Abstract
The magnitude of the genetic relatedness of the two antigenic subtypes of equine herpesvirus 1 (EHV-1) was determined by DNA-DNA reassociation kinetics. Denatured, labeled viral DNA from one EHV-1 subtype was allowed to reassociate in the presence or absence of the unlabeled heterologous viral DNA. The initial rate of reassociation of either labeled viral DNA was increased by the presence of the heterologous viral DNA to an extent indicating 10 to 20% homology between the two EHV-1 genomes. Similar estimates of the amount of homology between the genomes of the two EHV-1 subtypes were obtained by determining the maximum fraction of labeled viral DNA that could be made resistant to S1 nuclease by hybridization with a large molar excess of the unlabeled, heterologous viral DNA. Analysis of the thermal stability of the subtype 1-subtype 2 heteroduplex DNA indicated approximately 30% base pair mismatching within the hybrid DNA molecules. Cross-hybridization of 32P-labeled virion DNA to nitrocellulose blots of restriction endonuclease cleavage fragments of each EHV-1 subtype DNA indicated that the observed homology between the two viruses was nonuniformly distributed with the viral genome. No homology could be detected between the DNA of either EHV-1 subtype and that of a strain of equine cytomegalovirus (EHV-2). The data suggest that the two biotypes of EHV-1 have arisen by divergent evolution from a common progenitor herpesvirus.
Full text
PDF

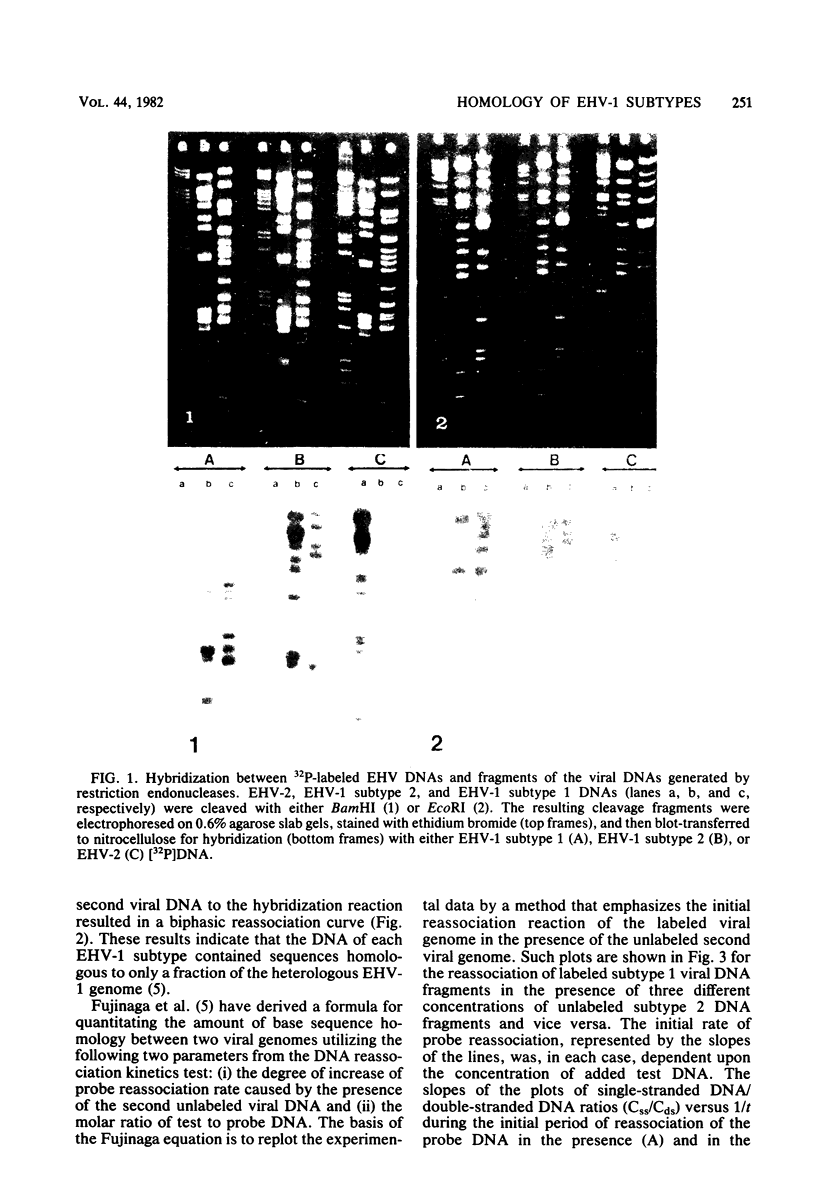
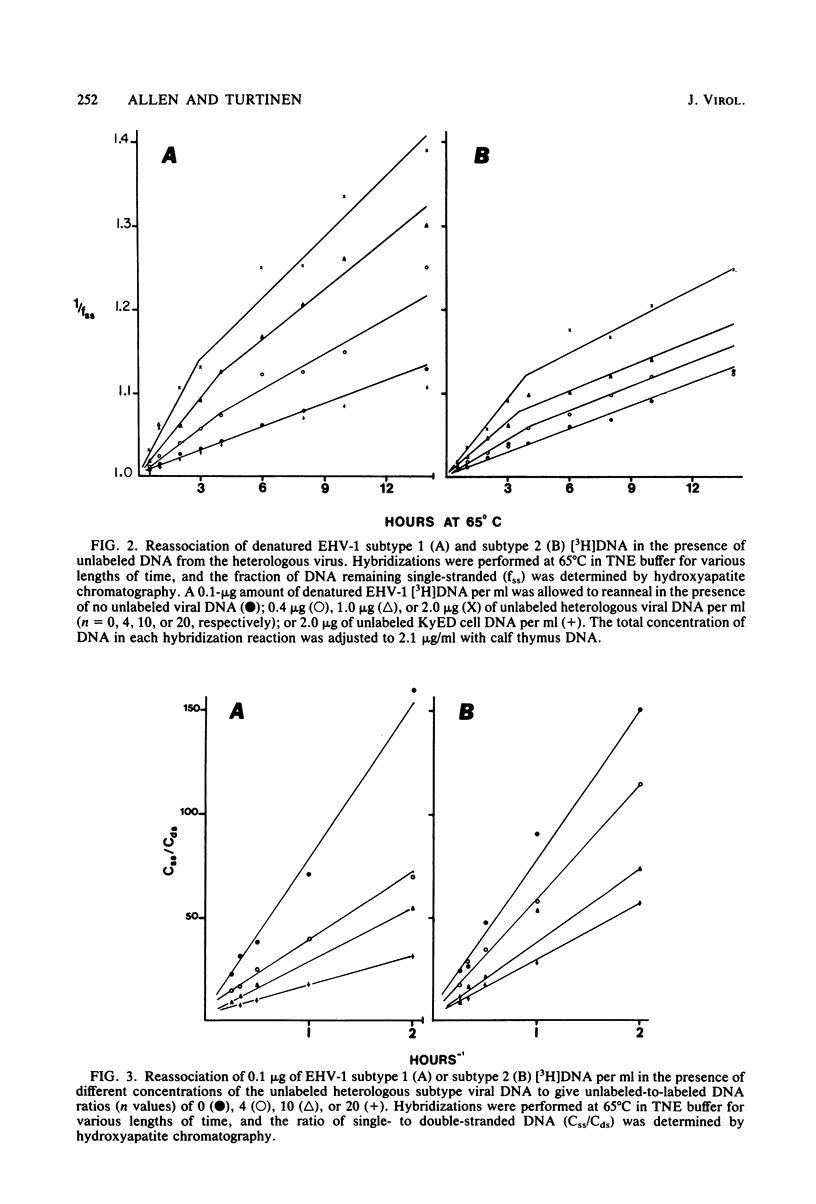



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allen G. P., O'Callaghan D. J., Randall C. C. Genetic relatedness of equine herpesvirus types 1 and 3. J Virol. 1977 Dec;24(3):761–767. doi: 10.1128/jvi.24.3.761-767.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows R., Goodridge D. Experimental studies on equine herpesvirus type 1 infections. J Reprod Fertil Suppl. 1975 Oct;(23):611–615. [PubMed] [Google Scholar]
- Fujinaga K., Sekikawa K., Yamazaki H. Method for determination of nucleotide sequence homology between viral genomes by DNA reassociation kinetics. J Virol. 1975 Mar;15(3):466–470. doi: 10.1128/jvi.15.3.466-470.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys A. J., Flavell R. A. A physical map of the DNA regions flanking the rabbit beta-globin gene. Cell. 1977 Oct;12(2):429–439. doi: 10.1016/0092-8674(77)90119-2. [DOI] [PubMed] [Google Scholar]
- Kieff E., Hoyer B., Bachenheimer S., Roizman B. Genetic relatedness of type 1 and type 2 herpes simplex viruses. J Virol. 1972 May;9(5):738–745. doi: 10.1128/jvi.9.5.738-745.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird C. D., McConaughy B. L., McCarthy B. J. Rate of fixation of nucleotide substitutions in evolution. Nature. 1969 Oct 11;224(5215):149–154. doi: 10.1038/224149a0. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- SHIMIZU T., ISHIZAKI R., ISHII S., KAWAKAMI Y., KAJI T., SUGIMURA K., MATUMOTO M. Isolation of equine abortion virus from natural cases of equine abortion in horse kidney cell culture. Jpn J Exp Med. 1959 Dec;29:643–649. [PubMed] [Google Scholar]
- Sabine M., Robertson G. R., Whalley J. M. Differentiation of sub-types of equine herpesvirus I by restriction endonuclease analysis. Aust Vet J. 1981 Mar;57(3):148–149. doi: 10.1111/j.1751-0813.1981.tb00495.x. [DOI] [PubMed] [Google Scholar]
- Sharp P. A., Pettersson U., Sambrook J. Viral DNA in transformed cells. I. A study of the sequences of adenovirus 2 DNA in a line of transformed rat cells using specific fragments of the viral genome. J Mol Biol. 1974 Jul 15;86(4):709–726. doi: 10.1016/0022-2836(74)90348-9. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Studdert M. J., Blackney M. H. Equine herpesviruses: on the differentiation of respiratory from foetal strains of type 1. Aust Vet J. 1979 Oct;55(10):488–492. doi: 10.1111/j.1751-0813.1979.tb00377.x. [DOI] [PubMed] [Google Scholar]
- Studdert M. J., Simpson T., Roizman B. Differentiation of respiratory and abortigenic isolates of equine herpesvirus 1 by restriction endonucleases. Science. 1981 Oct 30;214(4520):562–564. doi: 10.1126/science.6270790. [DOI] [PubMed] [Google Scholar]
- Turtinen L. W., Allen G. P., Darlington R. W., Bryans J. T. Serologic and molecular comparisons of several equine herpesvirus type 1 strains. Am J Vet Res. 1981 Dec;42(12):2099–2104. [PubMed] [Google Scholar]
- Wetmur J. G., Davidson N. Kinetics of renaturation of DNA. J Mol Biol. 1968 Feb 14;31(3):349–370. doi: 10.1016/0022-2836(68)90414-2. [DOI] [PubMed] [Google Scholar]
- Whalley J. M., Robertson G. R., Davison A. J. Analysis of the genome of equine herpesvirus type 1: arrangement of cleavage sites for restriction endonucleases EcoRI, BglII and BamHI. J Gen Virol. 1981 Dec;57(Pt 2):307–323. doi: 10.1099/0022-1317-57-2-307. [DOI] [PubMed] [Google Scholar]




