 and radiation injury10.
Several transcription factor binding sites in the 5′-flanking region of the TM promoter regulate TM expression 11;12. The SP1/KLF binding site (at -207), to which Kruppel-like factor 2 and 4 (KLF2 and KLF4) bind, appears particularly important for upregulating TM, and forced over-expression of KLF2 or KLF4 substantially increases TM expression 13;14. Despite the clear role for KLF2 and KLF4, however, several lines of evidence point to additional mechanisms. First, TM is down-regulated by inflammatory cytokines 13, some of which induce KLF4 15;16. Second, KLF2 is induced by mevastatin, simvastatin and lovastatin, but not by pravastatin 17;18, although pravastatin strongly up-regulates TM 19.
Conway and coworkers reported in 1994 that exposure of endothelial cells to heat shock increased the expression of TM, suggesting that heat shock proteins (HSPs), heat shock factors (HSFs), and/or heat shock elements (HSEs) may be involved in transcriptional regulation of TM 20. More recent data suggest that statin-induced improvement of endothelial function may be related to activation of heat shock factor 1 (HSF1)21;22. However, a firm relationship among HSF1, eNOS activity, and TM expression has not been established.
The present study clarifies how HSF1 participates in statin-induced upregulation of endothelial TM at the transcriptional level. Analogous mechanisms may apply to other, but not all pleiotropic effects of statins. Hence, these findings provide a potential basis for differential regulation of pleiotropic statin effects and thus may have wide-ranging implications for selective regulation of such effects for therapeutic purposes.
Materials and Methods
Cell culture and reagents
Cell cultures were performed as described previously23. Human umbilical vein endothelial cells (HUVECs) and human coronary artery endothelial cells (HCAECs) were cultured in EGM-2 and EGM-2 MV medium, respectively (Lonza, Walkersville, MD). Atorvastatin was from Pfizer (New York, NY), pravastatin from TRC (North York, Ontario, Canada), simvastatin from Spectrum Laboratory Product (Gardena, CA), and geldanamycin, 2-Phenyl-4,4,5,5-tetramethylimidazoline-1-oxyl 3-oxide (PTIO), 3-(2-hydroxy-2-nitroso-1-propylhydrazino)-1-propanamine (PAPA), 3-morpholinosydnonimine hydrochloride (SIN-1), mevalonate, PD98059, and U0126 from Sigma Chemical Co. (St Louis, MO). KNK437 was from EMD Biosciences (San Diego, CA). All experiments in the present study were performed at least 5 times. The bars showing the real-time PCR results and the densitometric data (Figures 2–4) represent means of 5 independent experiments ± standard deviations (SD). Statistical comparisons against vehicle-treated control samples were performed with Student’s t-test for samples with unequal variances. A two-sided significance level of 0.05 was used throughout.
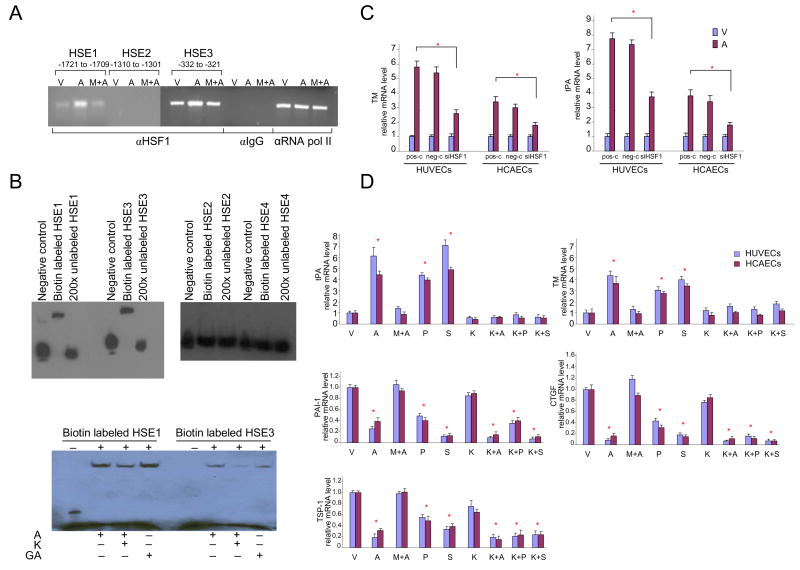
Figure 2. Statin-induced binding of HSF1 to HSE1 and HSE3 and influence of inhibition of HSF1 binding on pleiotropic statin effects.
A) Chromatin immunoprecipitation (ChIP) assay showing binding of HSF1 to HSE1 and HSE3.
HUVECs were treated with vehicle (V), atorvastatin (A), or pre-treated with mevalonate for 30 minutes before being treated with atorvastatin (M+A) for 6 hours and subjected to ChIP analysis. Immunoprecipitated DNA was isolated and amplified by PCR.
B) EMSA analysis confirming binding of HSF1 to HSE1 and HSE3 in the TM promoter. HUVECs were treated with atorvastatin (A), geldanamycin (GA), or pre-treated with KNK437 (K) for 60 minutes before being treated with atorvastatin for 6 hours. Nuclear extracts were then subjected to EMSA analysis.
C) Knockdown of HSF1 reduces the effect of atorvastatin on endothelial expression of TM and tPA.
Knockdown of HSF1 was performed with siRNA. After 48 hours, HUVECs and HCAECs were treated with vehicle or atorvastatin for 24 hours. TM and tPA mRNAs were quantified by real-time PCR.
D) Influence of inhibition of binding of HSF1 to HSE on pleiotropic statin effects. Real-time quantitative PCR analysis of RNA from HUVECs and HCAECs treated with vehicle (V), statins (atorvastatin, A; pravastatin, P; simvastatin, S); pre-treated with mevalonate for 30 minutes before being treated with atorvastatin (M+A); KNK437 (K), or pre-treated with KNK437 for 60 minutes before being treated with statins (atorvastatin, K+A; pravastatin, K+P; simvastatin, K+S) for 24 hours.
Atorvastatin: 1×10−5M; pravastatin: 4×10−5M; simvastatin: 1×10−5M; mevalonate: 5 ×10−4; KNK437:1×10−;4.
* p < 0.05 versus vehicle.
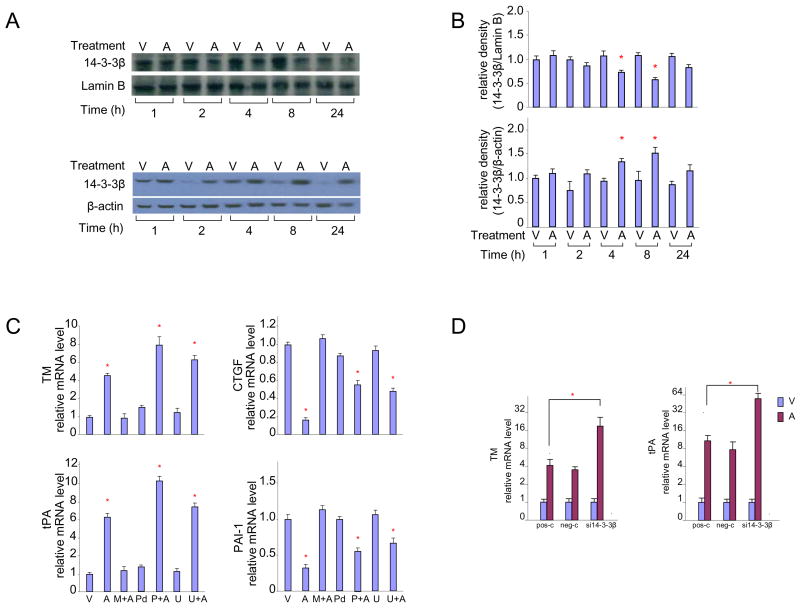
Figure 4. Involvement of the MARK pathway and 14-3-3β protein in statin-induced regulation of endothelial TM.
A) Decreased nuclear content and concomitant increase in cytoplasmic content of 14-3-3β protein after exposure to atorvastatin for 4–8 hours.
HUVEC treated with vehicle (V) or atorvastatin (A) for up to 24 hours. Changes in nuclear (top) and cytoplasmic (bottom) 14-3-3β protein levels were analyzed by western blot.
B) Densitometric analysis of blots shown in panel A.
C) Inhibition of MEK activation enhances the effect of atorvastatin on TM and tPA, but not onCTGFandPAI-1.
HUVECs were treated for 24 hours with vehicle (V), atorvastatin (A), pretreated with mevalonate for 30 minutes before atorvastatin treatment (M+A), PD98059 alone (Pd), U0126 alone (U), or pre-treated with PD98059 (P+A) or U0126 (U+A) for 30 minutes before atorvastatin treatment. TM, tPA, CTGF and PAI-1 mRNAs were quantified by real-time RT-PCR.
D) Knockdown of 14-3-3β augments the effect of atorvastatin on endothelial expression of TM and tPA.
siRNA was used to knock down the 14-3-3β gene. Forty-eight hours later, HUVECs or HCAECs were treated with vehicle or atorvastatin for 24 hours. TM and tPA mRNAs were quantified by real-time PCR.
Atorvastatin: 1×10−5M; mevalonate: 5×10−4M; PD98059 and U0126: 1×10−5M.
* p < 0.05 versus vehicle.
Reporter construct assays
TM reporter constructs were created by PCR amplification from a PAC clone containing the human TM gene (RP4-753D, BACPAC Resources, Oakland, CA) and inserted into the Nhe I and Hind III site of the pGL3.0 basic luciferase reporter vector (Promega, Madison, WI). The first untranscribed base -1 was defined as the -169 base from the ATG codon of the TM translation site. Luciferase reporter constructs and the pRL-TK vector (Promega) were co-introduced into cells by transient transfection using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). Adual-Luciferase reporter assay system (Promega) was used to measure firefly and Renilla luciferase activity of cell lysates. Firefly luciferase values were normalized to Renilla luciferase values. Site specific mutations of HSEs and SP1/KLF elements were created with the QuikChange Site Directed Mutagenesis Kit (Stratagene, La Jolla, CA) (supporting information).
RNAi
Interference transfections were performed with the silencer siRNA Starter Kit (Ambion, Austin, TX). The Ambion pre-designed 14-3-3β siRNA and HSF1 siRNA duplex oligonucleotide sense sequence were as follows: 5′-GCAGAAAACAGAGAGGAAUtt-3′ (14-3-3β, siRNA ID 41953) and 5′-GCUUCCACGUGUUCGACCAtt-3′ (HSF1, siRNA ID S6951), respectively. The Ambion negative control siRNA #1 (cat # 4611) was used for negative control, non-transfected cells were used as a positive control. Cells were incubated with siRNA for 48 hours and then treated with atorvastatin for 24 hours. Total RNA was extracted at 24 hours and analyzed by real time PCR.
Chromatin Immunoprecipitation Assay
Chromatin immunoprecipitation assays (ChIP) assays were performed with the ChIP-IT Enzymatic Kit (Active Motif, Carlsbad, CA). Briefly, cells were cross-linked with 1% formaldehyde and Chromatin was sheared with an enzymatic Shearing Cocktail. Chromatin containing DNA fragments with average size 500 bp were immunoprecipitated with anti-HSF1 monoclonal antibody (sc-13516 X, Santa Cruz Biotechnology, Santa Cruz, CA), with mouse IgG and anti-RNA polymerase II antibody (Active Motif) as negative and positive controls, respectively. DNA fragments were amplified using primers designed around the HSE sites in the TM 5′-flanking region. Primer sets were as follows: 5′-CTGATTCAGCCTAGGCAGC-3′ and 5′-GCTGAGTTGAGCTCTTCGCTG-3′ (HSE1, -1721 to -1709, PCR product 209bp); 5′-AGAAGGAGGACTCTGTGCTCCTA-3′ and 5′-CATAATGGTGAGAGGCAAAC-3′ (HSE2, -1310 to -1301, PCR product 168bp); 5′-GTCTTGCAGGTCCTGTGCAC-3′ and 5′-CTGTAACAAGACGACTGTC-3′ (HSE3, -332 to -321, PCR product 208bp); 5′-CAATCCGAGTATGCGGCATC-3′ and 5′-GATCTCGAGTTTATAAGTGCCCG-3′ (HSE4, - 71 to -54, PCR product 98bp). Positive control PCR primers amplified a region of the constitutively active glyceraldehyde-β-phosphate dehydrogenase (GAPDH) promoter, primer set was: 5′-TACTAGCGGTTTTACGGGCG-3′ and 5′-TCGAACAGGAGGAGCAGAGAGCGA-3′ (PCR product 166bp). Negative control primers amplified a region of genomic DNA between the GAPDH gene and the chromosome condensation-related SMC-associated protein (CNAP) gene, the sequences were: 5′-ATGGTTGCCACTGGGGATCT-3′ and 5′-TGCCAAAGCCTAGGGGAAGA-3′ (PCR product 174bp). PCR products were resolved on a 2% agarose gel and visualized by ethidium bromide staining.
Electrophoretic Mobility Shift Assays (EMSA)
Cells were treated with 1 × 10−5M atorvastatin for 6 hours, and nuclear extracts were prepared with the Nuclear Extraction Kit (Active Motif). EMSA analysis was performed with the LightShift Chemiluminescent EMSA Kit (Pierce, Rockford, IL). The following double-stranded oligonucleotides were used (only top strands are shown): 5′-TGTGCATTCCGGGAGCTTCAGACCC-3′ (HSE1, -1721 to -1709); 5′-TTAGCCCGAAACTTCTCCAACTTCC-3′ (HSE2, -1310 to -1301); 5′-CATGTATGAAAAGAAAGAAAGGAGGACC-3′ (HSE3, -332 to -321); 5′-GGCACTTCCTTCCTTTTCCCGAACGTCC-3′ (HSE4: -71 to -54). Oligonucleotides were labeled with the Biotin3′ End DNA Labeling Kit (Pierce).
Immunoprecipitation
Cells were lysed in modified RIPA lysis buffer (Upstate, Temecula, CA). Lysate (500μg total protein) was incubated with 2μg of HSP90 monoclonal antibody (SPA-835, Stressgen, British Columbia, Canada,) and 50μl of protein G-agarose beads (Upstate) overnight at 4°C on a rotating platform. Beads were washed extensively in lysis buffer, proteins were eluted with 2x Laemmli sample buffer, and the amount of bound protein was measured by western blotting.
Western blots
Total cytoplasmic and nuclear protein was isolated with the Nuclear Extract Kit (Active Motif) and run on 7 to 10% SDS/PAGE gels. After blotting to PVDF membranes (Invitrogen), membranes were blocked for 60 minutes at room temperature and incubated overnight at 4°C in buffer (TBS with 0.1% Tween 20 and 5% milk power) containing 1:1000 diluted Santa Cruz antibodies against HSF1 (sc-13516 X), 14-3-3β (sc-629), β-actin (sc-10731), Lamin B (sc-6217) or HSP90 (SPA-835, Stressgen). Detection of primary antibodies was preformed with HRP-conjugated goat anti-rat/rabbit, or rabbit anti-goat secondary antibody (Santa Cruz Biotechnology) diluted 1:2500. Immunoreactive bands were visualized with Chemiluminescent substrate (Pierce, Rockford, IL). All films presented in this manuscript were scanned in a Lexmark X73 (Lexmark International, Lexington, KY) scanner. The images were not enhanced or altered with any software. Densitometric analysis was performed using Quantity One software (Bio-Rad, Hercules, CA).
RNA analysis
Total RNA was isolated using Ultraspec reagent (Biotecx Laboratories, Houston, Texas) and cDNA was generated with the cDNA Archive Kit (Applied Biosystems, Foster, CA). Gene expression levels were measured with TaqMan real-time quantitative PCR using Applied Biosystems pre-designed primer/probe sets: TM, Hs00264920_s1; tPA, Hs00263492_m1; TSP-1, Hs00170236_m1; PAI-1, Hs00167155_m1; CTGF, Hs00170014_m1; and S27, Hs01378332_g1. PCR amplification and detection were carried out on an ABI Prism 7000 Sequence Detection Sytem (Applied Biosystems) and values were normalized to ribosomal protein S27 mRNA.
Results
Binding of HSF1 to HSE contributes to statin-induced TM up-regulation
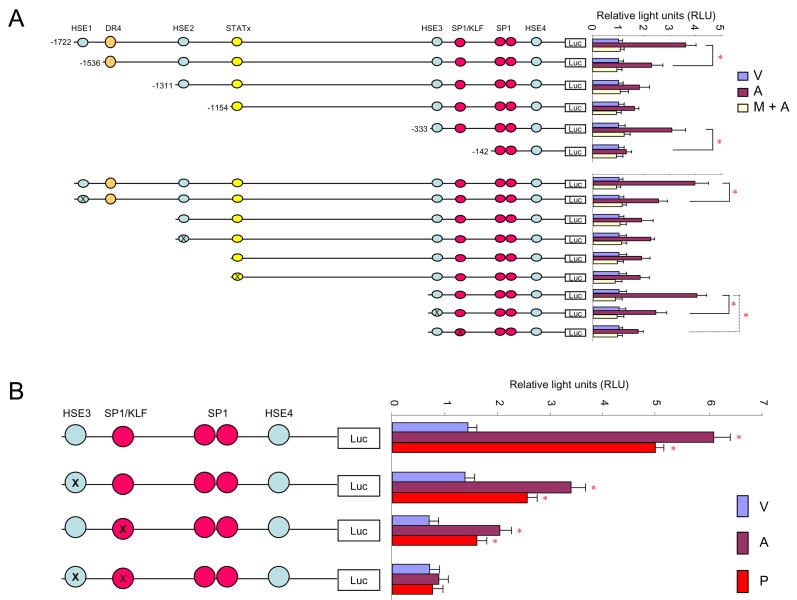
Analysis of the TM promoter with rVista 2.0 (Lawrence Livermore National Laboratory, www.dcode.org) revealed several potential transcription factor binding sites. Besides the 4 SP1 and SP1/KLF sites and one DR4 site described previously, one binding site for STATx (-1153 to -1142) and 4 HSE sites (HSE1: -1721 to -1709, HSE2: -1310 to -1301, HSE3: -332 to -321, and HSE4: -71 to -54) were identified. Primary HUVEC cultures were transiently transfected with reporter constructs containing a firefly luciferase gene under control of the human TM promoter. The constructs had various parts of the TM promoter either deleted or mutated. Cells were treated for 24 hours with vehicle, atorvastatin or pre-treated with mevalonate for 30 minutes before treatment with atorvastatin.
Deletion of HSE1 or HSE3 significantly reduced the atorvastatin-induced luciferase signal, whereas deletion of HSE2 or DR4 had no effect. Deletion of the sequence -333 to -1154, which contains the STATx binding site, increased the atorvastatin-induced luciferase signal. Interestingly, mutation of the STATx binding site did not change the luciferase signal significantly (Figure 1 A), suggesting the presence of a represser element distinct from the STATx site. Mutation of the SP1/KLF binding site on -207 partly reduced baseline and atorvastatin-induced luciferase reporter signal. Simultaneous deletion of HSE3 and SP1/KLF, however, completely abolished the effect of atorvastatin, as well as the effect of the non-KLF2 dependent pravastatin (Figure 1B).
Figure 1. Deletion and mutation analysis of the TM promoter region.
A) Deletion and mutation analysis of the TM promoter.
HUVECs were transiently transfected with luciferase reporter constructs with different parts of the human TM 5′-flanking region deleted. Cell were treated with vehicle (V), atorvastatin (A), or pre-treated with mevalonate for 30 minutes before being treated with atorvastatin (M+A) for 24 hours.
Deletion/mutation of HSE1 and HSE3, or of SP1/KLF element partly inhibits statin-induced TM upregulation.
B) Mutation analysis of HSE and/or SP1/KLF element.
HUVECs, transiently transfected with luciferase reporter constructs with HSE3, SP1/KLF, or both elements mutated, were treated with vehicle (V), atorvastatin (A) or pravastatin (P) for 24 hours.
Mutation of either HSE3 or SP1/KLF element only partly inhibited statin-induced TM upregulation, whereas, mutation of both elements together completely abolished statin-induced TM upregulation.
Relative light units (RLU) were determined.
Atorvastatin: 1×10−5M; mevalonate: 5×10−4M; pravastatin: 4×10−5M
* p < 0.05 versus vehicle.
ChIP assays confirmed binding of HSF1 to the TM promoter. Atorvastatin-induced binding of HSF1 to HSE1 and HSE3, but not to HSE2 or HSE4 (Figure 2A). EMSA analysis confirmed the results of the ChIP assay. Furthermore, EMSA showed statin-induced binding of HFS1 to HSE 1 and HSE3 to be decreased by pre-treatment with KNK437 (Figure 2B). KNK437 has low toxicity, is a highly selective inhibitor of HSF1 activity, does not inhibit constitutive expression of HSP, inhibits induced synthesis of HSP, and reduces formation of HSF1/HSE complexes 24. Atorvastatin did not induce binding of STAT1/STAT3 to the STATx site (data not shown).
To support that atorvastatin-induced binding of HSF1 to HSE was indeed involved in up-regulation of TM, we investigated the effects of siRNA HSF1 knockdown and KNK437 on statin-induced TM expression in HUVECs and HCAECs. HSF1 knockdown significantly reduced atorvastatin-induced TM and tPA expression (Figure 2C), but did not reverse the effect of statin on TSP-1, CTGF, or PAI-1 (data not shown). Similarly, KNK437 blocked statin-induced upregulation of TM and tPA, but not down-regulation of TSP-1, CTGF, or PAI-1 (Figure 2D). Taken together, these data are consistent with the notion that HSF1 binding to HSE is involved in the statin-induced regulation of certain genes, such as TM and tPA, but not in the regulation of others.
Atorvastatin induces NO dependent dissociation of HSF1 from HSP90 and nuclear translocation of HSF1
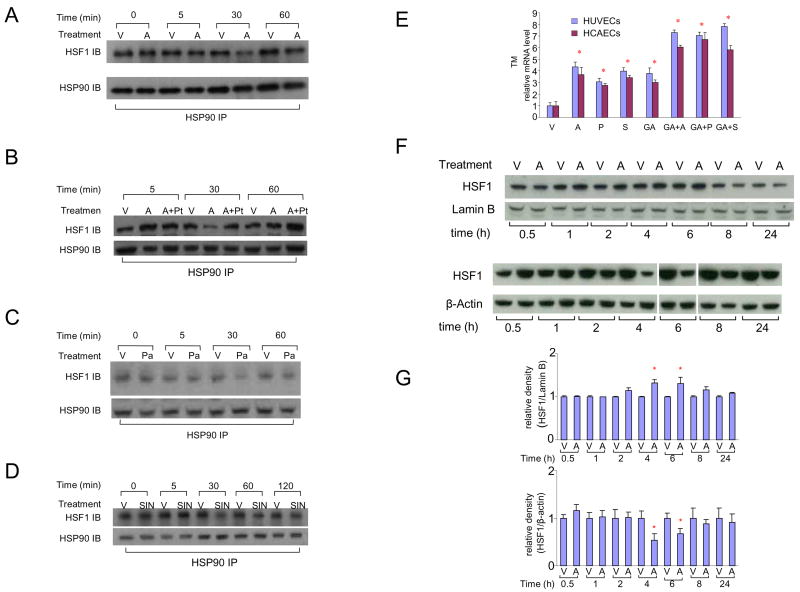
In normal cells, most HSF1 is retained in the cytoplasm as transcriptionally inactive monomers bound to heat shock protein 90 (HSP90) in a multichaperone complex. Various stimulants and cellular stresses induce dissociation of HSF1 from this complex, causing activation and nuclear translocation of HSF1 25;26. Significant reductions in the amount of HSF1 immunoprecipitated by the HSP90 antibody were observed after 30 minutes of atorvastatin treatment (Figure 3A), indicating rapid dissociation of HSF1 from HSP90. This effect was reversed by an NO scavenger (Figures 3B) and mimicked by slow and rapid NO donors (Figure 3C and 3D), indicating that statin-induced dissociation of HSF1 from HSP90 is NO-mediated.
Figure 3. Atorvastatin causes NO-dependent dissociation of HSF1 from HSP90 followed by nuclear translocation of HSF1.
A) Dissociation of HSF1 from HSP90 after 30 minutes exposure of endothelial cells to atorvastatin.
HUVECs were exposed to vehicle (V) or atorvastatin (A) for 0–60 minutes and anti-HSP90 immunoprecipitated protein was analyzed by anti-HSF1 and anti-HSP90 western blot.
B) Inhibition of atorvastatin-induced HSF1-HSP90 dissociation by co-exposure to an NO scavenger.
HUVECs were treated with vehicle (V), atorvastatin (A), or pre-treated with PTIO, an NO scavenger, for 30 minutes before being treated with atorvastatin (A+Pt) for up to 60 minutes. Immunoprecipitated protein was analyzed by western blot.
C) Exposure of endothelial cells to a slow NO donor causes HSF1-HSP90 dissociation. HUVECs were treated with vehicle (V), or PAPA (Pa), a slow NO donor, for up to 60 minutes. Immunoprecipitated protein was analyzed by western blot.
D) Exposure of endothelial cells to a rapid NO donor causes HSF1-HSP90 dissociation. HUVECs were treated with vehicle (V), or SIN-1 (SIN), a rapid NO donor, for up to 120 minutes. Immunoprecipitated protein was analyzed by western blot.
E) Activation of HSF1-HSP90 dissociation upregulates TM.
TM transcript levels in HUVECs and HCAECs treated with vehicle (V), statins (atorvastatin, A; pravastatin, P; simvastatin, S); geldanamycin (GA); GA combined with statins (atorvastatin, GA+A; pravastatin, GA+P; simvastatin: GA+S) for 24 hours.
F) Increase in nuclear HSF1 content and decrease in cytoplasmic HSF1 content after exposure of endothelial cells to atorvastatin for 4–6 hours.
Total cytoplasmic (bottom) and nuclear (top) proteins were harvested from HUVECs treated with vehicle (V) or atorvastatin (A) for up to 24 hours, and analyzed by western blots.
G) Densitometric analysis of blots shown in panel F.
Atorvastatin: 1×10−5M; pravastatin: 4×10−5M; simvastatin: 1×10−5M; geldanamycin: 2μg/ml; PTIO: 1×10−4M; PAPA: 1×10−5M; SIN-1: 1×10−5M.
* p < 0.05 versus vehicle.
To further support the putative effect of HSP90/HSF1 dissociation on HSE dependent TM expression, we used geldanamycin, a commonly used inhibitor of HSP90 reported to stimulate HSF1 dissociation from HSP90 25. EMSA confirmed binding of HSF1 to HSE1 and HSE3 after geldanamycin treatment (Figure 2B). Furthermore, geldanamycin alone or in combination with statins, significantly upregulated TM in HUVECs and HCAECs (Figure 3E).
The effect of atorvastatin on nuclear translocation of HSF1 in HUVECs was studied with western blotting. Decreased cytoplasmic levels and increased nuclear levels of HSF1 were found starting 4 hours after atorvastatin treatment (Figure 3F and 3G), consistent with rapid nuclear translocation of HSF1 after dissociation of the HSF1-HSP90 complex.
The MARK pathway and protein 14-3-3 are involved in statin-induced upregulation of endothelial TM
HSF1 transcriptional activation is negatively regulated by phosphorylation of serine residues 303 and 307 by mitogen activated protein kinase (MARK)27;28. Phosphorylation converts HSF1 to a form recognized and bound by the intracellular regulatory proteins, 14-3-329.
Western-blot analysis of 14-3-3 proteins in HUVECs revealed that atorvastatin substantially decreased nuclear content of 14-3-3β with a concomitant increase in cytoplasmic levels 4 to 8 hours after treatment (Figure 4A and 4B). Atorvastatin did not significantly change nuclear and cytoplasmic levels of 14-3-3ε and 14-3-3η) (data not shown).
To confirm that negative regulation of HSF1 by MARK and 14-3-3β does indeed reduce statin-induced TM upregulation, MEK inhibition and silencing of the 14-3-3β gene were used. HUVECs were pre-treated with inhibitors of MEK activation, PD98059 or U0126 30;31 for 30 minutes. MEK inhibition enhanced atorvastatin-induced upregulation of TM and tPA (Figure 4C), consistent with the notion that the MARK pathway acts to repress genes that are upregulated by statins. Moreover, siRNA knockdown of 14-3-3β enhanced atorvastatin-induced overexpression of TM and tPA (Figure 4D). Taken together, these results suggest that the MARK pathway, by phosphorylating HSF1 to allow subsequent association of HSF1 with 14-3-3β, counteracts the effects of atorvastatin on TM expression. Interestingly, down-regulation of TSP, CTGF and PAI-1 by atorvastatin was unaffected by MEK inhibition or silencing of 14-3-3β (data not shown), again indicating that downregulation of these genes by statins occurs via other pathways.
Discussion
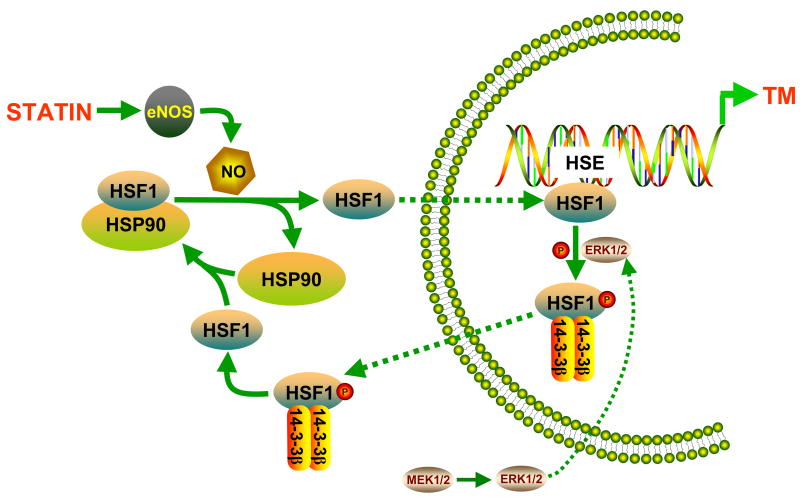
The present study shows that both KLF and HSF1 contribute to achieve maximal statin-induced activation of TM gene expression and that the mechanism involves binding of HSF1 to specific HSE sites in the TM promoter. The proposed pathway by which HSF1 contributes to statin-induced upregulation of TM is depicted in Figure 5. Statins increase production of NO by eNOS. NO induces dissociation of HSF1 from the HSP90-containing multichaperone complex. HSF1 subsequently translocates into the nucleus and binds to two of four HSE elements in the TM promoter, resulting in increased TM expression. Subsequent phosphorylation of HSF1 promotes its binding to protein 14-3-3β, facilitating nuclear export of HSF1 to end the stimulation of TM transcription. Our results suggest that upregulation of other endothelial genes by statins may be mediated via similar pathways, whereas downregulated genes are regulated by different mechanisms.
Figure 5. Mechanism by which HSF1 contributes to statin-induced upregulation of endothelial TM.
Statins cause NO-dependent dissociation of HSF1 from HSP90, nuclear translocation of HSF1, and activation of specific HSEs in the TM promoter. Erk-dependent phosphorylation of HSF1 and subsequent nuclear export of HSF1, facilitated by protein 14-3-3β, contributes to termination of statin-induced TM upregulation.
HSPs and HSF1 are presumed to play roles in the pathogenesis of and/or protection against atherosclerosis and other cardiovascular disorders 32. Interestingly, Uchiyama et al. have suggested the involvement of HSF1 in pleiotropic effects of statins, including upregulation of TM and downregulation of PAI-1, through upregulation of HSPs 21;22. Their findings are somewhat in conflict with data from our and other laboratories. We and others have shown that statins consistently down regulate heat shock proteins, notably HSP70 and HSP90, in endothelial cells and other cells 23;33. We show here that the TM promoter contains specific HSE sites and that HSF1 induces TM transcription in a direct manner by binding to these HSE sites. Moreover, in contrast to genes that are upregulated by statins, such as TM and tPA, downregulation of CTGF, TSP, and PAI-1 appears to be mediated by other mechanisms. This finding is also somewhat in contrast to Uchiyama et al., who reported that an HSE oligonucleotide inhibited simvastatin-induced downregulation of PAI-1 22. It is conceivable that the observed differences between their study and ours relate to differential inactivation/inhibition of nuclear versus cytoplasmic HSF1.
It is well known that statins increase the activity of eNOS, that NO triggers various physiological responses, and that many of the antiproliferative, anti-inflammatory and vasculoprotective effects of statins are mediated by this mechanism. In unstressed cells, HSF1 exists in the cytoplasm associated with HSP90. The HSP90-HSF1 complex dissociates in response to various cellular stresses25. We show here that NO causes dissociation of HSF1 from HSP90, a finding that may explain previous observations that NO induces nuclear translocation and functional activation of HSF1 34. Because HSP90 functions as part of a large complex with other proteins 35, NO effects may not only be directed towards HSF1 or HSP90, but also towards other proteins within the HSP90 complex. Interestingly, NO mediated activation of HSF1 was blocked by DTT, a disulfide reducing agent34. NO may induce S-nitrosylation of proteins, 2 sulfhydryl groups in close proximity may form a disulfide bond upon S-nitrosylation 36. The blocking effect of DTT on NO mediated activation of HSF1 indicates that protein S-nitrosylation may be involved in the mechanism of HSF1 activation. Further studies are needed to examine the effects of S-nitrosylation on HSF1 dissociation from HSP90.
In vivo, endothelial KLF2 is associated with resistance to atherogenesis. KLF2 is regulated by shear stress and atheroprotective wave forms up-regulate KLF2 37. Overexpression of KLF2 in vitro induces TM and eNOS expression in endothelial cells 38. Interestingly, KLF2 expression is induced by a number of statins, but not by pravastatin 17;18, despite the fact that pravastatin upregulates TM 19, suggesting that other transcription factor(s) besides KLF2 may induce TM expression.
Our data showed that when only the KLF2 binding site in the TM promoter was mutated, atorvastatin and pravastatin retained the ability to increase luciferase activity in HUVECs transfected with the reporter construct. Conversely, mutation of HSE3 alone did not completely abolish statin-induced luciferase activity. When both HSE3 and the SP1/KLF element were mutated, however, statin-induced luciferase activity was completely abolished, suggesting that both KLF2 and HSF1 participate in statin-dependent TM up-regulation.
While HSF1 binds to the distal (HSE1) and proximal (HSE3) HSE elements and both elements play a role in statin-induced TM promoter activity, the relative importance of HSE1 and HSE3 remains to be elucidated. The reporter construct assays revealed that sequence -333 to -1154 in the 5′-flanking region of the human TM gene may have a represser function, which appears not to be mediated via the STAT1/STAT3 binding site in this part of the TM promoter. It is conceivable that binding of HSF1 to the distal HSE (HSE1) is necessary to overrule the effect of that represser domain.
In conclusion, this study demonstrates that 1) statins upregulate endothelial TM by a mechanism that involves NO-dependent dissociation of HSF1 from HSP90, nuclear translocation of HSF1, and activation of specific HSEs in the TM promoter; 2) Erk-dependent phosphorylation of HSF1 and subsequent nuclear export of HSF1, facilitated by protein 14-3-3β, contributes to the termination of statin-induced TM upregulation; and 3) analogous HSF1-dependent mechanisms may apply to genes that are upregulated by statins, but not to genes that are down-regulated. These findings have broad implications and point to the opportunity to selectively regulate specific pleiotropic statin effects by modulating heat shock proteins.
Acknowledgments
Supported by grants from the National Institutes of Health (CA83719), the Veterans Administration, and the Defense Threat Reduction Agency (HDTRA1-07-C-0028).
Footnotes
Authorship
Contribution: Q.F., M. Berbee, and X.Q. performed experiments; Q.F., J.W., L.M.F., and M.H.-J. designed the research and interpreted data; and Q.F., M. Boerma, M. Berbee, and M.H.-J. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
References
- 1.Takemoto M, Liao JK. Pleiotropic effects of 3-hydroxy-3-methylglutaryl coenzyme a reductase inhibitors. Arterioscler Thromb Vasc Biol. 2001;21:1712–1719. doi: 10.1161/hq1101.098486. [DOI] [PubMed] [Google Scholar]
- 2.Laufs U, La FV, Plutzky J, Liao JK. Upregulation of endothelial nitric oxide synthase by HMG CoA reductase inhibitors. Circulation. 1998;97:1129–1135. doi: 10.1161/01.cir.97.12.1129. [DOI] [PubMed] [Google Scholar]
- 3.Dulak J, Loboda A, Jazwa A, Zagorska A, Dorler J, Alber H, Dichtl W, Weidinger F, Frick M, Jozkowicz A. Atorvastatin affects several angiogenic mediators in human endothelial cells. Endothelium. 2005;12:233–241. doi: 10.1080/10623320500476559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Essig M, Nguyen G, Prie D, Escoubet B, Sraer JD, Friedlander G. 3-Hydroxy-3-methylglutaryl coenzyme A reductase inhibitors increase fibrinolytic activity in rat aortic endothelial cells. Role of geranylgeranylation and Rho proteins. Circ Res. 1998;83:683–690. doi: 10.1161/01.res.83.7.683. [DOI] [PubMed] [Google Scholar]
- 5.Masamura K, Oida K, Kanehara H, Suzuki J, Horie S, Ishii H, Miyamori I. Pitavastatin-induced thrombomodulin expression by endothelial cells acts via inhibition of small G proteins of the Rho family. Arterioscler Thromb Vasc Biol. 2003;23:512–513. doi: 10.1161/01.ATV.0000060461.64771.F0. [DOI] [PubMed] [Google Scholar]
- 6.Shi J, Wang J, Zheng H, Ling W, Joseph J, Li D, Mehta JL, Ponnappan U, Lin P, Fink LM, Hauer-Jensen M. Statins increase thrombomodulin expression and function in human endothelial cells by a nitric oxide-dependent mechanism and counteract tumor necrosis factor alpha-induced thrombomodulin downregulation. Blood Coagul Fibrinolysis. 2003;14:575–585. doi: 10.1097/00001721-200309000-00010. [DOI] [PubMed] [Google Scholar]
- 7.Bernard GR, Vincent JL, Laterre PF, LaRosa SP, Dhainaut JF, Lopez-Rodriguez A, Steingrub JS, Garber GE, Helterbrand JD, Ely EW, Fisher CJ., Jr Efficacy and safety of recombinant human activated protein C for severe sepsis. N Engl J Med. 2001;344:699–709. doi: 10.1056/NEJM200103083441001. [DOI] [PubMed] [Google Scholar]
- 8.Spragg RG, Lewis JF, Walmrath HD, Johannigman J, Bellingan G, Laterre PF, Witte MC, Richards GA, Rippin G, Rathgeb F, Hafner D, Taut FJ, Seeger W. Effect of recombinant surfactant protein C-based surfactant on the acute respiratory distress syndrome. N Engl J Med. 2004;351:884–892. doi: 10.1056/NEJMoa033181. [DOI] [PubMed] [Google Scholar]
- 9.Saito H, Maruyama I, Shimazaki S, Yamamoto Y, Aikawa N, Ohno R, Hirayama A, Matsuda T, Asakura H, Nakashima M, Aoki N. Efficacy and safety of recombinant human soluble thrombomodulin (ART-123) in disseminated intravascular coagulation: results of a phase III, randomized, double-blind clinical trial. J Thromb Haemost. 2007;5:31–41. doi: 10.1111/j.1538-7836.2006.02267.x. [DOI] [PubMed] [Google Scholar]
- 10.Hauer-Jensen M, Fink LM, Wang J. Radiation injury and the protein C pathway. Crit Care Med. 2004;32:S325–S330. doi: 10.1097/01.ccm.0000126358.15697.75. [DOI] [PubMed] [Google Scholar]
- 11.Tazawa R, Hirosawa S, Suzuki K, Hirokawa K, Aoki N. Functional characterization of the 5′-regulatory region of the human thrombomodulin gene. J Biochem. 1993;113:600–606. doi: 10.1093/oxfordjournals.jbchem.a124089. [DOI] [PubMed] [Google Scholar]
- 12.Horie S, Ishii H, Matsumoto F, Kusano M, Kizaki K, Matsuda J, Kazama M. Acceleration of thrombomodulin gene transcription by retinoic acid: retinoic acid receptors and Sp1 regulate the promoter activity through interactions with two different sequences in the 5′-flanking region of human gene. J Biol Chem. 2001;276:2440–2450. doi: 10.1074/jbc.M004942200. [DOI] [PubMed] [Google Scholar]
- 13.Archipoff G, Beretz A, Freyssinet J, Klein-Soyer C, Brisson C, Cazenave J. Heterogeneous regulation of constitutive thrombomodulin or inducible tissue-factor activities on the surface of human saphenous-vein endothelial cells in culture following stimulation by interleukin-1, tumor necrosis factor, thrombin or phorbol ester. Biochem J. 1991;273:679–684. doi: 10.1042/bj2730679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin Z, Kumar A, Sen-Banerjee S, Staniszewski K, Parmar K, Vaughan DE, Gimbrone MA, Balasubramanian V, Garcia-Cardena G, Jain MK. Kruppel-like factor 2 (KLF2) regulates endothelial thrombotic function. Circ Res. 2005;96:e48–e57. doi: 10.1161/01.RES.0000159707.05637.a1. [DOI] [PubMed] [Google Scholar]
- 15.Sen-Banerjee S, Lin Z, Atkins GB, Greif DM, Rao RM, Kumar A, Feinberg MW, Chen Z, Simon DI, Luscinskas FW, Michel TM, Gimbrone MA, Garcia-Cardena G, Jain MK. KLF2 Is a novel transcriptional regulator of endothelial proinflammatory activation. J Exp Med. 2004;199:1305–1315. doi: 10.1084/jem.20031132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamik A, Lin Z, Kumar A, Balcells M, Sinha S, Katz J, Feinberg MW, Gerzsten RE, Edelman ER, Jain MK. Kruppel-like factor 4 regulates endothelial inflammation. J Biol Chem. 2007;282:13769–13779. doi: 10.1074/jbc.M700078200. [DOI] [PubMed] [Google Scholar]
- 17.Sen-Banerjee S, Mir S, Lin Z, Hamik A, Atkins GB, Das H, Benerjee P, Kumar A, Jain MK. Kruppel-like factor 2 as a novel mediator of statin effects in endothelial cells. Circulation. 2005;112:720–726. doi: 10.1161/CIRCULATIONAHA.104.525774. [DOI] [PubMed] [Google Scholar]
- 18.Parmar KM, Nambudiri V, Dai G, Larman B, Gimbrone MA, Garcia-Cardena G. Statins exert endothelial atheroprotective effects via the KLF2 transcription factor. J Biol Chem. 2005;280:26714–26719. doi: 10.1074/jbc.C500144200. [DOI] [PubMed] [Google Scholar]
- 19.Lin SJ, Chen YH, Lin FY, Hsieh LY, Wang SH, Lin CY, Wang YC, Ku HH, Chen JW, Chen YL. Pravastatin induces thrombomodulin expression in TNFalpha-treated human aortic endothelial cells by inhibiting Rac1 and Cdc42 translocation and activity. J Cell Biochem. 2007;101:642–653. doi: 10.1002/jcb.21206. [DOI] [PubMed] [Google Scholar]
- 20.Conway EM, Liu L, Nowakowski B, Steiner-Mosonyi M, Jackman RW. Heat shock of vascular endothelial cells induces an up-regulatory transcriptional response of the thrombomodulin gene that is delayed in onset and does not attenuate. J Biol Chem. 1994;269:22804–22810. [PubMed] [Google Scholar]
- 21.Uchiyama T, Atsuta H, Utsugi T, Oguri M, Hasegawa A, Nakamura T, Nakai A, Nakata M, Maruyama I, Tomura H, Okajima F, Tomono S, Kawazu S, Nagai R, Kurabayashi M. HSF1 and constitutively active HSF1 improve vascular endothelial function (heat shock proteins improve vascular endothelial function) Atherosclerosis. 2007;190:321–329. doi: 10.1016/j.atherosclerosis.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 22.Uchiyama T, Atsuta H, Utsugi T, Ohyama Y, Nakamura T, Nakai A, Nakata M, Maruyama I, Tomura H, Okajima F, Tomono S, Kawazu S, Nagai R, Kurarbayashi M. Simvastatin induces heat shock factor 1 in vascular endothelial cells. Atherosclerosis. 2006;188:265–273. doi: 10.1016/j.atherosclerosis.2005.10.045. [DOI] [PubMed] [Google Scholar]
- 23.Boerma M, Burton GR, Wang J, Fink LM, McGehee RE, Hauer-Jensen M. Comparative expression profiling in primary and immortalized endothelial cells: changes in gene expression in response to HMG-CoA reductase inhibition. Blood Coagul Fibrinolysis. 2006;17:173–180. doi: 10.1097/01.mbc.0000220237.99843.a1. [DOI] [PubMed] [Google Scholar]
- 24.Ohnishi K, Takahashi A, Yokota S, Ohnishi T. Effects of heat shock protein inhibitor KNK437 on heat sensitivity and heat tolerance in human squamous cell carcinoma cell lines differing in p53 status. Int J Radiat Biol. 2004;80:607–614. doi: 10.1080/09553000412331283470. [DOI] [PubMed] [Google Scholar]
- 25.Zou J, Quo Y, Guettouche T, Smith DF, Voellmy R. Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell. 1998;94:471–480. doi: 10.1016/s0092-8674(00)81588-3. [DOI] [PubMed] [Google Scholar]
- 26.Bharadwaj S, Ali A, Ovsenek N. Multiple components of the HSP90 chaperone complex function in regulation of heat shock factor 1 In vivo. Mol Cell Biol. 1999;19:8033–8041. doi: 10.1128/mcb.19.12.8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X, Khaleque MA, Zhao MJ, Zhong R, Gaestel M, Calderwood SK. Phosphorylation of HSF1 by MAPK-activated protein kinase 2 on serine 121, inhibits transcriptional activity and promotes HSP90 binding. J Biol Chem. 2006;281:782–791. doi: 10.1074/jbc.M505822200. [DOI] [PubMed] [Google Scholar]
- 28.Kline MP, Moromoto RI. Repression of the heat shock factor 1 transcriptional activation domain is modulated by constitutive phosphorylation. Mol Cell Biol. 1997;17:2107–2117. doi: 10.1128/mcb.17.4.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X, Grammatikakis N, Siganou A, Calderwood SK. Regulation of molecular chaperone gene transcription involves the serine phosphorylation, 14-3-3 epsilon binding, and cytoplasmic sequestration of heat shock factor 1. Mol Cell Biol. 2003;23:6013–6026. doi: 10.1128/MCB.23.17.6013-6026.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alessi DR, Cuenda A, Cohen P, Dudley DT, Saltiel AR. PD 098059 is a specific inhibitor of the activation of mitogen-activated protein kinase kinase in vitro and in vivo. J Biol Chem. 1995;270:27489–27494. doi: 10.1074/jbc.270.46.27489. [DOI] [PubMed] [Google Scholar]
- 31.Favata MF, Horiuchi KY, Manos EJ, Daulerio AJ, Stradley DA, Feeser WS, Van Dyk DE, Pitts WJ, Earl RA, Hobbs F, Copeland RA, Magolda RL, Scherle PA, Trzaskos JM. Identification of a novel inhibitor of mitogen-activated protein kinase kinase. J Biol Chem. 1998;273:18623–18632. doi: 10.1074/jbc.273.29.18623. [DOI] [PubMed] [Google Scholar]
- 32.Xu Q. Role of heat shock proteins in atherosclerosis. Arterioscler Thromb Vasc Biol. 2002;22:1547–1559. doi: 10.1161/01.atv.0000029720.59649.50. [DOI] [PubMed] [Google Scholar]
- 33.Pirillo A, Jacoviello C, Longoni C, Radaelli A, Catapano AL. Simvastatin modulates the heat shock response and cytotoxicity mediated by oxidized LDL in cultured human endothelial smooth muscle cells. Biochem Biophys Res Commun. 1997;231:437–441. doi: 10.1006/bbrc.1997.6117. [DOI] [PubMed] [Google Scholar]
- 34.Xu Q, Hu Y, Kleindienst R, Wick G. Nitric oxide induces heat-shock protein 70 expression in vascular smooth muscle cells via activation of heat shock factor 1. J Clin Invest. 1997;100:1089–1097. doi: 10.1172/JCI119619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Voellmy R, Boellmann F. Chaperone regulation of the heat shock protein response. Adv Exp Med Biol. 2007;594:89–99. doi: 10.1007/978-0-387-39975-1_9. [DOI] [PubMed] [Google Scholar]
- 36.Arnelle DR, Stamler JS. NO+, NO, and NO− donation by S-nitrosothiols: implications for regulation of physiological functions by S-nitrosylation and acceleration of disulfide formation. Arch Biochem Biophys. 1995;318:279–285. doi: 10.1006/abbi.1995.1231. [DOI] [PubMed] [Google Scholar]
- 37.Evinger AJ, Levin ER. Requirements for estrogen receptor alpha membrane localization and function. Steroids. 2005;70:361–363. doi: 10.1016/j.steroids.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 38.Dekker RJ, Boon RA, Rondaij MG, Kragt A, Volger OL, Elderkamp YW, Meijers JC, Voorberg J, Pennekoek H, Horrevoets AJ. KLF2 provokes a gene expression pattern that establishes functional quiescent differentiation of the endothelium. Blood. 2006;107:4354–4363. doi: 10.1182/blood-2005-08-3465. [DOI] [PubMed] [Google Scholar]