Abstract
Glossina fuscipes fuscipes Newstead 1910 (Diptera: Glossinidae) is the primary vector of human sleeping sickness in Kenya and Uganda. This is the first report on its population structure. A total of 688 nucleotides of mitochondrial ribosomal 16S2 and cytochrome oxidase I genes were sequenced. Twenty-one variants were scored in 79 flies from three geographically diverse natural populations. Four haplotypes were shared among populations, eight were private and nine were singletons. The mean haplotype and nucleotide diversities were 0.84 and 0.009, respectively. All populations were genetically differentiated and were at demographic equilibrium. In addition, a longstanding laboratory culture originating from the Central African Republic (CAR-lab) in 1986 (or before) was examined. Haplotype and nucleotide diversities in this culture were 0.95 and 0.012, respectively. None of its 27 haplotypes were shared with the East African populations. A first approximation of relative effective population sizes was Uganda > CAR-lab > Kenya. It was concluded that the structure of G. f. fuscipes populations in East Africa is localized.
Keywords: Glossina, breeding structure, genetic differentiation, tsetse
Introduction
The tsetse fly Glossina fuscipes fuscipes is classified with the palpalis group, the subgenus Nemorhina Robineau-Desvoidy (Gooding & Krafsur, 2005). Flies of the palpalis group generally occupy riverine and lacustrine habitats and are opportunistic feeders (Leak, 1998). Glossina f. fuscipes may be found within a large area of central Africa, from Cameroon, Gabon, Congo, Zaire and the Central African Republic eastwards to Uganda and western Kenya. Highly localized patches exist on the margins of Lake Victoria in Tanzania and small, isolated patches of G. f. fuscipes are also found in southwestern Ethiopia and southern Sudan (Rogers & Robinson, 2004). Glossina f. fuscipes are vectors of the trypanosomes (Kinetoplastida: Trypanosomatidae; Trypanosoma brucei gambiense and Trypanosoma bruce rhodesiense) that cause human African trypanosomiasis (HAT) or sleeping sickness, particularly in Uganda and western Kenya (Waiswa et al., 2006). Uganda is an active focus of HAT (Welburn et al., 2006), although G. f. fuscipes is principally zoophagic (Wamwiri et al., 2007). Trypanosome reservoirs there include cattle and other domestic livestock, as well as numerous wild mammals (Waiswa et al., 2003). Here, we examine patterns of mitochondrial diversity in G. f. fuscipes, with the objectives of estimating its population structure and the degree of population subdivision in East Africa. We also compare the East African population samples with the principal laboratory strain from which any others are derived. Because mitochondrial variation is single copy and inherited matrilineally, it shows divergence among lineages about four times as rapidly as autosomal loci such as microsatellites.
Materials and methods
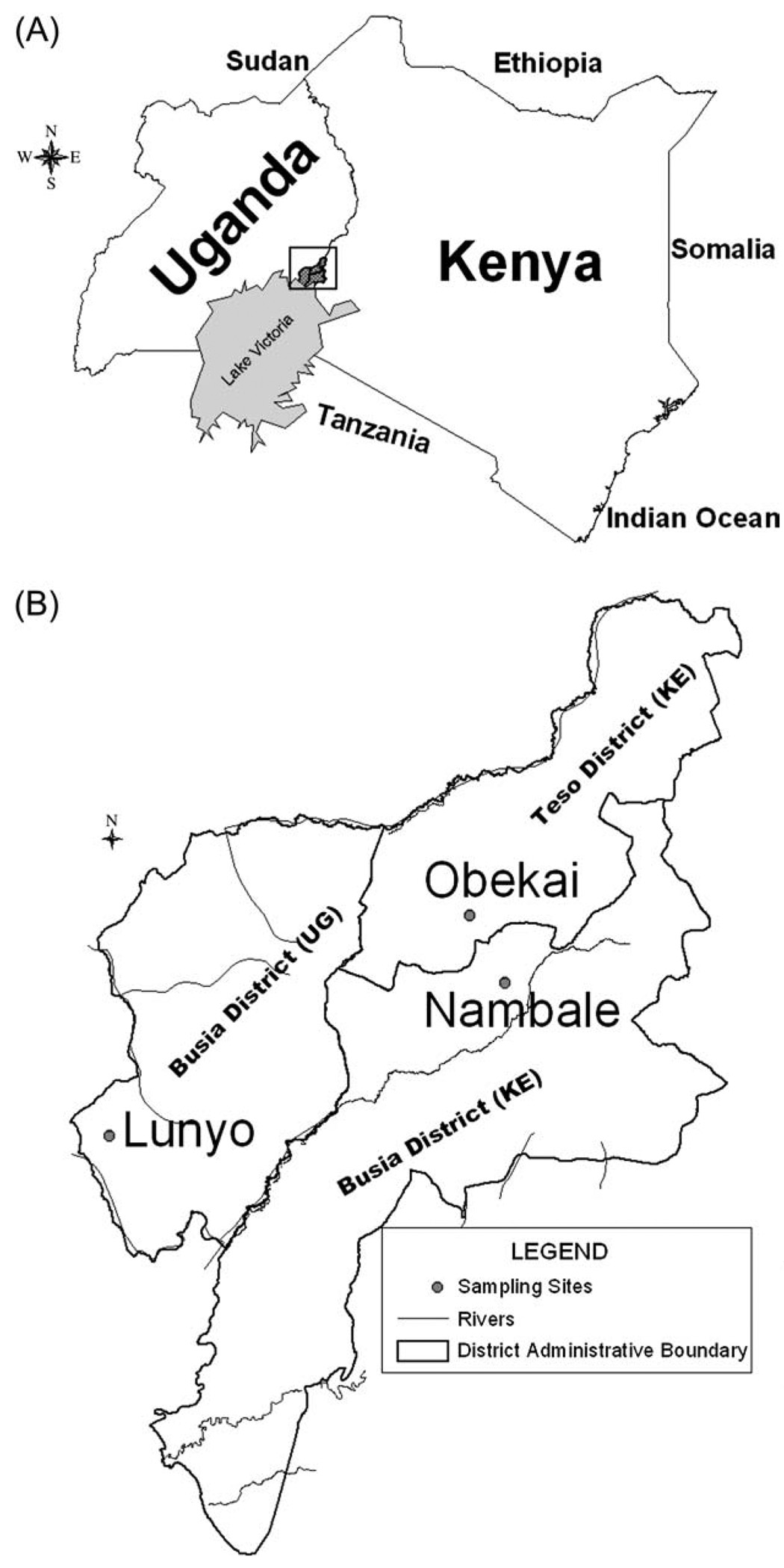
Tsetse flies were sampled in 2003 in Nambale, Busia District (34.24 °E, 0.46 °S) and, Chakol Obekai, in Teso District (34.21 °E, 0.51 °S), both in western Kenya, and in 2000 in Lunyo in eastern Uganda (33.91 °E, 0.33 °S), about 55 km from the Kenyan sampling sites (Fig. 1). The Kenyan districts are adjacent and the sampling sites c. 32 km apart. Since 1986, the joint laboratory of the Food and Agriculture Organization/International Atomic Energy Agency (FAO/IAEA) has maintained a culture of G. f. fuscipes that originated from the Central African Republic (CAR-lab). The conditions and year of its original establishment are unknown. Its numbers have varied from as few as 375 to as many as 11 000 females. Population size was approximately 11 000 females at the time of genetic sampling. All extant cultures of G. f. fuscipes are derived from the FAO/IAEA stock (A. S. Robinson and A. G. Parker, FAO/IAEA Laboratories, year?, personal communication to E.S.K.).
Fig. 1.
(A) Sampling region north of Lake Victoria. (B) Detail of sampling sites near the Uganda-Kenya border.
DNA was extracted using the Qiagen DNeasy Extraction Kit (Qiagen, Xxx, XX, Xxx). Subsequently, 300 bp of the ribosomal 16S (r16S2) and 440 bp of the cytochrome oxidase I (COI) mitochondrial genes were amplified in 50 µL consisting of 48 µL of 1.1x ReddyMix PCR Master Mix (ABGene, New York, NY, U.S.A.), 2 µL of DNA extract (approximately 80 ng), and 0.5 µm each of oligonucleotide primers N1-J-12585 (5′GGT CCC TTA CGA ATT TGA ATA TAT CCT3′) and LR-N-12866 (5′ACA TGA TCT GAG TTC AAA CCG G3′) for r16S2, and C1-J-1751 (5′GGA TCA CCT GAT ATA GCA TTC CC3′) and C1-N-2191 (5′CCC GGT AAA ATT AAA ATA TAA ACT TC3′) for COI (Simon et al., 1994). Thermocycling was performed in a PTC-100 (MJ Research, Woburn, MA, U.S.A.) thermocycler and consisted of 30 cycles at 93°C for 45 s, 52°C for 1 min and 72°C for 1 min. DNA sequencing was performed at the Iowa State University DNA Sequencing and Synthesis Facility using an ABI Model 3730 automated sequence analyser. BioEdit (Hall, 1999) was used to edit the nucleotide sequences. Sequence alignment was accomplished using ClustalX (Thompson et al., 1997).
Genetic statistics
Haplotype and nucleotide diversities were calculated according to Nei (1986) and Nei & Kumar (2000). Haplotype diversity refers to the probability that two randomly chosen flies will have different haplotypes. Nucleotide diversity πS is the average proportion of nucleotide differences between all pairs of sequences and is estimated over all possible pairwise comparisons. Genetic differentiation of populations was estimated in several ways. Student’s t-tests were used to test hypotheses that haplotype and nucleotide diversities were homogeneous. Raymond & Rousset’s (1995) exact test was also used; it is analogous to Fisher’s exact test. Departures from homogenous dispersion of haplotypes were estimated by FST = 1−(Hw/Hb), where Hw and Hb are the average pairwise nucleotide differences within and between samples, respectively (Hudson et al., 1992). As used here, FST measures the probability that two randomly chosen sequences from any two samples differ by one or more nucleotide substitutions. Analysis of molecular variance (amova) on haplotype frequencies was carried out using arlequin. amova also yields estimates of F. FST can be used to estimate gene flow in terms of the mean number of reproducing females per generation according to Wright’s island model.
Theta (θ) is the product of the effective population size 2Ne and the mutation rate µ. It was estimated from the average number of pairwise nucleotide differences among sequences (Tajima, 1989). The main underlying assumption is that of mutation-drift equilibrium. Its biological meaning here is a scaling factor relating the relative effective sizes Ne of the sampled populations. Effective size is the harmonic mean reproductive population size averaged over many generations.
Pairwise nucleotide differences (mismatch distributions) and frequencies were also used as the basis for tests of selective neutrality and demographic equilibrium. Tests used FS (Fu, 1997) and R2 (Ramos-Onsins & Rozas, 2002) statistics. If neutrality can be assumed and there is no genetic ‘hitchhiking’, these tests are the most powerful available to detect historical demographic expansions (Rogers & Harpending, 1992; Ramos-Onsins & Rozas, 2002). It should be noted, however, that the power to reject a null hypothesis when it is false varies with the interval between a population expansion and sampling and the magnitude of the expansion. The genetic statistics were calculated by using DnaSP Version 4.10.9 (Rozas et al., 2003) and arlequin Version 3.11 (Excoffier et al., 2005).
Results
The concatenated fragments of r16S2 and COI genes numbered 668 nucleotides, of which 59 were variable (8.8%) and 54 were shared between two or three populations. There were no alignment gaps or missing data. The haplotype distributions for each sample are set out in Table 1. A total of 48 mitochondrial haplotypes were recorded, 29 of which were singletons (60.4%). It can be seen at a glance that the East African G. f. fuscipes populations shared no haplotype with the laboratory culture (CAR-lab).
Table 1.
Glossina fuscipes fuscipes mitochondrial haplotype frequency distribution.
| Kenya |
Uganda |
Central African Republic |
|||
|---|---|---|---|---|---|
| Haplotype | Nambale | Chakol Obekai |
Lunyo | Laboratory | Totals |
| 1 | 1 | 1 | |||
| 2 | 13 | 5 | 18 | ||
| 3 | 1 | 1 | |||
| 4 | 1 | 1 | |||
| 5 | 4 | 4 | |||
| 6 | 1 | 1 | |||
| 7 | 1 | 1 | |||
| 8 | 1 | 1 | |||
| 9 | 1 | 1 | |||
| 10 | 2 | 2 | |||
| 11 | 1 | 1 | |||
| 12 | 2 | 2 | |||
| 13 | 21 | 3 | 24 | ||
| 14 | 3 | 3 | |||
| 15 | 1 | 1 | |||
| 16 | 2 | 2 | |||
| 17 | 1 | 2 | 3 | 6 | |
| 18 | 2 | 2 | |||
| 19 | 1 | 1 | |||
| 20 | 1 | 1 | |||
| 21 | 1 | 1 | |||
| 22 | 2 | 2 | |||
| 23 | 2 | 2 | |||
| 24 | 2 | 2 | |||
| 25 | 5 | 1 | 5 | ||
| 26 | 2 | 2 | |||
| 27 | 2 | 2 | |||
| 28 | 2 | 2 | |||
| 29 | 1 | 1 | |||
| 30 | 1 | 1 | |||
| 31 | 1 | 1 | |||
| 32 | 1 | 1 | |||
| 33 | 1 | 1 | |||
| 34 | 3 | 3 | |||
| 35 | 1 | 1 | |||
| 36 | 1 | 1 | |||
| 37 | 1 | 1 | |||
| 38 | 1 | 1 | |||
| 39 | 1 | 1 | |||
| 40 | 1 | 1 | |||
| 41 | 6 | 6 | |||
| 42 | 1 | 1 | |||
| 43 | 1 | 1 | |||
| 44 | 1 | 1 | |||
| 45 | 8 | 8 | |||
| 46 | 1 | 1 | |||
| 47 | 1 | 1 | |||
| 48 | 1 | 1 | |||
| n | 47 | 16 | 16 | 48 | 127 |
| Haplotypes, n | 9 | 7 | 10 | 27 | 48 |
| Variable Sites, n | 17 | 17 | 53 | 22 | – |
Natural populations
A total of 21 haplotypes were recorded in the field samples, of which four (19%) were shared, eight (38%) occurred in only one population and nine (43%) were singletons. Only one Kenyan haplotype was shared with the Ugandan G. f. fuscipes.
Twelve haplotypes were recorded in Nambale and Chakol Obekai, four of which were shared between them (Table 2). Ten haplotypes were recorded in the Ugandan samples, five (50%) of which were singletons and only one (10%) of which was also found in Nambale and Chakol Obekai. Average pairwise differences in mitochondrial gene sequence variation (Table 3) between the Kenyan samples did not differ significantly (P ~ 0.72, Student’s t-test). The Kenyan data were then pooled for analysis. Exact tests of differentiation based on haplotype frequencies, however, indicted that even the two Kenyan populations differed from each other significantly (P = 0.02 ± 0.002). By the same two tests, the Kenyan flies differed greatly from the Ugandan population sample (P < < 0.001). The pairwise estimate of FST (Table 2) between Kenyan samples did not differ from zero, in keeping with a null hypothesis of much gene flow between them. FST between Kenyan and Ugandan samples was 0.28, equivalent to the exchange of 1.3 reproducing females per generation, according to the simplifying assumptions of the island model of population structure. Haplotype diversities (Hs) varied from 0.72 in Nambale, Kenya, to 0.94 in Lunyo, Uganda (Table 3). Here, Hs is the probability that two randomly chosen flies from a sample will have different haplotypes; these probabilities differed significantly among populations. Nucleotide diversities per nucleotide πs were five times greater in eastern Uganda than in western Kenya. Mean pairwise nucleotide differences between populations θ were also very much greater in Uganda than in Kenya. The foregoing data suggest that the Ugandan population was very much larger than the Kenyan populations. Mean haplotype and nucleotide diversities over the three field samples were Hs = 0.84 (± 0.05) and πs = 0.009 (± 0.001), respectively.
Table 2.
Number of haplotypes shared (upper diagonal) and genetic differentiation (FST) among samples (lowest diagonal).
| Chakol | Nambale | Uganda | CAR-lab | |
|---|---|---|---|---|
| Chakol | 4 | 1 | 0 | |
| Nambale | −0.026 | 1 | 0 | |
| Uganda | 0.28 | 0.28 | 0 | |
| CAR-lab | 0.58 | 0.46 | 0.58 |
CAR-lab, Central African Republic laboratory strain.
Table 3.
Indices of molecular variation in Glossina fuscipes fuscipes including haplotype (HS) and nucleotide (πS) diversities. Kenyan samples were pooled for tests for selective neutrality (R2) and demographic equilibrium (Fs).
| Sample | n | Mean pairwise differences (θ) | R2* | FS† | HS‡ | πS§ |
|---|---|---|---|---|---|---|
| Chakol Obekai | 16 | 3.18 | 0.87 ± 0.06 | 0.005 ± 0.001 | ||
| Nambale | 47 | 3.22 | 0.094 | − 0.891 | 0.72 ± 0.05 | 0.005 ± 0.0006 |
| Uganda | 16 | 17.80 | 0.160 | 2.160 | 0.94 ± 0.04 | 0.027 ± 0.002 |
| CAR-lab | 48 | 8.36 | 0.189* | −7.611¶ | 0.95 ± 0.02 | 0.012 ± 0.0004 |
Test according to Ramos-Onsins & Rozas (2002).
Test according to Fu (1997).
Between field-collected samples: Student’s t = 13.12, P < 0.006.
Between field-collected samples: t = 1.99, P = 0.19.
P < 0.001.
CAR-lab, Central African Republic laboratory strain.
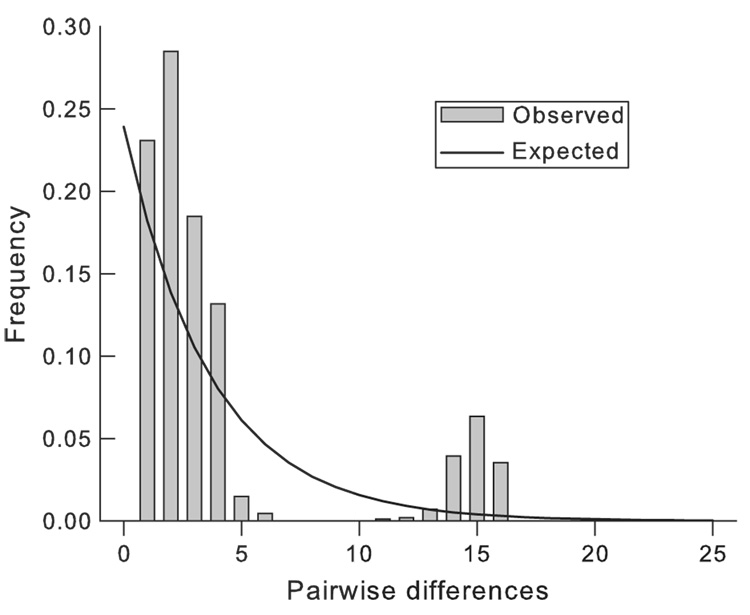
Tests for selective neutrality and demographic equilibrium in the East African populations included Fu’s FS and Ramos-Onsins and Rozas’ statistic R2 (Table 3). These tests can be used to detect recent demographic expansions or contractions because they cause ‘waves’ in mismatch distributions. Two such waves are apparent in Fig. 2. All tests were consistent, however, with the null hypothesis thereby providing no statistical support for demographic disequilibria arising from earlier bottlenecks or selective sweeps. Estimates of θ support the suggestion that the Ugandan population was about four times as large as the Kenyan populations (Table 3).
Fig. 2.
Mismatch distribution at mitochondrial loci in Kenyan Glossina fuscipes fuscipes.
Laboratory-cultured flies
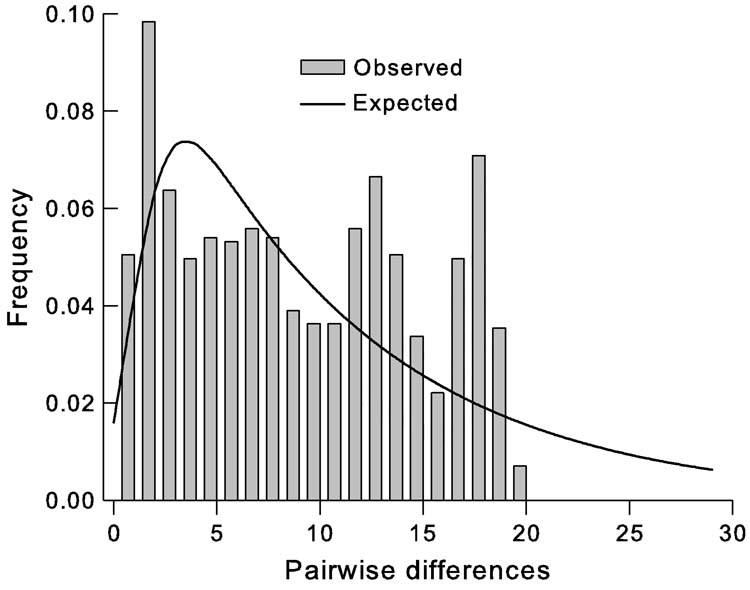
In CAR-lab flies, 27 haplotypes (lineages) were recorded, 20 of which were detected in the sample of 48 only once (Table 1). The proportion of singletons in the laboratory culture was much greater than that in the field populations (74% vs. 43%, respectively; χ2(2) = 4.81, P < 0.03). Diversities in the laboratory flies were similar to diversities in Ugandan populations, but the number of haplotypes was much greater. The number of lineages is consistent with a recent population expansion. The mismatch distribution shows clear departures from neutral expectations (Fig. 3) that are statistically significant according to the FS and R2 statistics (Table 3). The pattern of mitochondrial gene sequence variation is consistent with laboratory records that show the G. f. fuscipes culture was maintained for c. 115 generations at c. 325–1700 females until 2001, when it was increased to approximately 11 000 females (A. G. Parker, year?, personal communication to E.S.K.). Some eight generations later, genetic samples were taken.
Fig. 3.
Mismatch distribution at mitochondrial loci in laboratory-reared Glossina fuscipes fuscipes.
Relative effective population numbers
The estimate of θ for the laboratory culture (8.4; Table 3) provides a measure of relative population numbers (sizes), assuming mutation-drift equilibria and equal mutation rates. This number, 8.4, can be related to the effective numbers in the tsetse stock culture. There were approximately 11 000 females at the time of sampling, but the culture’s effective number is related to its harmonic mean over the 117 generations of its establishment. Thus, we estimate the laboratory culture Ne to be c. 1060. Kenya θ, at 3.2 (Table 3), was 0.38 that of CAR-lab. Uganda θ was 17.8, 2.13 times the laboratory culture estimate. We can then estimate Ne to a first approximation for the Kenyan and Ugandan G. f. fuscipes populations at 403 and 2258 female adults, respectively.
Discussion
Averaged over all field samples, mitochondrial diversity in G. f. fuscipes, at 0.84, was similar in magnitude to that found in Glossina morsitans morsitans Westwood (0.81), Glossina m. submorsitans in Ethiopia (0.84), Glossina pallidipes Austin in Tanzania (0.82), but not Glossina swynnertoni Austin (0.59), all in the morsitans group (Krafsur et al., 2000; Ouma et al., 2005, 2006; Marquez et al., 2006). The chief determinants of diversity are mutation rates and effective population numbers. The greater diversities in the Ugandan sample, only ~ 55 km from the Kenyan populations, suggested much larger effective population numbers. That inference is supported by the θ estimates that we use to estimate relative effective population sizes among the three populations. Why the great difference? Declining G. f. fuscipes numbers in Kenya must be considered, given high human populations on the Kenyan side of the border and ongoing loss of natural habitats. We obtained no statistical evidence for bottle-necks or recent population expansions because the neutrality tests were consistent with hypotheses of selective neutrality and demographic equilibrium.
Mitochondrial mutation rates are known to be substantial and inherited matrilineally, leading to rapid lineage sorting (Avise, 1994). Effective population numbers of tsetse flies are generally very much smaller than those of more common Diptera such as Drosophila spp., mosquitoes, house flies and other common, noxious species. Genetic drift is inversely proportional to effective population size. The chief demographic reason for comparatively smaller tsetse populations is their low reproductive rates – a single fed female must live nearly a month before she can deposit two mature larvae. The low reproductive rates are compensated, however, by longevities that greatly exceed those of other Diptera.
According to the exact test of Raymond & Rousset (1995), mitochondrial variation in the two Kenyan populations differed by more than chance alone. The populations were only c. 32 km apart and each was highly differentiated from the Ugandan population sample. This level of differentiation results from the patchy distribution of G. f. fuscipes populations that seem more typical of savannah-inhabiting tsetse flies whose demes seem to form islands (Krafsur, 2003). It can be assumed that the Kenyan and Ugandan populations are sympatric by virtue of sharing the same river drainage formation (the rivers Malaba and Sio).
In the riverine and lacustrine tsetse taxa, wet season dispersion from dry season refugia is thought to establish demes in which genetic drift leads to differentiation. Later, the retreat of flies to dry season refugia downstream can lead to mixing with survivors of formerly differentiated demes. Such a view is supported by the work of Cuisance et al. (1985), who used mark-release-recapture methods to demonstrate dispersals of Glossina palpalis gambiensis up to 21 km along gallery forests in Burkina Faso and related studies showed dispersals up to 5.1 km across savannahs and watersheds. Genetic support for mixing of genetically differentiated demes (the Wahlund effect, indicative of non-random mating within demes) can be provided by detecting significant heterozygote deficiencies in samples. Indeed, heterozygote deficiencies were detected at microsatellite loci in G. p. gambiensis in Burkina Faso (Solano et al., 1999) and Glossina p. palpalis in Cote d’Ivoire (Ravel et al., 2007). A deficiency of heterozygotes will also occur if two or more reproductively isolated cryptic species are sampled. Spatial examination of mitochondrial variation in G. p. gambiensis in Senegal and Mali demonstrated significant genetic differentiation among demes in different watersheds (Marquez et al., 2004). To these examples may be added G. f. fuscipes in East Africa, demes of which share little mitochondrial variation in common, even in the same watershed. Would a continuous cycle of annual mixing of autosomal genotypes and mitochondrial haplotypes, followed by dispersion and genetic drift, lead to the degree of spatial differentiation observed in palpalis group flies? It seems doubtful, but the question requires sophisticated simulation studies for proper evaluation. For now, it seems that palpalis group taxa show population structures that are not greatly different from those of morsitans group flies.
Experimental studies in Uganda have indicated high dispersal rates in male G. f. fuscipes, random movements of 338 m/day; for G. fuscipes and morsitans group tsetse; analysis suggested a mean displacement rate of 252 m/day (Rogers, 1977). Estimates of tsetse dispersal distances vary and are contentious because the underlying assumptions of random diffusion models do not always apply in the field (e.g. Hargrove, 1981). It is clear, however, that tsetse have the capacity to disperse far and wide. Such dispersal rates among the G. f. fuscipes populations sampled here would predict a rapid fusion of Kenyan and Ugandan populations. It seems, however, that forces of genetic drift in East African G. f. fuscipes are very much stronger than gene flow. Its dispersal tendencies seem to be either overestimated or thwarted by environmental circumstances, unapparent to us, in the habitats lying between the three sampled populations. It will be interesting to learn the population structure of this species in the central part of its range.
None of 49 composite haplotypes were shared between the IAEA CAR-lab sample and East African G. f. fuscipes. The CAR-lab flies had been in culture for more than 117 generations since the population’s establishment, but there was no detectable evidence of a genetic bottleneck that greatly reduced its original mitochondrial diversity. The pattern of variation in the G. f. fuscipes laboratory culture, at least 16 years after its establishment, indicates rapid population growth, already known from insectary records, and numerous (27) lineages, also indicative of rapid growth. The magnitude of mitochondrial diversity suggests that the colonization procedure was efficient in that existing genetic diversity was maintained. The much greater number of singular haplotypes in CAR-lab flies (20) than in Ugandan flies (9) testifies to highly effective insectary husbandry practices without which haplotypes in low frequency would be lost rapidly through drift. Careful husbandry is absolutely necessary in culturing tsetse flies because of their slow reproduction rates.
Acknowledgements
The authors thank Dr Alan Robinson and Andrew Parker of the Food and Agriculture Organization/International Atomic Energy Agency laboratory at Seibersdorf, Austria, for specimens and history of the Glossina fuscipes fuscipes culture, Joseph Sulo MD, who arranged the sampling of tsetse in Uganda, and Dr Steven Torr and an anonymous reviewer for trenchant critique of the manuscript. The research reported here was supported by a USPHS-National Institutes of Health grant (5R01-AI-052456).
References
- Avise JC. Molecular Markers, Natural History and Evolution. London: Chapman & Hall; 1994. pp. xxx – xxx. [Google Scholar]
- Cuisance D, Fevrier J, Dejardin J, Filledier J. Dispersion linéaire de Glossina palpalis gambiensis et de Glossina tachinoides dans une galerie forestière en zone soudano-guinéenne (Burkina-Faso) Revue d’ Élevage et de Médecine Vétérinaire des Pays Tropicaux. 1985;38:153–172. [Google Scholar]
- Excoffier L, Laval G, Schneider S. arlequin Version 3.0: an integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Fu YX. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997;147:915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooding RH, Krafsur ES. Tsetse genetics. Annual Review of Entomology. 2005;50:101–123. doi: 10.1146/annurev.ento.50.071803.130443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- Hargrove JW. Tsetse dispersal reconsidered. Journal of Animal Ecology. 1981;50:351–373. [Google Scholar]
- Hudson RR, Slatkin M, Maddison WP. Estimation of levels of gene flow from DNA sequence data. Genetics. 1992;132:583–589. doi: 10.1093/genetics/132.2.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krafsur ES, Madsen M, Wohlford DL, Mihok S, Griffiths NT. Population genetics of G. morsitans submorsitans Newstead. Bulletin of Entomological Research. 2000;90:329–335. doi: 10.1017/s0007485300000456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leak SGA. Tsetse Biology and Ecology. Wallingford, U.K: CABI Publishing; 1998. pp. xxx – xxx. [Google Scholar]
- Marquez JG, Vreysen MJB, Robinson AS, Bado S, Krafsur ES. Mitochondrial diversity analysis of Glossina palpalis gambiensis from Mali and Senegal. Medical and Veterinary Entomology. 2004;18:1–8. doi: 10.1111/j.0269-283X.2004.00508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquez JM, Malele II, Ouma JO, Krafsur ES. Glossina swynnertoni Austen: effective population size and gene flow estimated by mitochondrial diversity. Bulletin of Entomological Research. 2006;96:353–360. [PMC free article] [PubMed] [Google Scholar]
- Nei M. Molecular Evolutionary Genetics. New York: Columbia University Press; 1986. pp. xxx – xxx. [Google Scholar]
- Nei M, Kumar S. Molecular Evolution and Phylogenetics. New York: Oxford University Press Inc; 2000. pp. xxx – xxx. [Google Scholar]
- Ouma JO, Marquez JG, Krafsur ES. Macrogeographic structure of the tsetse fly, Glossina pallidipes (Diptera: Glossinidae) Bulletin of Entomological Research. 2005;95:437–447. doi: 10.1079/BER2005376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouma JO, Marquez GM, Krafsur ES. Patterns of genetic diversity and differentiation in the tsetse fly Glossina morsitans morsitans Westwood populations in East and southern Africa. Genetica. 2006;130:139–151. doi: 10.1007/s10709-006-9001-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Onsins SE, Rozas J. Statistical properties of new neutrality tests against population growth. Molecular Biology and Evolution. 2002;19:2092–2100. doi: 10.1093/oxfordjournals.molbev.a004034. [DOI] [PubMed] [Google Scholar]
- Ravel S, de Meeus T, Dujardin JP, et al. The tsetse fly Glossina palpalis palpalis is composed of several genetically differentiated small populations in the sleeping sickness focus of Bonon, Cote d’ Ivoire. Infection, Genetics and Evolution. 2007;7:116–125. doi: 10.1016/j.meegid.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Raymond M, Rousset F. An exact test for population differentiation. Evolution. 1995;49:1280–1283. doi: 10.1111/j.1558-5646.1995.tb04456.x. [DOI] [PubMed] [Google Scholar]
- Rogers AR, Harpending H. Population growth makes waves in the distribution of pairwise genetic differences. Molecular Biology and Evolution. 1992;9:552–569. doi: 10.1093/oxfordjournals.molbev.a040727. [DOI] [PubMed] [Google Scholar]
- Rogers DJ. Study of a natural population of Glossina fuscipes fuscipes Newstead and a model for fly movement. Journal of Animal Ecology. 1977;46:309–330. [Google Scholar]
- Rogers DJ, Robinson TP. Tsetse distribution. In: Maudlin I, Holmes PH, Miles MA, editors. The Trypanosomes. Wallingford, U.K: CABI Publishing; 2004. pp. 139–179. [Google Scholar]
- Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Simon C, Frati F, Beckenbach A, Crespi B, Liu H, Flook P. Evolution, weighting, and phylogenetic utility of mitochondrial gene sequence and a compilation of conserved polymerase chain reaction primers. Annals of the Entomological Society of America. 1994;87:651–701. [Google Scholar]
- Solano P, de La Rocque S, Cuisance D, Geoffroy B, de Meeus T, Cuny G, Duvallet G. Intraspecific variability in natural populations of Glossina palpalis gambiensis from West Africa, revealed by genetic and morphometric analyses. Medical and Veterinary Entomology. 1999;13:401–407. doi: 10.1046/j.1365-2915.1999.00189.x. [DOI] [PubMed] [Google Scholar]
- Tajima F. The effect of change in population size on DNA polymorphism. Genetics. 1989;123:597–601. doi: 10.1093/genetics/123.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TL, Plewniak F, Jeanmougin F, Higgins DG. The ClustalX Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waiswa C, Olaho-Mukani W, Katunguka-Rwakishaya E. Domestic animals as reservoirs for sleeping sickness in three endemic foci in southeastern Uganda. Annals Tropical Medicine and Parasitology. 2003a;97:149–155. doi: 10.1179/000349803235001688. [DOI] [PubMed] [Google Scholar]
- Waiswa C, Picozzsi K, Katunguka-Rwakishaya E, Olaho-Mukani W, Musoke RA, Welburn SC. Glossina fuscipes fuscipes in the trypanosomiasis endemic areas of southeastern Uganda: apparent density, trypanosome infection rates and host feeding preferences. Acta Tropica. 2003b;99:23–29. doi: 10.1016/j.actatropica.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Wamwiri FN, Nkwenguilila G, Clausen PH. Hosts of Glossina fuscipes and G. pallidipes in areas of western Kenya with endemic sleeping sickness, as determined using an egg-yolk (IgY) ELISA. Annals of Tropical Medicine and Parasitology. 2007;101:225–232. doi: 10.1179/136485907X156979. [DOI] [PubMed] [Google Scholar]
- Welburn SC, Coleman PG, Maudlin I, Fevre EM, Odiit M, Eisler MC. Crisis, what crisis? Control of Rhodesian sleeping sickness. Trends in Parasitology. 2006;22:123–128. doi: 10.1016/j.pt.2006.01.011. [DOI] [PubMed] [Google Scholar]