Abstract
Genetic association studies on schizophrenia (SZ) have been repeatedly performed over the last two decades, resulting in a consensus that results are generally inconsistent. This consensus has begun to change as a result of meta-analyses (e.g., (Glatt and Jonsson, 2006)). The SchizophreniaGene database (http://www.schizophreniaforum.org/res/sczgene/default.asp) has been a leader in meta-analyses of SZ association data, by dynamically and comprehensively cataloging all public genetic association studies, and preparing meta-analyses of case-control data. There are 19 “top” candidate genes from these analyses (access on December 20, 2007), showing the highest effect sizes and nominally significant associations of at least one variant in the meta-analyses of all ethnic samples or of samples of Caucasian ancestry. We selected 40 polymorphisms in 12 selected “top” genes for additional meta-analyses, which had at least one familial association data. We found gene-wide (correction for the number of meta-analyses for each gene) significant allelic association evidence for seven genes in the combined samples. The odds ratios (ORs) of the associated minor risk alleles range from 1.072 to 1.121, for DRD4, MTHFR, PPP3CC and TP53. For protective allele associations, the ORs are between 0.842 and 0.886, for DAO, IL1B, and SLC6A4. In population based sub-analyses, we found significant results in four genes in Asians (ORs between 1.084 and 1.309 for DRD4, GABRB2, PPP3CC, and TP53), and one gene in European (OR of 0.888 for SLC6A4). However, none of these associations survive experiment-wide correction for multiple testing. No significant heterogeneity between case-control and family-based study designs was detected in 35 out of 40 polymorphisms. Our results suggest eight potential SZ candidate genes and suggest that family data can reasonably be included in the meta-analysis of genetic associations.
Keywords: meta-analysis, schizophrenia, genetics, single nucleotide polymorphism, variable number tandem repeats
1. Introduction
Schizophrenia (SZ) is a genetically complex and heterogeneous psychiatric disorder. Genetic association studies on SZ have been repeatedly performed over the last two decades. There are more than 1,200 published reports in the PubMed database of the National Center for Biotechnology Information (NCBI), which are cataloged in the SchizophreniaGene (SZGene) database of the Schizophrenia Research Forum (http://www.schizophreniaforum.org/res/sczgene/default.asp). It would appear that very few consistent replications have been observed in these association studies. Clinical and genetic heterogeneity might account for the apparent inconsistency. Another possible cause is the very limited sample size in most individual studies, which thus have relatively low statistical power. Meta-analysis is a quantitative approach to systematically combing existing results from multiple individual studies and generating an overall conclusion; it has been shown as an effective method to estimate the genetic effects on common diseases with increased power (Ioannidis et al., 2007; Levinson, 2005; Munafo and Flint, 2004; Munafo, 2006).
SZGene has become the leading source of meta-analyses in SZ, by dynamically and comprehensively cataloging all known genetic association studies, and preparing meta-analyses of polymorphisms in candidate genes that have been examined in four or more sample sets (http://www.schizophreniaforum.org/res/sczgene/default.asp). There are 19 “top” candidate genes from these analyses (access on December 20, 2007), showing the highest effect sizes and nominally significant associations of at least one variant in the meta-analyses of all ethnic samples or of samples of Caucasian ancestry. However, these meta-analyses do not include family-based association studies at this time, and summary results in Asians are not presented either. This paper aims to maximize the number of individuals included, and to test for separate allelic associations in the two major population groups that have been studied, Asians and Europeans. We also investigated the influence of family-based data on meta-analysis of genetic association studies on SZ, by performing a testing of heterogeneity between case-control and family-based study designs. Of the 19 “top” SZ candidate genes, 13 had at least one family data set suitable for meta-analysis. Since we previously published a meta-analysis of G72/G30 (also known as D-amino acid oxidase activator, DAOA) in SZ and bipolar disorder (Shi et al., 2008), here we report our meta-analyses of 12 other genes, including AKT1 (v-akt murine thymoma viral oncogene homolog 1), COMT (catechol-O-methyltransferase, DAO (D-amino acid oxidase), DRD2 (dopamine receptor D2), DRD4 (dopamine receptor D4), DTNBP1 (dystrobrevin binding protein 1), GABRB2 (gamma-aminobutyric acid A receptor, beta), IL1B (interleukin1, beta proprotein), MTHFR (5,10-methylenetetrahydrofolate reductase), PPP3CC (protein phosphatase 3 [formerly 2B], catalytic), SLC6A4 (solute carrier family 6 member 4, also known as 5-hydroxytryptamine transporter, 5-HTT), and TP53 (tumor protein p53).
2. Methods
2.1. Literature searches
First, the association studies were extracted from the SZGene database (http://www.schizophreniaforum.org/res/sczgene/default.asp). Then, the literature was further retrieved by searching the PubMed database using the keywords “schizophrenia”, “gene” and symbols of the 12 genes or their full name. All association studies published in English before March 1, 2008 were analyzed. Additionally, all references cited in articles on association studies, reviews and/or meta-analyses (Supplementary 1 summarizes previous meta-analyses of genes COMT, DRD2, DRD4, DTNBP1, MTHFR, and SLC6A4) were examined to identify potential additional studies that might not be collected in SZGene or PubMed.
2.2. Inclusion Criteria
Polymorphisms (single nucleotide polymorphisms [SNPs] and variable number tandem repeats [VNTRs]) with meta-analyses in the SZGene database were re-analyzed. Studies included in our meta-analyses were based on the following criteria: (1) published in a peer-reviewed journal in English; (2) detailed description of the samples tested (including sample size, ancestry of samples, diagnostic criteria for schizophrenia); (3) data published or furnished by correspondence before March 2008. These data had to contain genetic polymorphism information (standard polymorphism name or position on chromosome), minor allele frequency in case and control groups in population-based studies or numbers of transmitted and un-transmitted minor alleles from heterozygous parents to affected offspring in family-based studies; (4) sample not duplicative of other reports. We used the data from the larger sample set if studies had overlapping subjects; (5) at least one family-based association data set available; and (6) genotypic distribution in the controls of population-based studies not inconsistent with Hardy-Weinberg Equilibrium (HWE).
2.3. Statistical Analyses
The term “data set” here refers to one polymorphism, studied in one case-control or family-based association sample. A single report could be considered as containing separate association data sets, when it included unrelated case-control and family-based data, or if it separately examined distinct samples such as German and Japanese. We integrated population-based and family-based association data set(s) into a single meta-analysis using a method recently developed independently by several groups (Cho et al., 2005; Kazeem and Farrall, 2005; Lohmueller et al., 2003). Briefly, counts of minor alleles in case and control groups in population-based studies were summarized in two-by-two tables. The minor alleles of each polymorphism are annotated in the SZGene database. For family-based studies, the number of each transmitted minor allele from heterozygous parents to affected offspring were treated as the number of occurrence of that “risk” or “protective” minor allele in cases. The controls were assumed from a very large population with equal numbers of each allele (to reflect the expected 50:50 transmission ratio from parents to offspring (Lohmueller et al., 2003)). The transmitted and un-transmitted minor alleles were summarized in two-by-one tables. For each table, the natural logarithms of odds ratios (Ln(ORs)) and standard errors (SEs) were calculated based on the allelic data (Kazeem and Farrall, 2005).
We tested heterogeneity between studies using Cochran’s chi-square-based Q-statistic and estimated the degree of heterogeneity with I2 (I2 = ((Q-(k-1))/Q)×100%, where k indicates number of studies). I2 ranges from 0% to 100%. It indicates the proportion of between-study variability in point estimates that was due to heterogeneity rather than sampling error (Higgins and Thompson, 2002; Huedo-Medina et al., 2006). An overall OR and 95% confidence interval (CI) was estimated under the Mantel-Haenszel’s fixed-effects model (MANTEL and HAENSZEL, 1959) if there was no evidence for heterogeneity (I2 < 50%), otherwise (I2 = 50%) under the DerSimonian-Laird random-effects model (DerSimonian and Laird, 1986). Significance of overall OR was examined using a Z-test. The nominally significant P values were adjusted using two Bonferroni corrections: 1) for each individual gene (to test gene-wide significance) when two or more polymorphisms in one gene were meta-analyzed or subgroup analyses were performed according to sample ancestry (Europeans and Asians), 2) for the whole experiment (40 meta-analyses for all populations and 120 for subpopulations were reviewed). Sensitivity analysis was analyzed by dropping one study in turn, recalculating the overall OR of the remaining studies, and testing its significance. This approach can determine whether a finding in the meta-analysis is due to the contribution of a single study. However, this type of analysis is largely influenced by the total number of studies included in the meta-analysis. Evidence for publication bias was assessed using Egger’s regression asymmetry test with a funnel plot of Ln(OR) against inverse SE in each study (Egger et al., 1997).
To detect potential effects of study design types (case-control versus family-based) or different populations (Europeans versus Asians), overall ORs and SEs were obtained for each subgroup, then heterogeneity of overall ORs between study designs or different populations were evaluated by means of a chi-square test with one degree of freedom, as described in detail elsewhere (Kazeem and Farrall, 2005). Meta-analyses were also carried out after excluding family-based association studies to visually evaluate the effects of such data. The statistical analyses were performed using the program Meta-analysis with Interactive eXplanations (MIX, version 1.61) (Bax et al., 2006). A significance level was set at P = 0.05 for all tests with exception of P = 0.1 for Egger’s test for publication bias.
Power was estimated using the Genetic Power Calculator (http://pngu.mgh.harvard.edu/~purcell/gpc/) (Purcell et al., 2003), under the assumption of a dominant model, with a disease prevalence of 1%, D' of 0.8 between risk allele and marker allele, and the real data from the meta-analyses (significance level adjusted by Bonferroni correction for each gene).
3. Results
3.1. Description of Studies
The procedure to retrieve studies for further meta-analysis is described in Supplementary Figure 1. In total, for 12 of the 19 “top” SZ candidate genes, we found 185 reports with 488 separate association data sets on 40 polymorphisms, which met our criteria for inclusion here (Supplementary Table 2). The number of published family-based association report(s) that have available data suitable for meta-analysis ranged from 1 (78–693 families) to 5 (1242 families). Seven hundred and seventeen data sets on 386 polymorphisms in these 12 genes were excluded from our analyses based on the study selection criteria (Supplementary Table 3). We did not include studies on DAOA, as we have recently published a meta-analysis on this gene using similar methods (Shi et al., 2008).
3.2. Meta-analyses
Table 1 shows the results of meta-analyses of 12 SZ candidate gene association studies. When we included all population sources in one analysis, with Bonferroni correction of each P value for the number of meta-analyzed polymorphisms in that gene, we found seven significant allelic associations (DAO, DRD4, IL1B, MTHFR, PPP3CC, SLC6A4, and TP53) (Table 1 and Supplementary Table 4). In population-based analyses, we found significant results in four genes in Asians (DRD4, GABRB2, PPP3CC, and TP53), and one gene in Europeans (SLC6A4) (Table 1 and Supplementary Table 4). The Forest plots of these significant meta-analyses are shown in Figure 1. However, none of the associations reached experiment-wide association, after correction for all the meta-analyses performed (Supplementary Table 4). For polymorphisms that revealed gene-wide significant results, there was evidence for significant heterogeneity between Asians and Europeans at rs1816072 in GABRB2 (P = 0.0009) and between case-control and family-based association studies at rs16944 in IL1B (P = 0.023) (Supplementary Table 5). Egger’s regression test did not detect significant publication bias (data not shown), except for the studies on the DTNP1 gene (rs2619539, P = 0.028; rs2619528, P = 0.049; rs760761, P = 0.026; rs1018381, P = 0.070).
Table 1.
Meta-analyses of allelic association studies for 12 schizophrenia candidate genes
| Gene | Polymorphism/ minor allelea | Populationb | Studiesc |
Minor allele frequencyd |
Family-based studies included
|
Family-based studies excluded
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fam | CC | Cases | Controls | I2 (%)e | OR (95% CI)f | P(Z)g | I2 (%)e | OR (95% CI)f | P(Z)g | |||
| AKT1 | rs2494732/G | All | 4 (683) | 6 | 0.445 (4195) | 0.433 (4416) | 36 | 1.065 (1.005–1.130) | 0.035 | 38 | 1.064 (0.999–1.131) | 0.050 |
| European | 2 (344) | 2 | 0.434 (2458) | 0.431 (2665) | 49 | 1.017 (0.943–1.097) | 0.660 | NA | NA | NA | ||
| Asian | 2 (339) | 4 | 0.461 (1737) | 0.435 (1706) | 0 | 1.144 (1.041–1.256) | 0.005 | 30 | 1.150 (1.040–1.271) | 0.006 | ||
| AKT1 | rs2498799/A | All | 2 (389) | 5 | 0.403 (2291) | 0.402 (2403) | 0 | 0.991 (0.912–1.078) | 0.838 | 0 | 0.977 (0.896–1.065) | 0.591 |
| European | 1 (265) | 1 | 0.225 (592) | 0.245 (659) | NA | NA | NA | NA | NA | NA | ||
| Asian | 1 (124) | 4 | 0.466 (1699) | 0.460 (1744) | 0 | 1.020 (0.927–1.121) | 0.689 | 0 | 1.006 (0.912–1.109) | 0.907 | ||
| AKT1 | rs3730358/T | All | 2 (389) | 6 | 0.133 (4205) | 0.137 (4249) | 0 | 0.987 (0.904–1.077) | 0.760 | 0 | 0.978 (0.895–1.069) | 0.630 |
| European | 1 (265) | 2 | 0.149 (2478) | 0.156 (2531) | 0 | 0.951 (0.854–1.060) | 0.363 | NA | NA | NA | ||
| Asian | 1 (124) | 4 | 0.111 (1727) | 0.109 (1718) | 0 | 1.058 (0.912–1.228) | 0.455 | 0 | 1.046 (0.898–1.218) | 0.565 | ||
| AKT1 | rs1130214/T | All | 2 (389) | 6 | 0.293 (4190) | 0.297 (4417) | 0 | 0.991 (0.926–1.061) | 0.796 | 0 | 0.989 (0.922–1.060) | 0.751 |
| European | 1 (265) | 2 | 0.300 (2456) | 0.301 (2662) | 0 | 0.998 (0.918–1.084) | 0.960 | NA | NA | NA | ||
| Asian | 1 (124) | 4 | 0.283 (1734) | 0.290 (1755) | 4 | 0.977 (0.867–1.101) | 0.703 | 25 | 0.972 (0.860–1.099) | 0.650 | ||
| AKT1 | rs3803300/A | All | 1 (124) | 5 | 0.352 (2389) | 0.353 (2447) | 51 | 0.965 (0.841–1.107) | 0.607 | 0 | 0.978 (0.864–1.109) | 0.732 |
| European | 0 | 1 | 0.095 (660) | 0.086 (707) | NA | NA | NA | NA | NA | NA | ||
| Asian | 1 (124) | 4 | 0.451 (1729) | 0.461 (1740) | 54 | 0.940 (0.808–1.094) | 0.422 | 62 | 0.922 (0.782–1.088) | 0.337 | ||
| COMT | rs737865/C | All | 1 (267) | 8 | 0.312 (5020) | 0.332 (7747) | 40 | 1.040 (0.983–1.100) | 0.173 | 36 | 1.044 (0.987–1.104) | 0.136 |
| European | 1 (267) | 5 | 0.318 (4060) | 0.340 (6638) | 61 | 1.035 (0.925–1.159) | 0.551 | 54 | 1.050 (0.944–1.169) | 0.368 | ||
| Asian | 0 | 3 | 0.287 (960) | 0.286 (1109) | NA | NA | NA | 0 | 1.004 (0.877–1.150) | 0.196 | ||
| COMT | rs4680/A | All | 4 (1153) | 40 | 0.418 (9762) | 0.428 (11607) | 11 | 1.006 (0.968–1.045) | 0.779 | 17 | 1.006 (0.966–1.047) | 0.777 |
| European | 2 (755) | 24 | 0.494 (5940) | 0.493 (7270) | 8 | 1.011 (0.964–1.061) | 0.646 | 11 | 1.017 (0.967–1.069) | 0.511 | ||
| Asian | 2 (398) | 15 | 0.298 (3760) | 0.317 (4284) | 8 | 1.003 (0.939–1.071) | 0.931 | 16 | 0.994 (0.927–1.065) | 0.862 | ||
| COMT | rs165599/G | All | 1 (267) | 8 | 0.359 (4810) | 0.362 (9709) | 54 | 1.058 (0.967–1.158) | 0.218 | 55 | 1.067 (0.977–1.167) | 0.035 |
| European | 1 (267) | 6 | 0.339 (4171) | 0.353 (8979) | 65 | 1.045 (0.935–1.169) | 0.437 | 67 | 1.059 (0.947–1.184) | 0.314 | ||
| Asian | 0 | 2 | 0.493 (639) | 0.469 (730) | NA | NA | NA | NA | NA | NA | ||
| DAO | rs4623951/C | All | 1 (124) | 4 | 0.296 (1509) | 0.334 (1461) | 0 | 0.842 (0.754–0.940) | 0.002h | 0 | 0.846 (0.758–0.948) | 0.004h |
| European | 0 | 2 | 0.378 (900) | 0.409 (905) | NA | NA | NA | NA | NA | NA | ||
| Asian | 1 (124) | 2 | 0.175 (609) | 0.212 (556) | 0 | 0.779 (0.643–0.944) | 0.011 | NA | NA | NA | ||
| DAO | rs2111902/C | All | 1 (113) | 8 | 0.391 (2517) | 0.368 (2960) | 72 | 1.055 (0.896–1.241) | 0.52 | 75 | 1.069 (0.902–1.267) | 0.444 |
| European | 0 | 5 | 0.305 (1364) | 0.304 (1812) | NA | NA | NA | 85 | 1.061 (0.792–1.422) | 0.692 | ||
| Asian | 1 (113) | 3 | 0.493 (1153) | 0.470 (1148) | 0 | 1.085 (0.969–1.216) | 0.157 | 0 | 1.096 (0.977–1.231) | 0.119 | ||
| DAO | rs3918346/T | All | 1 (113) | 8 | 0.351 (2521) | 0.330 (2966) | 78 | 1.037 (0.856–1.256) | 0.711 | 81 | 1.046 (0.855–1.281) | 0.661 |
| European | 0 | 5 | 0.237 (1365) | 0.238 (1814) | NA | NA | NA | 89 | 1.058 (0.734–1.523) | 0.763 | ||
| Asian | 1 (113) | 3 | 0.486 (1156) | 0.474 (1152) | 0 | 1.046 (0.934–1.172) | 0.44 | 0 | 1.051 (0.936–1.180) | 0.398 | ||
| DAO | rs3741775/G | All | 1 (113) | 8 | 0.420 (2514) | 0.445 (2959) | 79 | 0.910 (0.758–1.091) | 0.307 | 82 | 0.923 (0.763–1.117) | 0.411 |
| European | 0 | 5 | 0.469 (1363) | 0.465 (1812) | NA | NA | NA | 78 | 0.985 (0.788–1.232) | 0.895 | ||
| Asian | 1 (113) | 3 | 0.363 (1151) | 0.413 (1147) | 76 | 0.806 (0.600–1.083) | 0.152 | 84 | 0.824 (0.580–1.170) | 0.279 | ||
| DRD2 | rs1800497/T | All | 1 (90) | 10 | 0.216 (1124) | 0.223 (1218) | 86 | 0.833 (0.587–1.184) | 0.309 | 87 | 0.821 (0.553–1.219) | 0.328 |
| European | 1 (90) | 9 | 0.197 (912) | 0.199 (1024) | 73 | 0.908 (0.673–1.224) | 0.525 | 76 | 0.899 (0.630–1.281) | 0.555 | ||
| mixed | 0 | 1 | 0.300 (212) | 0.348 (194) | NA | NA | NA | NA | NA | NA | ||
| DRD2 | rs1801028/G | All | 1 (90) | 28 | 0.031 (5647) | 0.024 (7392) | 17 | 1.189 (1.014–1.395) | 0.033 | 19 | 1.184 (1.008–1.389) | 0.039 |
| European | 1 (90) | 15 | 0.025 (3887) | 0.020 (5499) | 39 | 1.158 (0.935–1.433) | 0.179 | 43 | 1.148 (0.925–1.423) | 0.210 | ||
| Asian | 0 | 11 | 0.037 (1443) | 0.029 (1585) | NA | NA | NA | 0 | 1.325 (0.992–1.771) | 0.057 | ||
| DRD2 | rs1799732/Del | All | 1 (78) | 15 | 0.122 (4509) | 0.116 (4466) | 77 | 0.882 (0.707–1.100) | 0.267 | 76 | 0.924 (0.745–1.146) | 0.471 |
| European | 1 (78) | 9 | 0.109 (3203) | 0.098 (3434) | 79 | 0.904 (0.657–1.244) | 0.534 | 77 | 0.990 (0.73–1.345) | 0.951 | ||
| Asian | 0 | 6 | 0.154 (1306) | 0.175 (1032) | NA | NA | NA | 66 | 0.843 (0.637–1.115) | 0.232 | ||
| DRD4 | rs1800955/C | All | 1 (90) | 7 | 0.420 (2128) | 0.398 (2206) | 0 | 1.113 (1.022–1.211) | 0.014h | 0 | 1.126 (1.032–1.230) | 0.008h |
| European | 1 (90) | 2 | 0.442 (258) | 0.436 (498) | 0 | 0.977 (0.809–1.180) | 0.811 | NA | NA | NA | ||
| Asian | 0 | 5 | 0.417 (1870) | 0.387 (1708) | NA | NA | NA | 0 | 1.150 (1.046–1.265) | 0.004h | ||
| DTNBP1 | rs760666/T | All | 2 (177) | 6 | 0.240 (1959) | 0.240 (2057) | 11 | 0.968 (0.877–1.069) | 0.521 | 0 | 0.995 (0.898–1.102) | 0.917 |
| European | 1 (41) | 6 | 0.240 (1959) | 0.240 (2057) | 0 | 0.988 (0.893–1.094) | 0.818 | 0 | 0.995 (0.898–1.102) | 0.917 | ||
| Mixed | 1 (136) | 0 | NA | NA | NA | NA | NA | NA | NA | NA | ||
| DTNBP1 | rs2619539/G | All | 3 (870) | 15 | 0.496 (6212) | 0.504 (6553) | 0 | 0.955 (0.911–1.001) | 0.057 | 0 | 0.960 (0.913–1.010) | 0.115 |
| European | 1 (41) | 10 | 0.452 (5085) | 0.463 (5401) | 0 | 0.945 (0.893–1.001) | 0.051 | 0 | 0.946 (0.894–1.002) | 0.057 | ||
| Asian | 1 (693) | 3 | 0.692 (1127) | 0.697 (1152) | 0 | 0.971 (0.879–1.073) | 0.563 | 0 | 0.992 (0.874–1.127) | 0.901 | ||
| DTNBP1 | rs3213207/G | All | 2 (177) | 17 | 0.100 (6889) | 0.108 (7098) | 29 | 0.915 (0.847–0.989) | 0.025 | 36 | 0.918 (0.848–0.993) | 0.034 |
| European | 1 (41) | 13 | 0.110 (5663) | 0.122 (5913) | 0 | 0.896 (0.826–0.972) | 0.008 | 0 | 0.895 (0.824–0.971) | 0.008 | ||
| Asian | 0 | 2 | 0.029 (156) | 0.040 (272) | NA | NA | NA | NA | NA | NA | ||
| DTNBP1 | rs1011313/T | All | 2 (177) | 17 | 0.107 (6681) | 0.100 (6596) | 10 | 1.084 (1.002–1.174) | 0.046 | 15 | 1.075 (0.992–1.165) | 0.080 |
| European | 1 (41) | 13 | 0.101 (5439) | 0.092 (5593) | 0 | 1.105 (1.009–1.210) | 0.031 | 5 | 1.103 (1.007–1.208) | 0.035 | ||
| Asian | 0 | 2 | 0.152 (982) | 0.156 (898) | NA | NA | NA | NA | NA | NA | ||
| DTNBP1 | rs2619528/A | All | 5 (1242) | 11 | 0.206 (4076) | 0.200 (4572) | 54 | 1.067 (0.939–1.213) | 0.320 | 66 | 1.103 (0.937–1.299) | 0.239 |
| European | 3 (413) | 8 | 0.206 (3549) | 0.204 (4172) | 63 | 1.116 (0.946–1.316) | 0.194 | 72 | 1.120 (0.929–1.350) | 0.234 | ||
| Asian | 1 (693) | 1 | 0.082 (267) | 0.097 (295) | NA | NA | NA | NA | NA | NA | ||
| DTNBP1 | rs2005976/A | All | 1 (693) | 9 | 0.197 (4675) | 0.205 (4672) | 21 | 0.947 (0.883–1.015) | 0.125 | 21 | 0.947 (0.883–1.015) | 0.157 |
| European | 0 | 9 | 0.197 (4675) | 0.205 (4672) | NA | NA | NA | 21 | 0.947 (0.883–1.015) | 0.157 | ||
| Asian | 1 (693) | 0 | NA | NA | NA | NA | NA | NA | NA | NA | ||
| DTNBP1 | rs760761/T | All | 1 (136) | 16 | 0.194 (5940) | 0.191 (6198) | 41 | 1.001 (0.938–1.068) | 0.982 | 44 | 0.997 (0.933–1.065) | 0.925 |
| European | 0 | 11 | 0.211 (4525) | 0.215 (4844) | NA | NA | NA | 36 | 0.972 (0.905–1.043) | 0.429 | ||
| Asian | 0 | 3 | 0.096 (1155) | 0.090 (1249) | NA | NA | NA | 43 | 1.132 (0.927–1.382) | 0.225 | ||
| DTNBP1 | rs2619522/C | All | 3 (870) | 14 | 0.185 (5679) | 0.179 (5941) | 47 | 1.008 (0.945–1.076) | 0.802 | 57 | 1.064 (0.941–1.203) | 0.321 |
| European | 1 (41) | 9 | 0.200 (4279) | 0.200 (4642) | 49 | 0.990 (0.920–1.066) | 0.794 | 55 | 1.036 (0.908–1.182) | 0.599 | ||
| Asian | 1 (693) | 3 | 0.095 (1141) | 0.088 (1194) | 39 | 1.069 (0.905–1.261) | 0.434 | 46 | 1.142 (0.931–1.401) | 0.202 | ||
| DTNBP1 | rs1018381/T | All | 3 (870) | 13 | 0.096 (4933) | 0.088 (4937) | 5 | 1.041 (0.949–1.142) | 0.395 | 16 | 1.064 (0.963–1.174) | 0.224 |
| European | 1 (41) | 11 | 0.089 (4439) | 0.086 (4553) | 23 | 1.065 (0.959–1.182) | 0.240 | 28 | 1.069 (0.962–1.187) | 0.217 | ||
| Asian | 1 (693) | 1 | 0.074 (310) | 0.068 (310) | NA | NA | NA | NA | NA | NA | ||
| DTNBP1 | rs909706/G | All | 3 (870) | 7 | 0.339 (3535) | 0.323 (3733) | 0 | 0.988 (0.928–1.052) | 0.712 | 0 | 1.006 (0.937–1.080) | 0.865 |
| European | 1 (41) | 6 | 0.356 (3350) | 0.353 (3383) | 0 | 1.009 (0.941–1.083) | 0.801 | 0 | 1.008 (0.939–1.082) | 0.827 | ||
| Asian | 1 (693) | 1 | 0.024 (185) | 0.030 (350) | NA | NA | NA | NA | NA | NA | ||
| DTNBP1 | rs2619538/T | All | 1 (136) | 10 | 0.372 (4817) | 0.363 (5172) | 40 | 1.036 (0.975–1.101) | 0.259 | 30 | 1.025 (0.964–1.090) | 0.437 |
| European | 0 | 6 | 0.440 (3562) | 0.437 (3909) | NA | NA | NA | 5 | 1.006 (0.942–1.074) | 0.858 | ||
| Asian | 0 | 2 | 0.037 (855) | 0.026 (938) | NA | NA | NA | NA | NA | NA | ||
| GABRB2 | rs187269/C | All | 2 (349) | 7 | 0.292 (1480) | 0.280 (1526) | 73 | 1.093 (0.890–1.342) | 0.399 | 78 | 1.057 (0.814–1.372) | 0.680 |
| European | 2 (349) | 3 | 0.328 (806) | 0.361 (758) | 44 | 0.955 (0.843–1.083) | 0.474 | 0 | 0.866 (0.746–1.004) | 0.057 | ||
| Asian | 0 | 4 | 0.249 (674) | 0.200 (768) | NA | NA | NA | 75 | 1.296 (0.883–1.903) | 0.186 | ||
| GABRB2 | rs252944/C | All | 2 (349) | 8 | 0.159 (1790) | 0.167 (1631) | 63 | 1.058 (0.862–1.299) | 0.589 | 64 | 0.989 (0.788–1.242) | 0.926 |
| European | 2 (349) | 4 | 0.129 (1135) | 0.150 (992) | 54 | 0.961 (0.763–1.210) | 0.733 | 0 | 0.843 (0.707–1.005) | 0.056 | ||
| Asian | 0 | 4 | 0.212 (655) | 0.193 (639) | NA | NA | NA | 70 | 1.250 (0.837–1.866) | 0.276 | ||
| GABRB2 | rs194072/C | All | 2 (349) | 8 | 0.170 (1808) | 0.181 (1613) | 65 | 1.060 (0.858–1.310) | 0.587 | 67 | 0.991 (0.782–1.257) | 0.942 |
| European | 2 (349) | 4 | 0.131 (1137) | 0.153 (991) | 58 | 0.955 (0.752–1.212) | 0.704 | 7 | 0.835 (0.702–0.994) | 0.042 | ||
| Asian | 0 | 4 | 0.236 (671) | 0.224 (622) | NA | NA | NA | 70 | 1.276 (0.850–1.915) | 0.241 | ||
| GABRB2 | rs1816072/C | All | 2 (349) | 8 | 0.414 (1863) | 0.420 (1614) | 69 | 1.012 (0.854–1.198) | 0.892 | 75 | 0.981 (0.801–1.203) | 0.856 |
| European | 2 (349) | 4 | 0.360 (1129) | 0.407 (995) | 39 | 0.881 (0.789–0.984) | 0.024 | 0 | 0.819 (0.723–0.928) | 0.0016h | ||
| Asian | 0 | 4 | 0.498 (734) | 0.441 (619) | NA | NA | NA | 32 | 1.309 (1.022–1.527) | 0.0006h | ||
| GABRB2 | rs1816071/G | All | 2 (349) | 8 | 0.359 (1847) | 0.375 (1619) | 67 | 1.001 (0.849–1.180) | 0.989 | 72 | 0.970 (0.797–1.181) | 0.764 |
| European | 2 (349) | 4 | 0.361 (1133) | 0.408 (993) | 35 | 0.886 (0.794–0.988) | 0.029 | 0 | 0.821 (0.725–0.930) | 0.002h | ||
| Asian | 0 | 4 | 0.356 (714) | 0.323 (626) | NA | NA | NA | 67 | 1.227 (0.895–1.683) | 0.203 | ||
| GABRB2 | rs6556547/T | All | 2 (349) | 8 | 0.123 (1788) | 0.121 (1581) | 75 | 1.081 (0.878–1.331) | 0.465 | 80 | 1.023 (0.807–1.298) | 0.851 |
| European | 2 (349) | 3 | 0.058 (794) | 0.078 (672) | 80 | 0.924 (0.669–1.276) | 0.630 | 0 | 0.722 (0.539–0.967) | 0.029 | ||
| Asian | 0 | 5 | 0.175 (994) | 0.152 (909) | NA | NA | NA | 80 | 1.220 (0.948–1.571) | 0.122 | ||
| IL1B | rs1143634/T | All | 1 (132) | 4 | 0.162 (1207) | 0.180 (1435) | 0 | 0.939 (0.810–1.089) | 0.406 | 0 | 0.946 (0.814–1.099) | 0.465 |
| European | 0 | 3 | 0.231 (791) | 0.245 (995) | NA | NA | NA | 0 | 0.945 (0.808–1.106) | 0.481 | ||
| Asian | 1 (132) | 1 | 0.032 (416) | 0.034 (440) | NA | NA | NA | NA | NA | NA | ||
| IL1B | rs16944/T | All | 1 (89) | 10 | 0.372 (1583) | 0.401 (2373) | 32 | 0.886 (0.805–0.974) | 0.013h | 5 | 0.867 (0.787–0.956) | 0.004h |
| European | 1 (89) | 6 | 0.332 (932) | 0.384 (1695) | 47 | 0.852 (0.754–0.962) | 0.01 | 8 | 0.820 (0.724–0.930) | 0.002h | ||
| Asian | 0 | 4 | 0.430 (651) | 0.445 (678) | NA | NA | NA | 0 | 0.942 (0.808–1.098) | 0.444 | ||
| MTHFR | rs1801131/C | All | 1 (267) | 10 | 0.308 (2428) | 0.297 (3690) | 38 | 1.065 (0.986–1.150) | 0.11 | 25 | 1.090 (1.007–1.181) | 0.034 |
| European | 0 | 8 | 0.329 (1963) | 0.309 (3203) | NA | NA | NA | 0 | 1.106 (1.014–1.205) | 0.022 | ||
| Asian | 1 (267) | 2 | 0.223 (465) | 0.224 (487) | 66 | 0.922 (0.689–1.234) | 0.587 | NA | NA | NA | ||
| MTHFR | rs1801133/T | All | 3 (416) | 16 | 0.348 (3874) | 0.317 (5210) | 52 | 1.121 (1.022–1.229) | 0.015h | 47 | 1.135 (1.065–1.210) | 0.0001h |
| European | 2 (149) | 11 | 0.329 (2533) | 0.304 (3927) | 57 | 1.107 (0.980–1.250) | 0.102 | 52 | 1.127 (1.005–1.264) | 0.041 | ||
| Asian | 1 (267) | 5 | 0.385 (1341) | 0.360 (1283) | 46 | 1.146 (1.033–1.272) | 0.010 | 38 | 1.195 (1.065–1.340) | 0.002h | ||
| PPP3CC | rs2461491/A | All | 3 (694) | 6 | 0.447 (6558) | 0.434 (6527) | 23 | 1.072 (1.022–1.239) | 0.004h | 0 | 1.070 (1.018–1.124) | 0.007h |
| European | 0 | 1 | 0.549 (1870) | 0.539 (2002) | NA | NA | NA | NA | NA | NA | ||
| Asian | 3 (694) | 5 | 0.406 (4688) | 0.388 (4525) | 28 | 1.084 (1.025–1.146) | 0.005h | 0 | 1.083 (1.021–1.149) | 0.008h | ||
| SLC6A4 | VNTR/allele 10 | All | 1 (266) | 9 | 0.279 (2043) | 0.302 (2336) | 49 | 0.882 (0.804–0.967) | 0.007h | 54 | 0.883 (0.751–1.038) | 0.133 |
| European | 1 (266) | 7 | 0.357 (1496) | 0.388 (1687) | 40 | 0.888 (0.807–0.978) | 0.016h | 48 | 0.882 (0.795–0.978) | 0.018 | ||
| Asian | 0 | 2 | 0.067 (547) | 0.080 (649) | NA | NA | NA | NA | NA | NA | ||
| TP53 | rs1042522/C | All | 1 (163) | 5 | 0.399 (1418) | 0.369 (1410) | 0 | 1.112 (1.001–1.236) | 0.048h | 0 | 1.130 (1.013–1.260) | 0.029h |
| European | 1 (163) | 2 | 0.278 (383) | 0.281 (443) | 0 | 0.969 (0.800–1.174) | 0.749 | NA | NA | NA | ||
| Asian | 0 | 3 | 0.443 (1035) | 0.409 (967) | NA | NA | NA | 0 | 1.181 (1.041–1.340) | 0.010h | ||
Polymorphsims and their minor alleles were annotated on the SchizophreniaGene of the Schizophrenia Research Forum (http://www.schizophreniaforum.org/res/sczgene/default.asp).
Meta-analyses were performed for samples from all populations, Europeans, and Asians, respectively.
Numbers of Family-based (Fam, numbers of families are shown in the parentheses) and Case-Control (CC) association studies included in the meta-analyses.
Minor allele frequencies based on case-control association data, numbers of cases and controls are shown in the parentheses.
I2 was employed to assess between-study heterogeneity. I2 > 50% indicates evidence for significant between-study heterogeneity.
Overall Odds Ratios (OR) and their 95% Confidence Intervals (CI) were based on the random-effects model if I2 = 50%, otherwise based on the fix-effects model (I2 < 50%).
P(Z), Z test was used to detect the significance of the overall OR. P-values < 0.05 are shown in bold type.
Associations remain significant after correction for the number of meta-analyses for each gene. Underlined are obvious different results (significant gene-wide associations appear or disappear) of meta-analyses including or excluding family-based association data in Caucasian and all populations. The results excluding family data are similar to those of "all excluding HWE deviations" in the Schizophrenia Research Forum. Note that meta-analyses were performed for polymorphisms with at least one family-based association study, except for those in the DAOA gene, which have been reported recently (Shi et al., Schizophr. Res. 2008; 98: 89–97). NA: not applicable.
Figure 1.
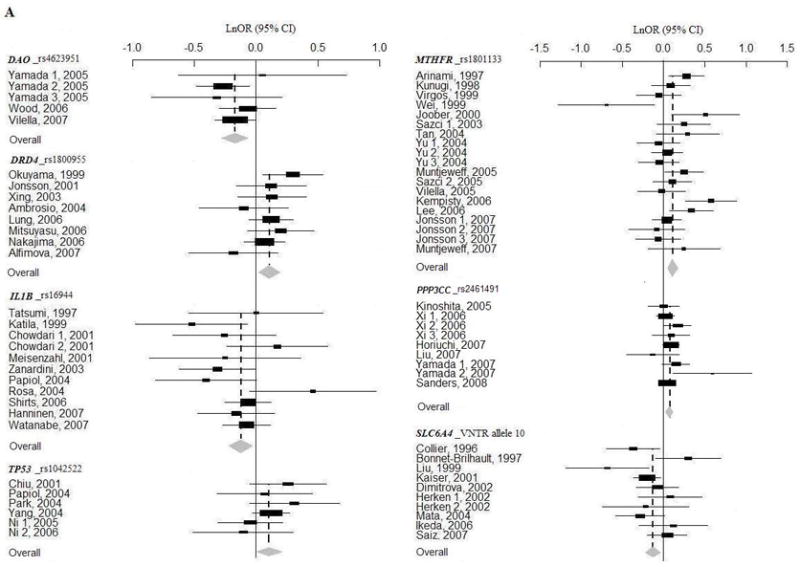
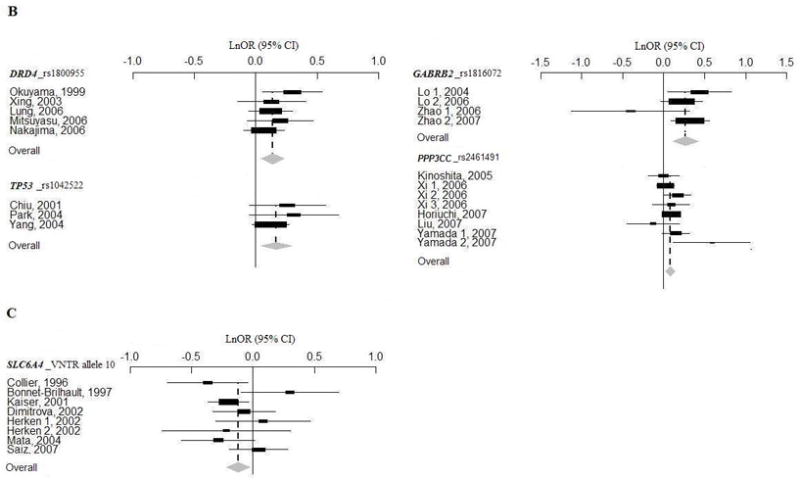
When family association data was removed, rs1816072 and rs1816071in GABRB2 and rs16944 in IL1B showed gene-wide significant disease associations in Europeans, while significant associations of 10 tandem repeats in SLC6A4 (allele 10) disappeared either in all combined populations or in Europeans (underlined results in Table 1). Whereas at significance level of P = 0.05, nominal associations in five polymorphisms were detected, three associations disappeared, and 17 remain in meta-analyses with case-control data (Table 1).
When meta-analyses were performed combing data sets from all the populations, sensitivity analysis indentified that the significant association with DAO was ascribed to a Japanese study with 570 cases and 570 controls (Yamada et al., 2005) (adjusted P = 0.132), and that the association with DRD4 was ascribed to a Japanese study with 252 cases and 269 controls (Okuyama et al., 1999) (adjusted P = 0.081). In population-based sub-analyses, sensitivity analysis found that the association with DRD4 was ascribed to a Japanese study with 252 cases and 269 controls (Okuyama et al., 1999) (adjusted P = 0.102), and that the association with SLC6A4 was ascribed to a study with 129 cases and 187 controls (Collier et al., 1996) (adjusted P = 0.192). Interestingly, when removing the data from a large study with 1870 cases and 2002 controls in European Americans (Sanders et al., 2008), we could detect four additional gene-wide significant associations (all populations: rs165599 in COMT and rs1801028 in DRD2; Europeans: rs1801028 in DRD2 and rs1011313 in DTNBP1, Supplementary Table 6).
3.3 Power estimation
The combined sample in our meta-analyses has generally low power to detect small genetic effects with OR less than 1.3 (data not shown). For example, a sample of approximately 1500 cases and 1500 controls has only 8% power to detect the highest OR of 1.2 for the SNP with MAF of 0.3 in the DAO gene in all populations at the experiment-wide significance level (P < 0.00125 for Bonferroni correction for 40 polymorphisms); a sample of 1000 cases has 5% power to detect OR of 0.4 in Asians at P < 0.00042 (Bonferroni correction for 120 tests based on 40 polymorphisms and sub-populations). To achieve 80% power, 7627 and 5400 cases and comparable controls are needed to detect above effect sizes, respectively.
4. Discussion
Our meta-analyses combing population- and family-based association studies identified significant association evidence across populations for genes DAO, DRD4, IL1B, MTHFR, PPP3CC, SLC6A4, and TP53. Where separate population analyses could be performed, we found that genes DRD4, GABRB2, PPP3CC, and TP53 had SNPs that were uniquely associated with disease risk in Asians and SLC6A4 in Europeans.
Empirical evidence from an analysis of 93 association studies has shown no significant heterogeneity between case-control and family-based study designs (Evangelou et al., 2006). Consistent with that evidence, we have not found significant heterogeneity between study designs for 35 out of 40 polymorphisms (Supplementary Table 5). The differences in the five polymorphisms may result from relatively small sample sizes of the family studies (numbers of families range from 78 to 267), or the limited power of the heterogeneity test of modest genetic effects itself. Therefore, it is reasonable to combine case-control and family association data for meta-analysis (Kazeem and Farrall, 2005; Evangelou et al., 2006).
Our results further suggest eight potential candidate genes that are involved in known biological pathways implicated in the pathophysiology of SZ. The DRD4 gene is in the dopaminergic signaling pathway, where abnormal dopamine transmission has been a prominent hypothesis for schizophrenia over the past half century (Carlsson and LINDQVIST, 1963; Carlsson, 1988; Carlsson and Carlsson, 2006; Toda and bi-Dargham, 2007; Winterer, 2006). SLC6A4, a serotonin transporter, plays a key role in serotonin neurotransmission, which has also implicated in the pathophysiology and antipsychotic treatment of schizophrenia (Breier, 1995; Iqbal and van Praag, 1995; Meltzer, 1989; Meltzer et al., 2003). GABRB2 is one of the most abundant GABA receptor subunits, which mediate inhibitory neurotransmission. GABAergic dysfunction in prefrontal cortex has been linked to the pathophysiology of SZ (Blum and Mann, 2002; Costa et al., 2004; Coyle, 2004; Guidotti et al., 2005). D-amino acid oxidase (DAO) modulates NMDA-receptor mediated signaling. Recently a NMDA receptor-mediated glutamate hypothesis of SZ has received accumulating evidence (Coyle, 1996; Coyle et al., 2003; Coyle, 2006; Lindsley et al., 2006; Moghaddam, 2003). PPP3CC may play a role in the downstream regulation of dopaminergic signal transduction (Greengard, 2001), as well as in the NMDA receptor-dependent synaptic plasticity (Groth et al., 2003). SZ has long been hypothesized as a neurodevelopmental disorder (Arnold et al., 2005; Weinberger, 1987). TP53 has shown a direct role in neurodevelopment (Culmsee and Mattson, 2005; Jacobs et al., 2006). MTHFR participates in the metabolism of folate and DNA methylation, thus may influence neurodevelopment (Greenblatt et al., 1994). IL1B also plays a role in neurodevelopment (Ashdown et al., 2006).
Our results should be interpreted with certain cautions. First, the genetic effects of associated alleles were modest or small (average allelic OR of 1.14 for “risk” genes and 0.87 for “protective” genes). Therefore, the power to detect such small genetic effects are relatively low, even the combined sample for several polymorphism has more than 20,000 individuals (data not shown). Second, most case-control studies we analyzed did not address population stratification using genomic control (Devlin and Roeder, 1999; Reich and Goldstein, 2001), structured association (Pritchard and Rosenberg, 1999; Pritchard et al., 2000), principal components analysis (Price et al., 2006), or other approaches with sub-structure/population informative loci, which would be an issue for the population-specific findings here (Cardon and Palmer, 2003; Freedman et al., 2004). Third, although no evidence for publication bias was detected for all the gene-wide significant associations, the power of the Egger’s regression test is generally low (Macaskill et al., 2001), and this method is further limited for some genes that were meta-analyzed with small numbers of studies (Egger et al., 1997). Fourth, none of the associations could survive correction for multiple testing for all the meta-analyses performed. However, experiment-wide Bonferroni correction may be overly conservative when tests are correlated (e.g., due to linkage disequilibrium of polymorphisms or gene-gene interaction), and there is no consensus on correction for multiple testing in a meta-analysis simultaneously testing a number of genes and polymorphisms (Bertram et al., 2007; Levinson, 2005; Lopez-Leon et al., 2007). Finally, we excluded association studies published in other languages other than in English, which may introduce publication bias in our meta-analyses.
We excluded studies with controls not in HWE, based on several reasons. First, violation of HWE in genetic association studies indicates genotyping errors, population stratification, or selection bias (Cardon and Palmer, 2003; Freedman et al., 2004; Hosking et al., 2004; Xu et al., 2002; Schaid and Jacobsen, 1999). Secondly, lack of HWE in a population implies continued selection, migration, mutation, and assorted mating (Salanti et al., 2005). Thirdly, studies departure from HWE have been proposed to be a potential source of heterogeneity across studies in meta-analysis (Attia et al., 2003; Salanti et al., 2005). However, recent empirical data suggest studies violating HWE may be included in meta-analysis except that the quality of those studies is highly doubtable (Attia et al., 2003; Minelli et al., 2007; Salanti et al., 2007). Family-based association studies were included in meta-analyses except that significant departure from HWE was reported. However, the type I error for classical chi-square goodness-of-fit HWE test in founders (parents) may be over inflated, because founders are not from a random sample from the general population due to typically ascertained families through the affected probands and/or due to potential disease association of the tested marker (Bourgain et al., 2004; Li and Li, 2008). Therefore, HWE assessing should be performed using all the family genotype data conditional on relatedness of the individuals, which can differentiate HWE departure caused by disease association from departure caused by other reasons such as genotyping errors (Bourgain et al., 2004; Li and Li, 2008).
In conclusion, our results further suggest eight susceptibility genes to schizophrenia with modest or small effects, and suggest that family data can reasonably be included in the meta-analysis of genetic associations.
Supplementary Material
Supplementary data associated with this article is available in the online version, at the Schizophrenia Research website.
Acknowledgments
This work was supported by the National Alliance for Research on Schizophrenia and Depression (NARSAD) Young Investigator Awards (to J Shi), Brain Research Foundation at the University of Chicago (to C Liu), NIMH MH065560-02, MH61613-05A1 (to ES Gershon), the Geraldi Norton Foundation and the Eklund Family. We thank those authors for kindly providing raw genotype data or describing their data in detail upon our request, and Dr. Judith A Badner at University of Chicago for helpful comments and discussions. We also thank the members (Lars Bertram, M.D; Nicole C. Allen, B.S.; Sachin Bagade, M.S.; Matthew B. McQueen, Sc.D.; Rudolph E. Tanzi, Ph.D.; Brit-Maren M. Schjeide, B.S.) of the Genetics and Aging Research Unit at the MassGeneral Institute for Neurodegenerative Diseases, Department of Neurology, Massachusetts General Hospital, Charlestown, MA. U.S.A., for their effort in establishing the SchizophreniaGene database (http://www.schizophreniaforum.org/res/sczgene/default.asp).
Role of the funding source Funding for this study was provided by NIH, NARSAD, Brain Research Foundation at the University of Chicago, the Geraldi Norton Foundation and the Eklund Family, which had no further role in study design, data collection and analysis, in the writing of the paper, and in the decision of submitting the paper for publication.
Footnotes
Contributors JS designed the study, wrote the protocol, searched the literature, performed the data-analysis, and drafted the manuscript. ESG and CL advised on data-analysis, participated in the design of the study, interpretation of the data, and revising the manuscript. All authors read and approved the final manuscript.
Conflict of interest The authors declare no financial conflicts.
References
- Arnold SE, Talbot K, Hahn CG. Neurodevelopment, neuroplasticity, and new genes for schizophrenia. Prog Brain Res. 2005;147:319–345. doi: 10.1016/S0079-6123(04)47023-X. [DOI] [PubMed] [Google Scholar]
- Ashdown H, Dumont Y, Ng M, Poole S, Boksa P, Luheshi GN. The role of cytokines in mediating effects of prenatal infection on the fetus: implications for schizophrenia. Mol Psychiatry. 2006;11:47–55. doi: 10.1038/sj.mp.4001748. [DOI] [PubMed] [Google Scholar]
- Attia J, Thakkinstian A, D'Este C. Meta-analyses of molecular association studies: methodologic lessons for genetic epidemiology. J Clin Epidemiol. 2003;56:297–303. doi: 10.1016/s0895-4356(03)00011-8. [DOI] [PubMed] [Google Scholar]
- Bax L, Yu LM, Ikeda N, Tsuruta H, Moons KG. Development and validation of MIX: comprehensive free software for meta-analysis of causal research data. BMC Med Res Methodol. 2006;6:50. doi: 10.1186/1471-2288-6-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet. 2007;39:17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- Blum BP, Mann JJ. The GABAergic system in schizophrenia. Int J Neuropsychopharmacol. 2002;5:159–179. doi: 10.1017/S1461145702002894. [DOI] [PubMed] [Google Scholar]
- Bourgain C, Abney M, Schneider D, Ober C, McPeek MS. Testing for Hardy-Weinberg equilibrium in samples with related individuals. Genetics. 2004;168:2349–2361. doi: 10.1534/genetics.104.031617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breier A. Serotonin, schizophrenia and antipsychotic drug action. Schizophr Res. 1995;14:187–202. doi: 10.1016/0920-9964(94)00043-8. [DOI] [PubMed] [Google Scholar]
- Cardon LR, Palmer LJ. Population stratification and spurious allelic association. Lancet. 2003;361:598–604. doi: 10.1016/S0140-6736(03)12520-2. [DOI] [PubMed] [Google Scholar]
- Carlsson A. The current status of the dopamine hypothesis of schizophrenia. Neuropsychopharmacology. 1988;1:179–186. doi: 10.1016/0893-133x(88)90012-7. [DOI] [PubMed] [Google Scholar]
- Carlsson A, Carlsson ML. A dopaminergic deficit hypothesis of schizophrenia: the path to discovery. Dialogues Clin Neurosci. 2006;8:137–142. doi: 10.31887/DCNS.2006.8.1/acarlsson. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson A, LINDQVIST M. Effect of chlopromazine or haploperidol on formation of 3 methoxytyramine and normetanephrine in mouce brain. Acta Pharmacol Toxicol (Copenh) 1963;20:140–144. doi: 10.1111/j.1600-0773.1963.tb01730.x. [DOI] [PubMed] [Google Scholar]
- Cho HJ, Meira-Lima I, Cordeiro Q, Michelon L, Sham P, Vallada H, Collier DA. Population-based and family-based studies on the serotonin transporter gene polymorphisms and bipolar disorder: a systematic review and meta-analysis. Mol Psychiatry. 2005;10:771–781. doi: 10.1038/sj.mp.4001663. [DOI] [PubMed] [Google Scholar]
- Collier DA, Arranz MJ, Sham P, Battersby S, Vallada H, Gill P, Aitchison KJ, Sodhi M, Li T, Roberts GW, Smith B, Morton J, Murray RM, Smith D, Kirov G. The serotonin transporter is a potential susceptibility factor for bipolar affective disorder. Neuroreport. 1996;7:1675–1679. doi: 10.1097/00001756-199607080-00030. [DOI] [PubMed] [Google Scholar]
- Costa E, Davis JM, Dong E, Grayson DR, Guidotti A, Tremolizzo L, Veldic M. A GABAergic cortical deficit dominates schizophrenia pathophysiology. Crit Rev Neurobiol. 2004;16:1–23. doi: 10.1615/critrevneurobiol.v16.i12.10. [DOI] [PubMed] [Google Scholar]
- Coyle JT. The glutamatergic dysfunction hypothesis for schizophrenia. Harv Rev Psychiatry. 1996;3:241–253. doi: 10.3109/10673229609017192. [DOI] [PubMed] [Google Scholar]
- Coyle JT. The GABA-glutamate connection in schizophrenia: which is the proximate cause? Biochem Pharmacol. 2004;68:1507–1514. doi: 10.1016/j.bcp.2004.07.034. [DOI] [PubMed] [Google Scholar]
- Coyle JT. Glutamate and schizophrenia: beyond the dopamine hypothesis. Cell Mol Neurobiol. 2006;26:365–384. doi: 10.1007/s10571-006-9062-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyle JT, Tsai G, Goff D. Converging evidence of NMDA receptor hypofunction in the pathophysiology of schizophrenia. Ann N Y Acad Sci. 2003;1003:318–327. doi: 10.1196/annals.1300.020. [DOI] [PubMed] [Google Scholar]
- Culmsee C, Mattson MP. p53 in neuronal apoptosis. Biochem Biophys Res Commun. 2005;331:761–777. doi: 10.1016/j.bbrc.2005.03.149. [DOI] [PubMed] [Google Scholar]
- DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- Egger M, Davey SG, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelou E, Trikalinos TA, Salanti G, Ioannidis JP. Family-based versus unrelated case-control designs for genetic associations. PLoS Genet. 2006;2:e123. doi: 10.1371/journal.pgen.0020123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman ML, Reich D, Penney KL, McDonald GJ, Mignault AA, Patterson N, Gabriel SB, Topol EJ, Smoller JW, Pato CN, Pato MT, Petryshen TL, Kolonel LN, Lander ES, Sklar P, Henderson B, Hirschhorn JN, Altshuler D. Assessing the impact of population stratification on genetic association studies. Nat Genet. 2004;36:388–393. doi: 10.1038/ng1333. [DOI] [PubMed] [Google Scholar]
- Glatt SJ, Jonsson EG. The Cys allele of the DRD2 Ser311Cys polymorphism has a dominant effect on risk for schizophrenia: Evidence from fixed- and random-effects meta-analyses. Am J Med Genet B Neuropsychiatr Genet. 2006;141:149–154. doi: 10.1002/ajmg.b.30273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblatt JM, Huffman LC, Reiss AL. Folic acid in neurodevelopment and child psychiatry. Prog Neuropsychopharmacol Biol Psychiatry. 1994;18:647–660. doi: 10.1016/0278-5846(94)90074-4. [DOI] [PubMed] [Google Scholar]
- Greengard P. The neurobiology of slow synaptic transmission. Science. 2001;294:1024–1030. doi: 10.1126/science.294.5544.1024. [DOI] [PubMed] [Google Scholar]
- Groth RD, Dunbar RL, Mermelstein PG. Calcineurin regulation of neuronal plasticity. Biochem Biophys Res Commun. 2003;311:1159–1171. doi: 10.1016/j.bbrc.2003.09.002. [DOI] [PubMed] [Google Scholar]
- Guidotti A, Auta J, Davis JM, Dong E, Grayson DR, Veldic M, Zhang X, Costa E. GABAergic dysfunction in schizophrenia: new treatment strategies on the horizon. Psychopharmacology (Berl) 2005;180:191–205. doi: 10.1007/s00213-005-2212-8. [DOI] [PubMed] [Google Scholar]
- Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- Hosking L, Lumsden S, Lewis K, Yeo A, McCarthy L, Bansal A, Riley J, Purvis I, Xu CF. Detection of genotyping errors by Hardy-Weinberg equilibrium testing. Eur J Hum Genet. 2004;12:395–399. doi: 10.1038/sj.ejhg.5201164. [DOI] [PubMed] [Google Scholar]
- Huedo-Medina TB, Sanchez-Meca J, Marin-Martinez F, Botella J. Assessing heterogeneity in meta-analysis: Q statistic or I2 index? Psychol Methods. 2006;11:193–206. doi: 10.1037/1082-989X.11.2.193. [DOI] [PubMed] [Google Scholar]
- Ioannidis JP, Patsopoulos NA, Evangelou E. Heterogeneity in meta-analyses of genome-wide association investigations. PLoS ONE. 2007;2:e841. doi: 10.1371/journal.pone.0000841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal N, van Praag HM. The role of serotonin in schizophrenia. Eur Neuropsychopharmacol. 1995;5(Suppl):11–23. doi: 10.1016/0924-977x(95)00027-m. [DOI] [PubMed] [Google Scholar]
- Jacobs WB, Kaplan DR, Miller FD. The p53 family in nervous system development and disease. J Neurochem. 2006;97:1571–1584. doi: 10.1111/j.1471-4159.2006.03980.x. [DOI] [PubMed] [Google Scholar]
- Kazeem GR, Farrall M. Integrating case-control and TDT studies. Ann Hum Genet. 2005;69:329–335. doi: 10.1046/j.1529-8817.2005.00156.x. [DOI] [PubMed] [Google Scholar]
- Levinson DF. Meta-analysis in psychiatric genetics. Curr Psychiatry Rep. 2005;7:143–151. doi: 10.1007/s11920-005-0012-9. [DOI] [PubMed] [Google Scholar]
- Li M, Li C. Assessing departure from Hardy-Weinberg equilibrium in the presence of disease association. Genet Epidemiol. 2008 doi: 10.1002/gepi.20335. [DOI] [PubMed] [Google Scholar]
- Lindsley CW, Shipe WD, Wolkenberg SE, Theberge CR, Williams DL, Jr, Sur C, Kinney GG. Progress towards validating the NMDA receptor hypofunction hypothesis of schizophrenia. Curr Top Med Chem. 2006;6:771–785. doi: 10.2174/156802606777057599. [DOI] [PubMed] [Google Scholar]
- Lohmueller KE, Pearce CL, Pike M, Lander ES, Hirschhorn JN. Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat Genet. 2003;33:177–182. doi: 10.1038/ng1071. [DOI] [PubMed] [Google Scholar]
- Lopez-Leon S, Janssens AC, Gonzalez-Zuloeta Ladd AM, Del-Favero J, Claes SJ, Oostra BA, van Duijn CM. Meta-analyses of genetic studies on major depressive disorder. Mol Psychiatry. 2007 doi: 10.1038/sj.mp.4002088. [DOI] [PubMed] [Google Scholar]
- Macaskill P, Walter SD, Irwig L. A comparison of methods to detect publication bias in meta-analysis. Stat Med. 2001;20:641–654. doi: 10.1002/sim.698. [DOI] [PubMed] [Google Scholar]
- MANTEL N, HAENSZEL W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- Meltzer HY. Clinical studies on the mechanism of action of clozapine: the dopamine-serotonin hypothesis of schizophrenia. Psychopharmacology (Berl) 1989;99(Suppl):S18–S27. doi: 10.1007/BF00442554. [DOI] [PubMed] [Google Scholar]
- Meltzer HY, Li Z, Kaneda Y, Ichikawa J. Serotonin receptors: their key role in drugs to treat schizophrenia. Prog Neuropsychopharmacol Biol Psychiatry. 2003;27:1159–1172. doi: 10.1016/j.pnpbp.2003.09.010. [DOI] [PubMed] [Google Scholar]
- Minelli C, Thompson JR, Abrams KR, Thakkinstian A, Attia J. How should we use information about HWE in the meta-analyses of genetic association studies? Int J Epidemiol. 2007 doi: 10.1093/ije/dym234. [DOI] [PubMed] [Google Scholar]
- Moghaddam B. Bringing order to the glutamate chaos in schizophrenia. Neuron. 2003;40:881–884. doi: 10.1016/s0896-6273(03)00757-8. [DOI] [PubMed] [Google Scholar]
- Munafo MR. Candidate gene studies in the 21st century: meta-analysis, mediation, moderation. Genes Brain Behav. 2006;5(Suppl 1):3–8. doi: 10.1111/j.1601-183X.2006.00188.x. [DOI] [PubMed] [Google Scholar]
- Munafo MR, Flint J. Meta-analysis of genetic association studies. Trends Genet. 2004;20:439–444. doi: 10.1016/j.tig.2004.06.014. [DOI] [PubMed] [Google Scholar]
- Okuyama Y, Ishiguro H, Toru M, Arinami T. A genetic polymorphism in the promoter region of DRD4 associated with expression and schizophrenia. Biochem Biophys Res Commun. 1999;258:292–295. doi: 10.1006/bbrc.1999.0630. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Rosenberg NA. Use of unlinked genetic markers to detect population stratification in association studies. Am J Hum Genet. 1999;65:220–228. doi: 10.1086/302449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Cherny SS, Sham PC. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19:149–150. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- Reich DE, Goldstein DB. Detecting association in a case-control study while correcting for population stratification. Genet Epidemiol. 2001;20:4–16. doi: 10.1002/1098-2272(200101)20:1<4::AID-GEPI2>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Salanti G, Higgins JP, Trikalinos TA, Ioannidis JP. Bayesian meta-analysis and meta-regression for gene-disease associations and deviations from Hardy-Weinberg equilibrium. Stat Med. 2007;26:553–567. doi: 10.1002/sim.2575. [DOI] [PubMed] [Google Scholar]
- Salanti G, Sanderson S, Higgins JP. Obstacles and opportunities in meta-analysis of genetic association studies. Genet Med. 2005;7:13–20. doi: 10.1097/01.gim.0000151839.12032.1a. [DOI] [PubMed] [Google Scholar]
- Sanders AR, Duan J, Levinson DF, Shi J, He D, Hou C, Burrell GJ, Rice JP, Nertney DA, Olincy A, Rozic P, Vinogradov S, Buccola NG, Mowry BJ, Freedman R, Amin F, Black DW, Silverman JM, Byerley WF, Crowe RR, Cloninger CR, Martinez M, Gejman PV. No significant association of 14 candidate genes with schizophrenia in a large European ancestry sample: implications for psychiatric genetics. Am J Psychiatry. 2008;165:497–506. doi: 10.1176/appi.ajp.2007.07101573. [DOI] [PubMed] [Google Scholar]
- Schaid DJ, Jacobsen SJ. Biased tests of association: comparisons of allele frequencies when departing from Hardy-Weinberg proportions. Am J Epidemiol. 1999;149:706–711. doi: 10.1093/oxfordjournals.aje.a009878. [DOI] [PubMed] [Google Scholar]
- Shi J, Badner JA, Gershon ES, Liu C. Allelic association of G72/G30 with schizophrenia and bipolar disorder: A comprehensive meta-analysis. Schizophr Res. 2008;98:89–97. doi: 10.1016/j.schres.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda M, bi-Dargham A. Dopamine hypothesis of schizophrenia: making sense of it all. Curr Psychiatry Rep. 2007;9:329–336. doi: 10.1007/s11920-007-0041-7. [DOI] [PubMed] [Google Scholar]
- Weinberger DR. Implications of normal brain development for the pathogenesis of schizophrenia. Arch Gen Psychiatry. 1987;44:660–669. doi: 10.1001/archpsyc.1987.01800190080012. [DOI] [PubMed] [Google Scholar]
- Winterer G. Cortical microcircuits in schizophrenia--the dopamine hypothesis revisited. Pharmacopsychiatry. 2006;39(Suppl 1):S68–S71. doi: 10.1055/s-2006-931498. [DOI] [PubMed] [Google Scholar]
- Xu J, Turner A, Little J, Bleecker ER, Meyers DA. Positive results in association studies are associated with departure from Hardy-Weinberg equilibrium: hint for genotyping error? Hum Genet. 2002;111:573–574. doi: 10.1007/s00439-002-0819-y. [DOI] [PubMed] [Google Scholar]
- Yamada K, Ohnishi T, Hashimoto K, Ohba H, Iwayama-Shigeno Y, Toyoshima M, Okuno A, Takao H, Toyota T, Minabe Y, Nakamura K, Shimizu E, Itokawa M, Mori N, Iyo M, Yoshikawa T. Identification of multiple serine racemase (SRR) mRNA isoforms and genetic analyses of SRR and DAO in schizophrenia and D-serine levels. Biol Psychiatry. 2005;57:1493–1503. doi: 10.1016/j.biopsych.2005.03.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data associated with this article is available in the online version, at the Schizophrenia Research website.


