Abstract
The genetic, biochemical and molecular bases of human cardiac disease have been the focus of extensive research efforts for many years. Early animal models of cardiovascular disease used pharmacologic or surgical interventions, or took advantage of naturally occurring genetic abnormalities and the data obtained were largely correlative. The inability to directly alter an organism’s genetic makeup and cellular protein content and accurately measure the results of that manipulation precluded rigorous examination of true cause-effect and structure-function relationships. Directed genetic manipulation in the mouse gave researchers the ability to modify and control the mammalian heart’s protein content, resulting in the rational design of models that could provide critical links between the mutated or absent protein and disease. Two techniques that have proven particularly useful are transgenesis, which involves the random insertion of ectopic genetic material of interest into a “host” genome, and gene targeting, which utilizes homologous recombination at a pre-selected locus. Initially, transgenesis and gene targeting were used to examine systemic loss-of-function and gain-of-function, respectively, but further refinements in both techniques have allowed for investigations of organ-specific, cell type-specific, developmental stage-sensitive and dose-dependent effects. Genetically engineered animal models of pediatric and adult cardiac disease have proven that, when used appropriately, these tools have the power to extend mere observation to the establishment of true causative proof. We illustrate the power of the general approach by showing how genetically engineered mouse models can define the precise signaling pathways that are affected by the gain-of-function mutation that underlies Noonan syndrome. Increasingly precise and modifiable animal models of human cardiac disease will allow researchers to determine not only pathogenesis, but also guide treatment and the development of novel therapies.
Keywords: Cardiac function, Mouse, Genetics, Transgenesis, Gene targeting
1. Introduction
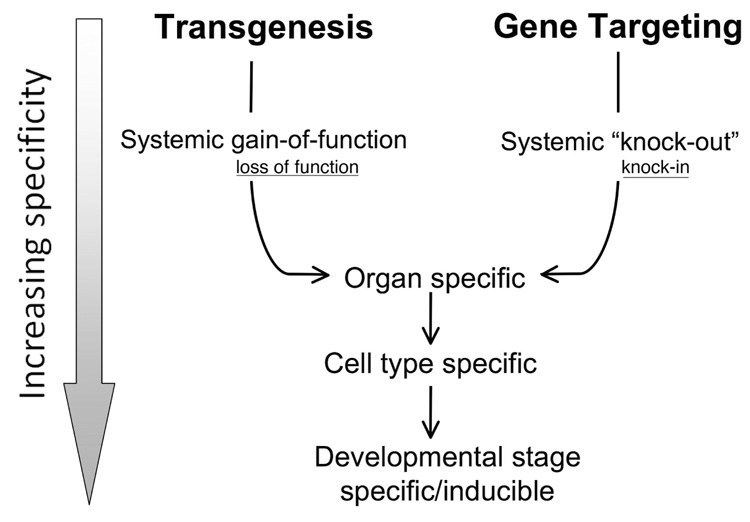
The last 15 years have been a particularly fruitful era for cardiovascular research. This is largely due to the development of technologies allowing the precise manipulation of the mammalian genome such that cause-and-effect and structure-function relationships of the cardiac proteins can be established. With the advent of genetic engineering via insertional transgenesis and gene targeting, it became possible to introduce known mutations into the mammalian genome and determine the subsequent effect(s) at the molecular, biochemical, cellular and whole organ levels, allowing hypotheses to be formulated and tested in a manner that simply had not been possible before. The advances made have largely come about as the tools of genetic manipulation have become more precise, moving from systemic expression or modification to cell type specificity, inducibility and, in some cases, reversibility (Fig. 1).
FIG. 1.
Evolution of genetic manipulation. Traditionally, transgenesis and gene targeting have been used to affect gain-of-function and knockouts, respectively. Systemic application has been gradually refined to the point where organ/cell specific, inducible and extinguishable systems are now commonly used. While transgenesis is traditionally associated with gain-of-function mutations as they are usually dominant, and gene targeting has been more frequently used to produce null alleles (knock-outs) rather than defined point mutations (knock-in), transgenesis can produce loss-of-function as well and gene targeting can result in a hypo- or hyperallele. These points are represented by the smaller, underlined print.
Gene targeting and insertional transgenesis (hereafter referred to simply as transgenesis), as the terms are commonly used today, are two very different technologies. However both result in permanent changes in the mouse genome such that new lines can be established, propagated and disseminated to multiple laboratories for intensive study of different issues. Both technologies have been extremely useful in developing models that test the effects of mutations associated with human cardiovascular disease. Using defined genetic mutations, one can determine the function of a particular protein by either ablating it [1, 2] or expressing it at high levels [3, 4] at different developmental times [5, 6]. One can even define the function of particular post-translational modifications by incorporating amino acid residues that can serve as mimetics for the post-translational modifications.
Literally hundreds of new animal models directed at studying different aspects of cardiovascular function and pathology have been made. Broad areas of investigation include heart function and failure, hypertrophy and dilation, activation/inactivation of particular signaling pathways and the structure-function relationships of proteins critical for normal cardiac function. Loss of function and gain of function are opposite, but complementary, approaches and in combination they are a formidable experimental paradigm for understanding normal and abnormal cardiac function and development. This review focuses on the strengths and weaknesses of the different approaches. Below we outline the rationale and development of ever more precise methods for manipulating the mammalian genome so that the mechanistic bases for cardiac function and the development of cardiovascular pathologies can be dissected and new therapeutic avenues identified and developed.
2. Transgenesis
The process of transgenesis, as described in mice in the early 1980s [7, 8], involves the injection of DNA into the pronucleus of a fertilized egg [9]. After a limited number of ex vivo divisions, the embryo is implanted into a pseudopregnant female and the resulting offspring are screened for the presence of the transgene. While the technique has been widely used and resulted in seminal advances to our understanding of critical proteins’ structure-function relationships as well as the molecular bases for normal and abnormal cellular function, transgenesis remains a rather blunt instrument for studying these processes. The methodology used in the vast majority of transgenic-based experiments precludes the ability to control the site of transgene insertion as well as the number of transgene copies inserted into the host genome. In addition, the endogenous gene and its protein product are still present and this may confound unambiguous interpretation of the data. Because transgenesis involves the random insertion of what can be a large DNA sequence into the genome, the potential for insertional mutagenesis must be also taken into account. While these potentially mutagenic effects have serendipitously led to important new insights [10, 11], they can also cloud the interpretation of data, and the possibility of insertional mutagenesis and variable copy number make the generation and maintenance of multiple lines necessary. Difficulties in controlling these events mandate asking if a phenotype is due to the mutant protein or is merely an artifact secondary to abnormally high protein expression disturbing other parts of one or more signaling pathways or cellular processes. Overall, the heart is extremely tolerant of high levels of expression of cardiac motor proteins [12, 13], however, overexpression of certain proteins can result in severe cardiac pathology [14]. Reporter genes and epitope tags, while ultimately useful in model analyses, must be used appropriately as both can affect a phenotype independent of their attached transgene [15, 16].
Despite these important and very real disadvantages, numerous transgenic experiments directed at studying normal and abnormal cardiovascular development are currently being reported, indicating that many investigators (and scientific reviewers) are able to tolerate the above shortcomings and take advantage of the technique’s strengths. Carefully planned experiments can overcome many, but not all of the limitations, and transgenesis remains a widely used technique for studying cardiovascular function and pathogenesis. The advantages of transgenesis are outlined below.
2.1 Cell type specificity
Early transgenic experiments used strong promoters (cytoplasmic actin, CMV, etc.) to ubiquitously drive transgene expression in essentially all cells and tissues [17, 18], complicating organ specific interpretation of any resulting phenotype. The importance of organ or cell type-specific promoters was quickly appreciated and, with respect to the heart, the development of cardiac-specific promoters greatly enhanced the use of transgenesis in examining abnormalities in the cardiac contractile apparatus [19–23]. The ideal promoter for studying cardiac development and pathology is cardiac-cell type specific and able to drive physiologically relevant levels of expression while being copy number dependent. This allows for the generation of multiple lines with varying degrees of expression; thus the dose dependent response of the system to transgene expression also can be determined. The ideal promoter-transgene construct is position independent via inclusion of “insulators” that are able to shield the construct from either activating or silencing effects exerted by the surrounding chromosomal neighborhood [24]. In initial experiments aimed at defining a cardiac-specific promoter, transcriptional cassettes characterized in vitro were found to be relatively inefficient in directing expression in transgenic hearts [23, 25]. Direct cardiac injection and testing of putative promoters in transgenic mice were subsequently used to characterize a number of cardiac-specific promoters [20, 22, 26, 27]. For cardiomyocyte-specific expression in the adult heart, the α-myosin heavy chain (MHC) promoter has proven to be the most useful due to its ability to drive high levels of cardiomyocyte-specific expression [12, 13, 28] in a copy number dependent and position independent manner [24]. As noted above, these latter two characteristics necessitate the insulation of the transgene’s sequences from the chromatin context into which it inserts. Without sufficient insulation, the chromatin milieu can have a significant impact on transgene expression [29] in that a relatively inactive chromatin neighborhood can effectively override a strong promoter and vice versa.Unfortunately, with the exception of the α-MHC promoter, most cardiac-specific promoters as currently used for transgenesis appear to lack appropriate insulating sequences [19, 24].
2.2 Gain and loss of function
The endogenous gene remains active in a transgenic animal. Thus, studies done in this system are usually directed towards gain of function, in which the transgene encodes a dominant protein, as endogenous protein would prevent expression of a phenotype if the transgene contained a recessive mutation. While still a major limitation of transgenesis, recent methods have broadened the use of transgenesis to loss of function models as well. For example, expression of proteins that inhibit translocation of protein kinase Cε [30] or proteins containing catalytically inactive domains [31], which can act in a dominant negative manner can essentially mimic a loss of function of a protein of interest.
2.3 Inducibility and reversibility
It has become increasingly evident that temporal gene expression leading to targeted protein production is critical for understanding a protein’s physiological role [32]. For example, α-MHC is transiently activated in the early heart tube but quickly becomes restricted to the atria as compartmentalization occurs during fetal development. Thus, in most cases, transgene expression driven by the α-MHC promoter is minimal in the ventricles during cardiac development. Approximately two days before birth, α-MHC transcription is reactivated in the ventricle in response to endogenous fetal thyroid hormone production. The promoter is also often down-regulated in the adult heart in response to cardiac disease [33, 34]. To drive expression in the embryonic/fetal ventricle, the β-MHC promoter is used and this promoter, in turn, is inactivated as α-MHC transcription is initiated in the ventricle [34]. Thus, it is sometimes possible to direct temporally-specific transgene transcription simply by taking advantage of the known transcriptional activity of endogenous cardiac promoters. However, when more precise temporal expression is desired or reversible transgene activation/inactivation needed, various inducible systems have been developed in which transgene expression can be controlled via drug intervention. The use of tetracycline, ecdysone, or RU486 to activate/deactivate transgene transcription has been used effectively to target transgene expression to precise developmental stages or immediately preceding or after various surgical interventions [35–37]. The most widely used system, which utilizes tetracycline (tet), allows one to reversibly induce expression by the addition or removal of tetracycline or its analog, doxycycline (dox). The system, although technically more difficult and cumbersome than normal transgenesis, has been used to activate transgene expression at a particular developmental stage, allow a phenotype to develop, and then shut off expression and see if the heart trends back toward normalcy [38–40]. Clearly, an inducible system could be useful in testing new therapies in that one could delineate the effect of the therapy’s administration, the consequences of its removal and the impact of timing of administration at different developmental times and during different pathogenic stages. Inducible technology adds the ability for precise temporal control to the spatial control afforded by organ or cell-specific expression.
2.3 Relative cost and time
Relative to gene targeting (see below) transgenesis offers a cost effective and relatively quick alternative. The end cost is often one-tenth to one-fifth that of gene targeting and in experienced laboratories creation of suitable transgenic F1 cohorts can be accomplished in roughly 10 weeks compared to months or even, if technical difficulties are encountered, years of development necessary for a gene targeted cohort.
3. Gene Targeting
3.1 Advantages of gene targeting versus transgenesis
In an effort to circumvent some of the shortcomings inherent to transgenesis and to make more precise genetic manipulations in a mammalian genome, techniques were developed to allow for homologous recombination at a pre-selected locus in the mouse. In gene targeting, totipotent embryonic stem cells (ESC) are genetically modified by electroporating the targeting construct into the cells and then selecting, based on drug resistance cassettes built into the construct, the targeted lines that have undergone the correct (and exceedingly rare) homologous recombination event. Surviving colonies are then grown under selective pressure, isolated and tested to confirm the presence of the targeted gene. These cells are, in turn, injected into an early blastocyst and the resulting embryo implanted into a pseudopregnant female. Males with the highest amount of chimerism are selected based on coat color and out-crossed to confirm germline transmission.
Depending on the nature of the experiment, the advantages of gene targeting over transgenesis can be significant. Homologous recombination eliminates the insertional randomness of transgenesis. As the targeting event is usually directed towards creating a null allele, the endogenous protein is not made and the resulting “knockout” can be studied to understand the consequences of the loss of this protein. Recessive alterations can also be bred to homozygosity [41] assuming the animal is viable. Heterozygotes and homozygotes can be compared to assess gene dosage effects. Large fragments can be inserted to eliminate the effects of copy number and diminish the effects of the adjacent chromatin [42]. Additionally, everything from an entire gene to a single amino acid residue can be targeted and replaced to produce “knockin” animals, allowing point mutations to be precisely created in the genome [43–46] under the control of the endogenous promoter. This methodology has proven to be extremely powerful and can be used to determine structure/function relationships or a mutation’s effect(s) over an organism’s entire lifespan. Mutations identified in human cardiac disease can be created in the mouse, allowing for direct translation to an animal model.
3.2 Conditional gene targeting
Similar to transgenesis, it quickly became clear that systemic gene targeting was too blunt an instrument for the precise study of a mutation’s effects on the cardiovascular system. This led to the development of cardiac-specific gene targeting utilizing DNA recombinases, with the Cre-loxP system being most commonly used [47, 48] although other, similarly based recombinase systems such as Flp/FRT may also be useful [49]. By driving Cre expression with a cardiac-specific promoter such as the α-MHC [50] or myosin light chain 2V sequence [29, 51] one can restrict homologous recombination to the cardiomyocytes when the Cre-expressing line is bred to a mouse carrying the appropriate DNA fragment flanked by loxP sites, which are substrates for the recombinase. In addition, constructs driving Cre can be engineered such that Cre production can be induced by pharmacological means. Thus the targeting event can occur at a defined time in development and only within a certain cell population, allowing for the study of events that are selectively lethal during particular times in development. Multiple Cre constructs have been developed that are able to spatially restrict the targeting event to the heart while also allowing for temporal control [52].
3.3 Disadvantages of gene targeting
The main drawbacks to gene targeting involve time and expense. Basic gene targeting is time consuming even in experienced hands and organ- and temporal-control makes creation of the mutant mice even more time intensive. The hope is that publicly available libraries and central resources will become more complete so that investigators don’t need to spend months or even years making the basic mouse. While these efforts are underway [53] they are in their infancy and currently of only limited use.
While gene targeting via homologous recombination is not as random or uncontrolled as transgenesis, one is still inserting and/or replacing large parts of a gene or even an entire gene, and contextual effects of the targeted locus may present. Additionally, the presence of necessary selection cassettes carrying drug resistance markers such as the neo locus can often result in strong interference in efficient transcription of the targeted site. For this reason, current targeting strategies most often flank these parts of the initial targeting construct with loxp or flp sites so that these extraneous sequences can be removed by cre or frt expression, respectively after the initial targeting construct is selected. Inclusion of the neo locus in the first targeting constructs might explain some conflicting data generated by different labs who targeted the same gene. This and subtle contextual differences can often lead to conflicting data. For example, in a carefully documented case of a single “knockout” carried out by three separate groups, systemic ablation of the muscle basic loop helix gene MRF4 resulted in 3 different phenotypes ranging from complete lethality to complete viability [54–56]. These results illustrate the impact of precisely where and how in the genome the homologous recombination event is targeted. Because many genes are closely linked with members of the same pathway or family, alterations in chromosomal context induced by the targeting event may be particularly confounding.
While the vast majority of gene targeting is directed towards “knockout” events, removal of the entire gene is often impractical and complicated by effects on neighboring genes. Thus, targeting is often restricted to particular domains, transcriptional sites or translational sites. This approach leaves portions of the endogenous gene intact and opens the possibility of partial or alternative transcripts that could result in peptides. Screening for these partial peptides is difficult and normal controls are often insufficient to exclude the existence of these partial alleles. Exhaustively screening for the presence of partially active/altered hypoalleles is rarely done, and thus much of the data that currently exist may be compromised to an unknown degree.
4. Dissecting a monogenic syndrome using genetic modification
4.1 General considerations
Both transgenesis and gene targeting have been productively applied to the study of monogenic diseases that are largely organ- or cell type specific-based. Yet many diseases, even those that are monogenic, often present as a combination of pathogenic phenomena across many different cell types and organ systems. Dissecting the underlying pathogenesis of these syndromes across the multiple organs that show involvement has proven difficult or impossible. The investigator ideally should be able to define the involvement of multiple organs as due to a direct effect of the mutant protein (or its absence) or to pathogenic sequelae that are secondary to the involvement of the primary cell type or organ. Often, a combinatorial approach using both transgenesis and gene targeting can be used most effectively to define the pathogenic underpinnings of the resulting syndrome by directing loss- and gain-of-function approaches to the spectrum of cell and tissue types that are involved, but in isolation from one another. A combination of these methodologies thus allows the investigator to apply a reductionist approach to the spectrum of pathologies by restricting expression of the mutation or gene ablation to a single cell type. One can then follow the disease’s course in that cell/organ as well as in others to determine if there is a primary effect in the organ(s) involved or if the pathology is due to secondary epiphenomena brought about as a result of a primary organ or cell type being affected.
4.2 Noonan syndrome
Noonan syndrome (NS) was first described in 1968 in a series of 19 patients [57] and is now recognized as an autosomal dominant disorder affecting ~1/2000 live births. It is characterized most commonly by facial dysmophia, short stature and cardiac anomalies. Deafness, motor delay, mild developmental delay, bleeding diatheses and other hematologic abnormalities can also be part of the syndrome although they are less frequent [58, 59].
Germline, missense mutations in the gene PTPN11 have been identified in ~50% of NS patients. PTPN11 encodes the protein tyrosine phosphatase, SHP2, which is ubiquitously expressed throughout development in many different cell types, including cardiomyocytes. SHP2 is highly conserved between species and contains two SH2 domains, a single protein tyrosine phosphatase (PTP) domain and a C-terminal tail containing phosphorylation sites. SHP2 demonstrates low basal catalytic activity because of an association between the amino terminal-SH2 and PTP domains, resulting in a “closed” conformation (Fig. 2). SHP2 binding to the phosphotyrosine motifs of certain activators (e.g. Gab 1) results in a change to an open conformation in which the PTP domain is accessible to suitable substrates. In general, PTPs provide a key link between multiple growth factors, hormones and cytokines and downstream signaling targets involved in a wide variety of cellular processes. This is supported by the fact that certain PTPN11 mutations mimic the loss of essential receptor tyrosine kinases [60, 61].
FIG. 2.
SHP2 demonstrates low basal catalytic activity due to an association between the N-SH2 and PTP domains resulting in a “closed” conformation. Association with a SHP2 binding protein results in a change to an open conformation in which the PTP domain is accessible to suitable substrates. NS mutations result in a perpetually open conformation, leading to gain-of-function of the PTP domain.
Although mutations have been defined throughout the coding region, multiple mutations are clustered in a 30 residue region near the amino terminal SH2-PTP interaction site, resulting in a loss of auto inhibition and increased PTP activity. This gain-of-function for at least some NS mutations was confirmed in vitro [62]. Multiple experiments have now established that SHP2 exerts its downstream effects mainly through the RAS/MAPK (mitogen-activated protein kinase) cascade, which is involved in activities ranging from cell proliferation and differentiation to survival. Given SHP2’s ubiquitous nature, it is not surprising that it can be a key regulator in the development, function and disease states of multiple organs including the gastrointestinal [63], endocrine [64, 65], hematologic [66], nervous, musculoskeletal [67] and cardiovascular systems. Both in vitro and in vivo studies implicate sustained MAPK activation as a major downstream effector in NS [68] and recent studies implicating gain-of-function mutations in other genes that encode members of the RAS pathway (KRAS, SOS1) suggest that NS is a disorder of excessive RAS activation [69–71]
4.2.1 Mouse models with altered SHP2 signaling
Given that SHP2 has a ubiquitous expression pattern and can affect multiple important signaling pathways, it is not surprising that mutations in PTPN11 have a syndromic presentation. Interestingly, gain and loss of function mutations in PTPN11 each cause distinct syndromes that include reproducible cardiac defects. LEOPARD syndrome (LS; Lentigines, EKG abnormalities, Ocular hypertelorism, valvar Pulmonary stenosis, Abnormal genitalia, Retarded growth, and Deafness) is a rare, autosomal dominant syndrome that shares many clinical features with NS and is also associated with PTPN11 mutations [72]. PTPN11 mutations associated with this syndrome occur in the PTP domain’s catalytic core and result in decreased, rather than increased, SHP2 activity in vitro [73–75]. However, the hypothesis that haploinsufficiency causes the constellation of findings in LS is contradicted by the fact that knockout mice lacking a single allele do not exhibit the anomalies associated with LS [61]. Recent data indicate that LS-associated alleles may exert a dominant negative effect on downstream ERK activation [74].
It is an intriguing question as to how gain and loss of function mutations in a single protein can produce similar syndromes, in particular syndromes with similar cardiac phenotypes. In NS, the most common cardiac manifestation is valvar pulmonary stenosis, but atrial/atrioventricular/ventricular septal defects, mitral valve abnormalities and hypertrophic cardiomyopathy are also seen. In LS, the major cardiac phenotype, in addition to pulmonary stenosis, is hypertrophic cardiomyopathy. Initially, systemic knock-in murine models of NS-associated human mutations were used to examine mutant SHP2’s effects on immune function and myocyte calcium handling [76, 77]. Araki and colleagues generated a knock-in mouse expressing the NS-associated mutation D16G in PTPN11 [78]. Homozygous mutants failed to come to term and the embryos demonstrated severe cardiac anomalies including a variety of septal defects, enlarged AV valve primordia and markedly thinned myocardium. The heterozygotes showed a dose-dependent response, including decreased viability, NS-like dysmorphia and cardiac defects similar to the homozygous animals, but less severe with no myocardial thinning. The endocardial cushions demonstrated a cell and pathway specific increase in ERK activation. These results support a causal role of SHP2 gain-of-function mutation in cardiac pathogenesis and valve abnormalities.
The developmental stage specific effects of mutant SHP2 expression in the heart were studied via transgenesis using the α- and β-MHC promoters [79]. In the mouse, β-MHC expression in the ventricles predominates prenatally, but, via thyroid hormone regulation, is silenced at birth and α-MHC is transcribed. Utilizing the developmental stage specific activity of these two endogenous promoters, transgenic mice were created in which the SHP2 gain-of-function mutation Q79R and normal wild type (WT) protein were expressed under the control of either the β- or α-MHC promoters to yield cardiomyocyte-specific expression in fetal and postnatal ventricles, respectively. Multiple lines were also created to examine dose-dependent effects and both the severity and frequency of cardiac defects were dose dependent. Strikingly, although mutant SHP2 was expressed at high levels in the postnatal ventricular cardiomyocytes, no pathology at either the molecular, cellular, biochemical or gross anatomical levels could be detected. In contrast, prenatal expression of the Q79R (driven by the β-MHC promoter) resulted in 38.8% lethality by 8 months of age and these animals had a cardiac phenotype similar to that of human NS in addition to exhibiting ventricular noncompaction.
Molecular examination of multiple downstream signaling pathways in those hearts showed hyperphosphorylation of the ERK1/2 branch in β-MHC Q79R, which was consistent with previous in vitro data [80]. ERK1/2 signaling in the mouse is thought to play a transient and dynamic role in early cardiac development [81] and the two isoforms appear to have some degree of autonomy based on data obtained with ERK1 and ERK2 knockout mice [82, 83]. To test the necessity of ERK pathway activation for pathogenic presentation, β-MHC Q79R mice were crossed with homozygote erk1- and heterozygote erk2- knockout mice (homozygote erk2- knockout mice die in utero). Both β-MHC Q79R × erk1−/− and β-MHC Q79R erk2+/− embryos showed substantial rescue of the β-MHC Q79R phenotype, with minimal ventricular noncompaction and infrequent ventricular septal defects, indicating that the upregulation of both ERK isoforms is necessary for mediating the effects of enhanced SHP2 signaling. Postnatal examination showed complete rescue of ventricular morphology and function implying that even partial correction of inappropriate ERK1 or ERK2 hyperphosphorylation during cardiac development can normalize postnatal cardiac anatomy and function.
These studies demonstrate that selective overexpression of mutant SHP2 in cardiomyocytes during embryogenesis is sufficient to cause cardiac pathology and demonstrate the utility of both transgenesis and gene targeting in the examination of specific protein alterations and their downstream effects in normal and pathologic cardiac development. The data suggest that it is the balance of SHP2 activity that dictates a normal phenotype and that deviation from a certain range, whether increased or decreased activity, results in syndromes that share some clinical features. Further investigation is needed to define fully the exact mechanisms by which seemingly opposite initial disturbances in protein production can produce similar end results.
A murine systemic knock-in expressing D16G, an NS-associated PTPN11 mutation identified in humans [78] has also yielded valuable insights into the underlying molecular mechanisms in NS. Endocardial cushion defects were evident, implicating the importance of controlled SHP2 expression in endocardial cell development. PTPN11D16G/+ mutants exhibited the involvement of other organ systems that, in some cases, mimicked human NS presentation. These animals had proportionate short stature and craniofacial anomalies, including increased length/width ratio and inter canthal distance, in addition to a broader, shorter snout mimicking the “triangular face” seen in humans. In addition, these animals developed a mild myeloproliferative disease and exhibited liver pathology. Molecular analyses revealed that activation of downstream signaling pathways was cell-type specific. This model highlights the utility of systemic genetic manipulation in recapitulating human monogenic syndromes, but also underscores the need for more precise genetic manipulation to determine the individual pathways involved in each cell type that is affected.
4.2.2 Altered SHP2 signaling in a progenitor cell population: genesis of a syndrome
The above studies establish the importance of SHP2 in cardiac development after cells have differentiated into cardiomyocytes and are producing the cardiac protein complement. However, questions regarding the mechanism by which SHP2 overexpression causes a reproducible syndrome involving a constellation of findings in multiple organ systems remain unanswered. One possibility is that each organ’s population of cells is affected independently due to SHP2 expression and the pathology develops in these separate groups after migration and differentiation. The ubiquity of SHP2 throughout the body could certainly explain the multi-system organ involvement. Another possibility is that the involvement of multiple organ systems is a secondary effect of compromised function in a rather limited number of organs. A third possibility is that SHP2 overexpression affects a population of progenitor cells, altering their differentiation, migration, survival and/or function. Thus, some or all of the organs to which these cells contribute would be altered as well.
Neural crest cells (NCCs) are just such a group of progenitor cells. NCCs are multipotential cells that undergo an epithelial-to-mesenchymal transition and subsequently delaminate from the dorsal neural tube or neural folds [84]. In general, NCCs are divided into cranial and truncal populations and are involved in the formation of a wide variety of organs including the nervous system (autonomic and sensory), melanocytes, endocrine/paracrine cells, craniofacial structures, connective tissue and heart [85]. A subpopulation of the cranial population, termed cardiac neural crest cells (cardiac NCCs) materially participate in remodeling of the pharyngeal arch arteries, septation of the outflow tract, closure of the ventricular septum, formation of the cardiac ganglia, valve development and myocardial function [86]. Normal induction, migration, differentiation, proliferation, and survival of NCCs is dependent upon a variety of transcriptional and environmental modifiers including Wnts, fibroblast growth factors, bone morphogenic growth proteins and retinoic acid [87, 88]. An in depth discussion of cardiac NCCs is beyond the scope of this review, but cardiac NCCs have been the subject of a number of excellent, recent reviews [89–92].
By linking a reporter gene downstream of a stop signal that can be excised by Cre, the Cre-lox technology has been used to track NCC cells in cell lineage studies [93–95]. That is, a construct is created such that cre expression is dependent upon a promoter that is active in pre- or post-migratory NCC and, when expression occurs, reporter gene expression is subsequently activated with the resultant cell population and its descendents irreversibly marked (Fig. 3A). In this manner one can follow the progenitor’s contribution to different cell populations and organs or tissues. We have recently developed a series of constructs to explore both gain and loss of function of SHP2 in pre-migratory NCC and the cells’ contributions to the heart (Fig. 3B), cranium and other organ systems in an attempt to define the cell and molecular underpinnings of the syndromic presentation in NS [79, 96].
FIG. 3.
(A) NCC lineage studies can be conducted by creating a construct in which cre expression is dependent upon a promoter (Wnt1) that is active in pre-migratory NCC and, when expression occurs, reporter gene expression is subsequently activated with the resultant cell population and its descendents irreversibly marked. (B) X-gal/nuclear fast stained heart at ED12.5 from a mouse carrying the construct as in (A). There is a robust population of NCCs that have migrated to the outflow tract cushions and towards the AV canals. Note that in the intercalated outflow tract cushion (located in the top part of the picture) the NCCs are infrequent whereas the cells are abundant in the other two cushions that are proximal to the aorticopulmonary septation.
Cell lineage studies conducted using Cre-lox technology to track NCC contribution to early heart and great vessel formation revealed that the cells delaminate and begin their migration by embryonic day (ED)8.5, with the cells reaching the heart and great vessel primordium by ED9.5. Arch remodeling and aortiocopulmonary (AP) septation occur from ED10.5 - ED15.5 and ED11.5 to ED13.5, respectively. Of direct relevance to the phenotypic presentation in NS, the mapping studies also demonstrated that NCCs are a major contributor to specific portions of the craniofacial bones and cartilage, particularly those bones responsible for formation of the frontal skull, face and nose [97, 98]. Using the same construct that showed NCC contribution to the outflow tract (Fig. 3B), we confirmed these data and showed that NCCs are major contributors to the frontal bones, nasal cartilage and bones, alisphenoid and squamosal bones and interperiatal bone as well as others (Nakamura et al, unpublished data). This evidence of NCC contribution to early cardiac and craniofacial development, two systems consistently involved in NS, make NCC-specific models of particular interest to investigators looking into the pathogenesis of NS and similar syndromes.
A variety of promoters have been used to successfully drive cre recombinase in an NCC specific manner [93–95]. In the near future, these constructs can be used to create NCC-specific, SHP2 gain- and loss-of-function mutant animals to determine if the craniofacial and cardiac phenotypes of NS and LS are recapitulated, in order to test the hypothesis that altered SHP2 function in premigratory NCCs is the common mechanism responsible for the craniofacial and cardiac defects seen in these syndromes. Cre-lox technology can be used to examine the effects of the gain-of-function of SHP2 by crossing loxP Q79R mutants (a gain-of-function mutation, see 4.2) with animals in which cre-recombinase is driven by the Wnt1 promoter, which is only expressed in pre-migratory NCC. The role of SHP2 in normal development of the systems associated with NS can be examined via NCC-specific loss of SHP2 function: a line of mice in which lox P sites have been placed in the C- and N-SH2 domains of SHP2 can be crossed with Wnt1-cre animals. Offspring of these animals would exhibit NCC-specific expression of a mutant SHP2 protein lacking catalytic activity. Preliminary data (Nakamura et al, unpublished data) indicate that both a loss of function and gain-of-function via expression of an NS mutation affect skull vault and facial development. As these studies progress it will be interesting to define the pathways that are affected downstream of altered SHP2 activity and see if they are different in the different NCC descendents.
5. Future directions
Despite the inherent limitations of transgenesis and gene targeting, both methodologies will continue to play a major role in cardiovascular research in the next decade as the drive continues to identify new therapeutic targets using the mouse. The past 20 years have seen significant improvements in the precision of both gain- and loss-of-function approaches and precision will continue to improve as appropriate promoters and transcriptional patterns are more rigorously defined. Initiatives to create banks of gene targeted animals (European Conditional Mouse Mutagenesis-www.eucomm.org/info/; International Gene Trap Consortium-www.genetrap.org; NIH Knock Out Mouse Project- www.nih.gov/science/models/mouse/knockout/) will accelerate discovery by eliminating some of the tedious and expensive groundwork involved in these types of experiments.
While the optimization of current techniques of genetic manipulation continues to progress, others methods of manipulating the cell proteome are also being developed and should be applicable to the cardiovascular system. Basic studies have provided extensive data on the physiological importance of post-transcriptional gene silencing, termed RNA interference (RNAi) [99–101]. RNAi is a phenomenon in which small, double stranded RNAs (short interfering RNA; siRNA) interact with messenger RNA (mRNA) containing homologous sequences in a sequence-specific manner. This interaction results in the formation of an RNA-induced silencing complex (RISC) and activation of Argonaute-2 (Argo2), which cleaves the mRNA, which is then degraded by cellular RNAse. One can envision placing expression of an appropriate RNAi under the control of an inducible promoter, resulting in the ability to carry out an inducible, reversible gene knockdown at different developmental times or during unique physiological stresses. Recent work involving adenovirus-mediated gene silencing demonstrated proof of principle for gene silencing in the cardiovascular system [102] although formidable technical problems involving delivery, efficacy, duration of action, and, with respect to human studies, safety, still need to be addressed. Similar considerations apply to the newly characterized microRNAs as well, which clearly play major roles in cardiac development [103] and in mediating the heart’s response to different stressors [104].
Despite its drawbacks, the mouse will remain the animal model of choice for gain and loss of function studies directed at understanding human heart function, disease and failure. The challenge will be to extend reductionist approaches and the study of monogenic disease to understanding the complex phenotypes due to multi-system interactions during organ specific and system-wide pathogenesis. Our ability to precisely and reversibly manipulate single and multiple gene activity in a cell type specific fashion will underpin those studies.
References
- 1.Robbins J. Gene targeting. The precise manipulation of the mammalian genome. Circ Res. 1993;73:3–9. doi: 10.1161/01.res.73.1.3. [DOI] [PubMed] [Google Scholar]
- 2.Mortensen RM. Double knockouts. Production of mutant cell lines in cardiovascular research. Hypertension. 1993;22:646–651. doi: 10.1161/01.hyp.22.4.646. [DOI] [PubMed] [Google Scholar]
- 3.Nabel EG, Plautz G, Nabel GJ. Gene transfer into vascular cells. J Am Coll Cardiol. 1991;17:189B–194B. doi: 10.1016/0735-1097(91)90957-b. [DOI] [PubMed] [Google Scholar]
- 4.Freimuth P. Protein overexpression in mammalian cell lines. Genet Eng (N Y) 2007;28:95–104. doi: 10.1007/978-0-387-34504-8_6. [DOI] [PubMed] [Google Scholar]
- 5.Heine HL, Leong HS, Rossi FM, McManus BM, Podor TJ. Strategies of conditional gene expression in myocardium: an overview. Methods Mol Med. 2005;112:109–154. doi: 10.1007/978-1-59259-879-3_8. [DOI] [PubMed] [Google Scholar]
- 6.Albanese C, Hulit J, Sakamaki T, Pestell RG. Recent advances in inducible expression in transgenic mice. Semin Cell Dev Biol. 2002;13:129–141. doi: 10.1016/s1084-9521(02)00021-6. [DOI] [PubMed] [Google Scholar]
- 7.Brinster RL, Chen HY, Trumbauer M, Senear AW, Warren R, Palmiter RD. Somatic expression of herpes thymidine kinase in mice following injection of a fusion gene into eggs. Cell. 1981;27:223–231. doi: 10.1016/0092-8674(81)90376-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gordon K, Ruddle FH. Gene transfer into mouse embryos. Dev Biol (N Y 1985) 1986;4:1–36. doi: 10.1007/978-1-4613-2143-9_1. [DOI] [PubMed] [Google Scholar]
- 9.Palmiter RD, Brinster RL. Transgenic mice. Cell. 1985;41:343–345. doi: 10.1016/s0092-8674(85)80004-0. [DOI] [PubMed] [Google Scholar]
- 10.Supp DM, Brueckner M, Kuehn MR, Witte DP, Lowe LA, McGrath J, et al. Targeted deletion of the ATP binding domain of left-right dynein confirms its role in specifying development of left-right asymmetries. Development. 1999;126:5495–5504. doi: 10.1242/dev.126.23.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Supp DM, Witte DP, Potter SS, Brueckner M. Mutation of an axonemal dynein affects left-right asymmetry in inversus viscerum mice. Nature. 1997;389:963–966. doi: 10.1038/40140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gulick J, Hewett TE, Klevitsky R, Buck SH, Moss RL, Robbins J. Transgenic remodeling of the regulatory myosin light chains in the mammalian heart. Circ Res. 1997;80:655–664. doi: 10.1161/01.res.80.5.655. [DOI] [PubMed] [Google Scholar]
- 13.Sanbe A, Gulick J, Hanks MC, Liang Q, Osinska H, Robbins J. Reengineering inducible cardiac-specific transgenesis with an attenuated myosin heavy chain promoter. Circ Res. 2003;92:609–616. doi: 10.1161/01.RES.0000065442.64694.9F. [DOI] [PubMed] [Google Scholar]
- 14.James J, Osinska H, Hewett TE, Kimball T, Klevitsky R, Witt S, et al. Transgenic over-expression of a motor protein at high levels results in severe cardiac pathology. Transgenic Res. 1999;8:9–22. doi: 10.1023/a:1008894507995. [DOI] [PubMed] [Google Scholar]
- 15.Chien KR. To Cre or not to Cre: the next generation of mouse models of human cardiac diseases. Circ Res. 2001;88:546–549. doi: 10.1161/01.res.88.6.546. [DOI] [PubMed] [Google Scholar]
- 16.Huang WY, Aramburu J, Douglas PS, Izumo S. Transgenic expression of green fluorescence protein can cause dilated cardiomyopathy. Nat Med. 2000;6:482–483. doi: 10.1038/74914. [DOI] [PubMed] [Google Scholar]
- 17.Babinet C, Morello D, Renard JP. Transgenic mice. Genome. 1989;31:938–949. doi: 10.1139/g89-165. [DOI] [PubMed] [Google Scholar]
- 18.Field LJ. Transgenic mice in cardiovascular research. Annu Rev Physiol. 1993;55:97–114. doi: 10.1146/annurev.ph.55.030193.000525. [DOI] [PubMed] [Google Scholar]
- 19.Hunter JJ, Tanaka N, Rockman HA, Ross J, Jr, Chien KR. Ventricular expression of a MLC-2v-ras fusion gene induces cardiac hypertrophy and selective diastolic dysfunction in transgenic mice. J Biol Chem. 1995;270:23173–23178. doi: 10.1074/jbc.270.39.23173. [DOI] [PubMed] [Google Scholar]
- 20.Hunter JJ, Zhu H, Lee KJ, Kubalak S, Chien KR. Targeting gene expression to specific cardiovascular cell types in transgenic mice. Hypertension. 1993;22:608–617. doi: 10.1161/01.hyp.22.4.608. [DOI] [PubMed] [Google Scholar]
- 21.Knotts JJ, Rindt H, Neumann J, Robbins J. In vivo regulation of the mouse beta myosin heavy chain gene. J Biol Chem. 1994;269:31275–31282. [PubMed] [Google Scholar]
- 22.Robbins J, Palermo J, Rindt H. In vivo definition of a cardiac specific promoter and its potential utility in remodeling the heart. Ann N Y Acad Sci. 1995;752:492–505. doi: 10.1111/j.1749-6632.1995.tb17458.x. [DOI] [PubMed] [Google Scholar]
- 23.Subramaniam A, Jones WK, Gulick J, Wert S, Neumann J, Robbins J. Tissue-specific regulation of the alpha-myosin heavy chain gene promoter in transgenic mice. J Biol Chem. 1991;266:24613–24620. [PubMed] [Google Scholar]
- 24.Robbins J. Remodeling the cardiac sarcomere using transgenesis. Annu Rev Physiol. 2000;62:261–287. doi: 10.1146/annurev.physiol.62.1.261. [DOI] [PubMed] [Google Scholar]
- 25.Buttrick PM, Kaplan ML, Kitsis RN, Leinwand LA. Distinct behavior of cardiac myosin heavy chain gene constructs in vivo. Discordance with in vitro results. Circ Res. 1993;72:1211–1217. doi: 10.1161/01.res.72.6.1211. [DOI] [PubMed] [Google Scholar]
- 26.Buttrick PM, Kass A, Kitsis RN, Kaplan ML, Leinwand LA. Behavior of genes directly injected into the rat heart in vivo. Circ Res. 1992;70:193–198. doi: 10.1161/01.res.70.1.193. [DOI] [PubMed] [Google Scholar]
- 27.Knotts S, Rindt H, Robbins J. Position independent expression and developmental regulation is directed by the beta myosin heavy chain gene's 5' upstream region in transgenic mice. Nucleic Acids Res. 1995;23:3301–3309. doi: 10.1093/nar/23.16.3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soonpaa MH, Field LJ. Survey of studies examining mammalian cardiomyocyte DNA synthesis. Circ Res. 1998;83:15–26. doi: 10.1161/01.res.83.1.15. [DOI] [PubMed] [Google Scholar]
- 29.Chen J, Kubalak SW, Chien KR. Ventricular muscle-restricted targeting of the RXRalpha gene reveals a non-cell-autonomous requirement in cardiac chamber morphogenesis. Development. 1998;125:1943–1949. doi: 10.1242/dev.125.10.1943. [DOI] [PubMed] [Google Scholar]
- 30.Mochly-Rosen D, Wu G, Hahn H, Osinska H, Liron T, Lorenz JN, et al. Cardiotrophic effects of protein kinase C epsilon: analysis by in vivo modulation of PKCepsilon translocation. Circ Res. 2000;86:1173–1179. doi: 10.1161/01.res.86.11.1173. [DOI] [PubMed] [Google Scholar]
- 31.De Windt LJ, Lim HW, Bueno OF, Liang Q, Delling U, Braz JC, et al. Targeted inhibition of calcineurin attenuates cardiac hypertrophy in vivo. Proc Natl Acad Sci U S A. 2001;98:3322–3327. doi: 10.1073/pnas.031371998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Syed F, Odley A, Hahn HS, Brunskill EW, Lynch RA, Marreez Y, et al. Physiological growth synergizes with pathological genes in experimental cardiomyopathy. Circ Res. 2004;95:1200–1206. doi: 10.1161/01.RES.0000150366.08972.7f. [DOI] [PubMed] [Google Scholar]
- 33.Dorn GW, 2nd, Robbins J, Ball N, Walsh RA. Myosin heavy chain regulation and myocyte contractile depression after LV hypertrophy in aortic-banded mice. Am J Physiol. 1994;267:H400–H405. doi: 10.1152/ajpheart.1994.267.1.H400. [DOI] [PubMed] [Google Scholar]
- 34.Izumo S, Nadal-Ginard B, Mahdavi V. All members of the MHC multigene family respond to thyroid hormone in a highly tissue-specific manner. Science. 1986;231:597–600. doi: 10.1126/science.3945800. [DOI] [PubMed] [Google Scholar]
- 35.Bujard H. Controlling genes with tetracyclines. J Gene Med. 1999;1:372–374. doi: 10.1002/(SICI)1521-2254(199909/10)1:5<372::AID-JGM61>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 36.Gossen M, Bonin AL, Freundlieb S, Bujard H. Inducible gene expression systems for higher eukaryotic cells. Curr Opin Biotechnol. 1994;5:516–520. doi: 10.1016/0958-1669(94)90067-1. [DOI] [PubMed] [Google Scholar]
- 37.Mansuy IM, Winder DG, Moallem TM, Osman M, Mayford M, Hawkins RD, et al. Inducible and reversible gene expression with the rtTA system for the study of memory. Neuron. 1998;21:257–265. doi: 10.1016/s0896-6273(00)80533-4. [DOI] [PubMed] [Google Scholar]
- 38.Freundlieb S, Baron U, Bonin AL, Gossen M, Bujard H. Use of tetracycline-controlled gene expression systems to study mammalian cell cycle. Methods Enzymol. 1997;283:159–173. doi: 10.1016/s0076-6879(97)83014-5. [DOI] [PubMed] [Google Scholar]
- 39.Freundlieb S, Schirra-Muller C, Bujard HA. A tetracycline controlled activation/repression system with increased potential for gene transfer into mammalian cells. J Gene Med. 1999;1:4–12. doi: 10.1002/(SICI)1521-2254(199901/02)1:1<4::AID-JGM4>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 40.Russell LK, Mansfield CM, Lehman JJ, Kovacs A, Courtois M, Saffitz JE, et al. Cardiac-specific induction of the transcriptional coactivator peroxisome proliferator-activated receptor gamma coactivator-1alpha promotes mitochondrial biogenesis and reversible cardiomyopathy in a developmental stage-dependent manner. Circ Res. 2004;94:525–533. doi: 10.1161/01.RES.0000117088.36577.EB. [DOI] [PubMed] [Google Scholar]
- 41.Capecchi MR. Altering the genome by homologous recombination. Science. 1989;244:1288–1292. doi: 10.1126/science.2660260. [DOI] [PubMed] [Google Scholar]
- 42.Bronson SK, Plaehn EG, Kluckman KD, Hagaman JR, Maeda N, Smithies O. Single-copy transgenic mice with chosen-site integration. Proc Natl Acad Sci U S A. (93) 1996;93:9067–9072. doi: 10.1073/pnas.93.17.9067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Askew GR, Doetschman T, Lingrel JB. Site-directed point mutations in embryonic stem cells: a gene-targeting tag-and-exchange strategy. Mol Cell Biol. 1993;13:4115–4124. doi: 10.1128/mcb.13.7.4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hasty P, Ramirez-Solis R, Krumlauf R, Bradley A. Introduction of a subtle mutation into the Hox-2.6 locus in embryonic stem cells. Nature. 1991;350:243–246. doi: 10.1038/350243a0. [DOI] [PubMed] [Google Scholar]
- 45.Lewis J, Yang B, Detloff P, Smithies O. Gene modification via "plug and socket" gene targeting. J Clin Invest. 1996;97:3–5. doi: 10.1172/JCI118403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Valancius V, Smithies O. Testing an "in-out" targeting procedure for making subtle genomic modifications in mouse embryonic stem cells. Mol Cell Biol. 1991;11:1402–1408. doi: 10.1128/mcb.11.3.1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marth JD. Recent advances in gene mutagenesis by site-directed recombination. J Clin Invest. 1996;97:1999–2002. doi: 10.1172/JCI118634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rajewsky K, Gu H, Kuhn R, Betz UA, Muller W, Roes J, et al. Conditional gene targeting. J Clin Invest. 1996;98:600–603. doi: 10.1172/JCI118828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoshioka J, Imahashi K, Gabel SA, Chutkow WA, Burds AA, Gannon J, et al. Targeted deletion of thioredoxin-interacting protein regulates cardiac dysfunction in response to pressure overload. Circ Res. 2007;101:1328–1338. doi: 10.1161/CIRCRESAHA.106.160515. [DOI] [PubMed] [Google Scholar]
- 50.Holtwick R, van Eickels M, Skryabin BV, Baba HA, Bubikat A, Begrow F, et al. Pressure-independent cardiac hypertrophy in mice with cardiomyocyte-restricted inactivation of the atrial natriuretic peptide receptor guanylyl cyclase-A. J Clin Invest. 2003;111:1399–1407. doi: 10.1172/JCI17061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Agah R, Frenkel PA, French BA, Michael LH, Overbeek PA, Schneider MD. Gene recombination in postmitotic cells. Targeted expression of Cre recombinase provokes cardiac-restricted, site-specific rearrangement in adult ventricular muscle in vivo. J Clin Invest. 1997;100:169–179. doi: 10.1172/JCI119509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sohal DS, Nghiem M, Crackower MA, Witt SA, Kimball TR, Tymitz KM, et al. Temporally regulated and tissue-specific gene manipulations in the adult and embryonic heart using a tamoxifen-inducible Cre protein. Circ Res. 2001;89:20–25. doi: 10.1161/hh1301.092687. [DOI] [PubMed] [Google Scholar]
- 53.Williams RW, Flaherty L, Threadgill DW. The math of making mutant mice. Genes Brain Behav. 2003;2:191–200. doi: 10.1034/j.1601-183x.2003.00017.x. [DOI] [PubMed] [Google Scholar]
- 54.Braun T, Arnold HH. Inactivation of Myf-6 and Myf-5 genes in mice leads to alterations in skeletal muscle development. Embo J. 1995;14:1176–1186. doi: 10.1002/j.1460-2075.1995.tb07101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Patapoutian A, Yoon JK, Miner JH, Wang S, Stark K, Wold B. Disruption of the mouse MRF4 gene identifies multiple waves of myogenesis in the myotome. Development. 1995;121:3347–3358. doi: 10.1242/dev.121.10.3347. [DOI] [PubMed] [Google Scholar]
- 56.Zhang W, Behringer RR, Olson EN. Inactivation of the myogenic bHLH gene MRF4 results in up-regulation of myogenin and rib anomalies. Genes Dev. 1995;9:1388–1399. doi: 10.1101/gad.9.11.1388. [DOI] [PubMed] [Google Scholar]
- 57.Noonan JA. Hypertelorism with Turner phenotype. A new syndrome with associated congenital heart disease. Am J Dis Child. 1968;116:373–380. doi: 10.1001/archpedi.1968.02100020377005. [DOI] [PubMed] [Google Scholar]
- 58.Mendez HM, Opitz JM. Noonan syndrome: a review. Am J Med Genet. 1985;21:493–506. doi: 10.1002/ajmg.1320210312. [DOI] [PubMed] [Google Scholar]
- 59.Shah N, Rodriguez M, Louis DS, Lindley K, Milla PJ. Feeding difficulties and foregut dysmotility in Noonan's syndrome. Arch Dis Child. 1999;81:28–31. doi: 10.1136/adc.81.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chan RJ, Feng GS. PTPN11 is the first identified proto-oncogene that encodes a tyrosine phosphatase. Blood. 2007;109:862–867. doi: 10.1182/blood-2006-07-028829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Neel BG, Gu H, Pao L. The 'Shp'ing news: SH2 domain-containing tyrosine phosphatases in cell signaling. Trends Biochem Sci. 2003;28:284–293. doi: 10.1016/S0968-0004(03)00091-4. [DOI] [PubMed] [Google Scholar]
- 62.Keilhack H, David FS, McGregor M, Cantley LC, Neel BG. Diverse biochemical properties of Shp2 mutants. Implications for disease phenotypes. J Biol Chem. 2005;280:30984–30993. doi: 10.1074/jbc.M504699200. [DOI] [PubMed] [Google Scholar]
- 63.Bard-Chapeau EA, Yuan J, Droin N, Long S, Zhang EE, Nguyen TV, et al. Concerted functions of Gab1 and Shp2 in liver regeneration and hepatoprotection. Mol Cell Biol. 2006;26:4664–4674. doi: 10.1128/MCB.02253-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang EE, Chapeau E, Hagihara K, Feng GS. Neuronal Shp2 tyrosine phosphatase controls energy balance and metabolism. Proc Natl Acad Sci U S A. 2004;101:16064–16069. doi: 10.1073/pnas.0405041101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ke Y, Lesperance J, Zhang EE, Bard-Chapeau EA, Oshima RG, Muller WJ, et al. Conditional deletion of Shp2 in the mammary gland leads to impaired lobulo-alveolar outgrowth and attenuated Stat5 activation. J Biol Chem. 2006;281:34374–34380. doi: 10.1074/jbc.M607325200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mohi MG, Neel BG. The role of Shp2 (PTPN11) in cancer. Curr Opin Genet Dev. 2007;17:23–30. doi: 10.1016/j.gde.2006.12.011. [DOI] [PubMed] [Google Scholar]
- 67.Saxton TM, Ciruna BG, Holmyard D, Kulkarni S, Harpal K, Rossant J, et al. The SH2 tyrosine phosphatase shp2 is required for mammalian limb development. Nat Genet. 2000;24:420–423. doi: 10.1038/74279. [DOI] [PubMed] [Google Scholar]
- 68.Edouard T, Montagner A, Dance M, Conte F, Yart A, Parfait B, et al. How do Shp2 mutations that oppositely influence its biochemical activity result in syndromes with overlapping symptoms? Cell Mol Life Sci. 2007;64:1585–1590. doi: 10.1007/s00018-007-6509-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tartaglia M, Pennacchio LA, Zhao C, Yadav KK, Fodale V, Sarkozy A, et al. Gain-of-function SOS1 mutations cause a distinctive form of Noonan syndrome. Nat Genet. 2007;39:75–79. doi: 10.1038/ng1939. [DOI] [PubMed] [Google Scholar]
- 70.Schubbert S, Zenker M, Rowe SL, Boll S, Klein C, Bollag G, et al. Germline KRAS mutations cause Noonan syndrome. Nat Genet. 2006;38:331–336. doi: 10.1038/ng1748. [DOI] [PubMed] [Google Scholar]
- 71.Roberts AE, Araki T, Swanson KD, Montgomery KT, Schiripo TA, Joshi VA, et al. Germline gain-of-function mutations in SOS1 cause Noonan syndrome. Nat Genet. 2007;39:70–74. doi: 10.1038/ng1926. [DOI] [PubMed] [Google Scholar]
- 72.Tartaglia M, Gelb BD. Noonan syndrome and related disorders: genetics and pathogenesis. Annu Rev Genomics Hum Genet. 2005;6:45–68. doi: 10.1146/annurev.genom.6.080604.162305. [DOI] [PubMed] [Google Scholar]
- 73.Hanna N, Montagner A, Lee WH, Miteva M, Vidal M, Vidaud M, et al. Reduced phosphatase activity of SHP-2 in LEOPARD syndrome: consequences for PI3K binding on Gab1. FEBS Lett. 2006;580:2477–2482. doi: 10.1016/j.febslet.2006.03.088. [DOI] [PubMed] [Google Scholar]
- 74.Kontaridis MI, Swanson KD, David FS, Barford D, Neel BG. PTPN11 (Shp2) mutations in LEOPARD syndrome have dominant negative, not activating, effects. J Biol Chem. 2006;281:6785–6792. doi: 10.1074/jbc.M513068200. [DOI] [PubMed] [Google Scholar]
- 75.Tartaglia M, Martinelli S, Stella L, Bocchinfuso G, Flex E, Cordeddu V, et al. Diversity and functional consequences of germline and somatic PTPN11 mutations in human disease. Am J Hum Genet. 2006;78:279–290. doi: 10.1086/499925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ohtani T, Ishihara K, Atsumi T, Nishida K, Kaneko Y, Miyata T, et al. Dissection of signaling cascades through gp130 in vivo: reciprocal roles for STAT3- and SHP2-mediated signals in immune responses. Immunity. 2000;12:95–105. doi: 10.1016/s1074-7613(00)80162-4. [DOI] [PubMed] [Google Scholar]
- 77.Hagiwara Y, Miyoshi S, Fukuda K, Nishiyama N, Ikegami Y, Tanimoto K, et al. SHP2-mediated signaling cascade through gp130 is essential for LIF-dependent I CaL, [Ca2+]i transient, and APD increase in cardiomyocytes. J Mol Cell Cardiol. 2007;43:710–716. doi: 10.1016/j.yjmcc.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 78.Araki T, Mohi MG, Ismat FA, Bronson RT, Williams IR, Kutok JL, et al. Mouse model of Noonan syndrome reveals cell type- and gene dosage-dependent effects of Ptpn11 mutation. Nat Med. 2004;10:849–857. doi: 10.1038/nm1084. [DOI] [PubMed] [Google Scholar]
- 79.Nakamura T, Colbert M, Krenz M, Molkentin JD, Hahn HS, Dorn GW, 2nd, et al. Mediating ERK 1/2 signaling rescues congenital heart defects in a mouse model of Noonan syndrome. J Clin Invest. 2007;117:2123–2132. doi: 10.1172/JCI30756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Krenz M, Yutzey KE, Robbins J. Noonan syndrome mutation Q79R in Shp2 increases proliferation of valve primordia mesenchymal cells via extracellular signal-regulated kinase 1/2 signaling. Circ Res. 2005;97:813–820. doi: 10.1161/01.RES.0000186194.06514.b0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Corson LB, Yamanaka Y, Lai KM, Rossant J. Spatial and temporal patterns of ERK signaling during mouse embryogenesis. Development. 2003;130:4527–4537. doi: 10.1242/dev.00669. [DOI] [PubMed] [Google Scholar]
- 82.Saba-El-Leil MK, Vella FD, Vernay B, Voisin L, Chen L, Labrecque N, et al. An essential function of the mitogen-activated protein kinase Erk2 in mouse trophoblast development. EMBO Rep. 2003;4:964–968. doi: 10.1038/sj.embor.embor939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pages G, Guerin S, Grall D, Bonino F, Smith A, Anjuere F, et al. Defective thymocyte maturation in p44 MAP kinase (Erk 1) knockout mice. Science. 1999;286:1374–1377. doi: 10.1126/science.286.5443.1374. [DOI] [PubMed] [Google Scholar]
- 84.Morales AV, Barbas JA, Nieto MA. How to become neural crest: from segregation to delamination. Semin Cell Dev Biol. 2005;16:655–662. doi: 10.1016/j.semcdb.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 85.Crane JF, Trainor PA. Neural crest stem and progenitor cells. Annu Rev Cell Dev Biol. 2006;22:267–286. doi: 10.1146/annurev.cellbio.22.010305.103814. [DOI] [PubMed] [Google Scholar]
- 86.Stoller JZ, Epstein JA. Cardiac neural crest. Semin Cell Dev Biol. 2005;16:704–715. doi: 10.1016/j.semcdb.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 87.Maschhoff KL, Baldwin HS. Molecular determinants of neural crest migration. Am J Med Genet. 2000;97:280–288. doi: 10.1002/1096-8628(200024)97:4<280::aid-ajmg1278>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 88.Li J, Molkentin JD, Colbert MC. Retinoic acid inhibits cardiac neural crest migration by blocking c-Jun N-terminal kinase activation. Dev Biol. 2001;232:351–361. doi: 10.1006/dbio.2001.0203. [DOI] [PubMed] [Google Scholar]
- 89.Snider P, Olaopa M, Firulli AB, Conway SJ. Cardiovascular development and the colonizing cardiac neural crest lineage. ScientificWorldJournal. 2007;7:1090–1113. doi: 10.1100/tsw.2007.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hutson MR, Kirby ML. Model systems for the study of heart development and disease. Cardiac neural crest and conotruncal malformations. Semin Cell Dev Biol. 2007;18:101–110. doi: 10.1016/j.semcdb.2006.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hutson MR, Kirby ML. Neural crest and cardiovascular development: a 20-year perspective. Birth Defects Res C Embryo Today. 2003;69:2–13. doi: 10.1002/bdrc.10002. [DOI] [PubMed] [Google Scholar]
- 92.Brown CB, Baldwin HS. Neural crest contribution to the cardiovascular system. Adv Exp Med Biol. 2006;589:134–154. doi: 10.1007/978-0-387-46954-6_8. [DOI] [PubMed] [Google Scholar]
- 93.Lee M, Brennan A, Blanchard A, Zoidl G, Dong Z, Tabernero A, et al. P0 is constitutively expressed in the rat neural crest and embryonic nerves and is negatively and positively regulated by axons to generate non-myelin-forming and myelin-forming Schwann cells, respectively. Mol Cell Neurosci. 1997;8:336–350. doi: 10.1006/mcne.1996.0589. [DOI] [PubMed] [Google Scholar]
- 94.Jiang X, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cardiac neural crest. Development. 2000;127:1607–1616. doi: 10.1242/dev.127.8.1607. [DOI] [PubMed] [Google Scholar]
- 95.Brown CB, Feiner L, Lu MM, Li J, Ma X, Webber AL, et al. PlexinA2 and semaphorin signaling during cardiac neural crest development. Development. 2001;128:3071–3080. doi: 10.1242/dev.128.16.3071. [DOI] [PubMed] [Google Scholar]
- 96.Nakamura T, Colbert MC, Robbins J. Neural crest cells retain multipotential characteristics in the developing valves and label the cardiac conduction system. Circ Res. 2006;98:1547–1554. doi: 10.1161/01.RES.0000227505.19472.69. [DOI] [PubMed] [Google Scholar]
- 97.Jiang X, Iseki S, Maxson RE, Sucov HM, Morriss-Kay GM. Tissue origins and interactions in the mammalian skull vault. Dev Biol. 2002;241:106–116. doi: 10.1006/dbio.2001.0487. [DOI] [PubMed] [Google Scholar]
- 98.Chai Y, Jiang X, Ito Y, Bringas P, Jr, Han J, Rowitch DH, et al. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development. 2000;127:1671–1679. doi: 10.1242/dev.127.8.1671. [DOI] [PubMed] [Google Scholar]
- 99.Mello CC, Conte D., Jr Revealing the world of RNA interference. Nature. 2004;431:338–342. doi: 10.1038/nature02872. [DOI] [PubMed] [Google Scholar]
- 100.Luther HP. Role of endogenous antisense RNA in cardiac gene regulation. J Mol Med. 2005;83:26–32. doi: 10.1007/s00109-004-0613-5. [DOI] [PubMed] [Google Scholar]
- 101.Fire AZ. Gene silencing by double-stranded RNA. Cell Death Differ. 2007;14:1998–2012. doi: 10.1038/sj.cdd.4402253. [DOI] [PubMed] [Google Scholar]
- 102.Kasahara H, Aoki H. Gene silencing using adenoviral RNAi vector in vascular smooth muscle cells and cardiomyocytes. Methods Mol Med. 2005;112:155–172. doi: 10.1007/978-1-59259-879-3_9. [DOI] [PubMed] [Google Scholar]
- 103.Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, et al. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 104.van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci U S A. 2006;103:18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]