Abstract
Human carbonic anhydrase II (HCA II), among the fastest enzymes known, catalyzes the reversible hydration of CO2 to . The rate-limiting step of this reaction is believed to be the formation of an intramolecular water wire and transfer of a proton across the active site cavity from a zinc-bound solvent to a proton shuttling residue (His64). X-ray crystallographic studies have shown this intramolecular water wire to be directly stabilized through hydrogen bonds via a small well-defined set of amino acids, namely, Tyr7, Asn62, Asn67, Thr199, and Thr200. Furthermore, X-ray crystallographic and kinetic studies have shown that the mutation of tyrosine 7 to phenylalanine, Y7F HCA II, has the effect of increasing the proton transfer rate by 7-fold in the dehydration direction of the enzyme reaction compared to wild-type (WT). This increase in the proton transfer rate is postulated to be linked to the formation of a more directional, less branched, water wire. To evaluate this proposal, molecular dynamics simulations have been employed to study water wire formation in both the WT and Y7F HCA II mutant. These studies reveal that the Y7F mutant enhances the probability of forming small water wires and significantly extends the water wire lifetime, which may account for the elevated proton transfer seen in the Y7F mutant. Correlation analysis of the enzyme and intramolecular water wire indicates that the Y7F mutant significantly alters the interaction of the active site waters with the enzyme while occupancy data of the water oxygens reveals that the Y7F mutant stabilizes the intramolecular water wire in a manner that maximizes smaller water wire formation. This increase in the number of smaller water wires is likely to elevate the catalytic turnover of an already very efficient enzyme.
1. Introduction
Many biologically relevant enzymes catalyze proton transfer (PT) reactions, yet few are as fast and efficient or have been as extensively characterized as carbonic anhydrase (CA).1-18 CA catalyzes the interconversion of carbon dioxide to bicarbonate and an excess proton at a rate near the diffusion controlled limit (Equation 1).2,19-21
| (1) |
Ubiquitous in nature, CA is responsible for CO2 transport and the regulation of pH in biological life from bacterial organisms to plants and animals. The PT event in HCA II is unique in that it represents the boundary between proton “transfer”, the proton exchange between a single donor and acceptor group, and proton “transport”, the proton exchange through several water molecules via Grotthuss shuttling.14,22-24 The PT event in CA is believed to proceed by means of an enzyme stabilized intramolecular water wire. Understanding how CA facilitates the formation of this intramolecular water wire and the stabilization of the PT event will also offer insight into the fundamental nature of PT in biological systems.
Human carbonic anhydrase II (HCA II) is among the fastest of the expressed catalytically active CA isozymes which together are fundamental to CO2 transport, fluid and acid secretion and pH buffering in physiology.25,26 The rate-limiting step in HCA II's efficient catalysis, at high buffer concentrations, is the PT between the zinc-bound water/hydroxide and His64, which is connected through an intramolecular water wire spanning 8−10 Å.6,19,27 The rate-limiting step is one of the two distinctly separate steps involved in HCA II's catalysis, which is described by ping-pong kinetics. The initial step in the hydration direction, involves the nucleophilic attack of the zinc-bound hydroxide by the carbonyl carbon of CO2, generating a bicarbonate ion (Equation 2).
| (2) |
The subsequent step (Equation 3) involves the PT between the zinc-bound water and an exogenous buffer, thereby regenerating the initial zinc-bound hydroxide species and priming the catalytic cycle.4,19,28,29
| (3) |
The PT event in HCA II between the zinc-bound solvent and the exogenous buffer can be subdivided into two PT events. The first of these PT events is from the zinc-bound solvent to His64 (Equation 4) and the second is the PT event from the protonated His64 to an exogenous buffer (denoted as B in Equation 5).
| (4) |
| (5) |
Which of these two events is the rate-limiting step depends on the exogenous buffer concentration. When the exogenous buffer concentration is high the rate limiting step is given by equation 4 where the PT event is between the zinc-bound solvent and His64 by means of the intramolecular water wire. 30 Conversely when the exogenous buffer concentration is low the rate-limiting step is given by equation 5 where the PT event is between the protonated His64 and an exogenous buffer.2 His64 is believed to be the proton acceptor/donor in equations 4 and 5 based on mutation studies showing that when His64 has been mutated to an alanine (H64A HCA II) this leads to a significant reduction in the catalytic turnover, kcat , for the hydration of CO2.31,32 In the case where the exogenous buffer concentration is not the limiting factor the role of the intramolecular water wire and the orientation of His64 becomes paramount in understanding the PT event. Numerous experimental and theoretical studies have focused on the nature of the intramolecular water wire and a consensus has been reached that two to four water molecules are involved.5,6,9,33-37 Several residues including Tyr7, Asn62, Asn67, Thr199, and Thr200 are believed to stabilize the intramolecular water wire. Therefore, a mutation of any of these critical amino acids is believed to influence the observed rate of PT.
It is of note that recent theoretical simulations using the SCC-DFTB semiempirical method37,38 have suggested that hydroxide transport and not the transport of an excess proton is fundamental to the rate limiting reaction in HCA II.39,40 These simulations have also theorized that the orientation of His64 and the intramolecular water wire are not important in the rate limiting step of catalysis. These theoretical simulations propose a new mechanistic interpretation to the rate limiting step yet; they do not disprove the standard Grotthuss proton hopping mechanism. While the intramolecular water wire may not be the only limiting factor in the PT event it is clearly crucial to the standard Grotthuss hopping mechanism. In addition to the pKa change between the donor/acceptor (diabatic free energy difference between the donor/acceptor) the orientation of His64 and the direct effect of His64 on the distribution/stabilization of water wire sizes/lifetimes in the active site are likely to be fundamental to the rate limiting PT event in HCA II.12,41
This work presents classical molecular dynamics (MD) simulations of the fully solvated wild type (WT) HCA II and mutant tyrosine 7 to phenylalanine (Y7F HCA II, PDB accession # 2NXR)1. From X-ray crystallographic studies and MD simulations of WT HCA II it has been postulated that Tyr7 is important in stabilizing the W3a water in the observed ordered intramolecular water wire of the active site cavity (Figure 1).9,12 Due to the proximity of Tyr7 to His64 the effect of the Y7F mutation on the intramolecular water wire, correlated system movements, and the orientation of His64 were investigated. Correlated motion analysis is effective at indicating long range interactions and regions of coupling in the enzyme. These correlated motions are believed to be mediated by hydrogen bonds and changes in the correlated motions often reflect alterations in hydrogen bonding patterns.42 In this study, correlated motions of the active site waters and enzyme residues indicate persistent hydrogen bonding patterns. Thus, changes in correlated motions reveal rearrangements of the hydrogen bonded networks in the active site. Correlation and other analyses of the proton acceptor (His64) and the intramolecular water wire also provide insight into the catalytic process in the WT and Y7F mutant. As in our previous investigation of His64 orientation12, the simulations presented here will include the hydration direction (zinc-bound water and neutral His64) and the dehydration direction (zinc-bound hydroxide and protonated His64) with His64 in both the “in” and “out” orientations.
Figure 1.
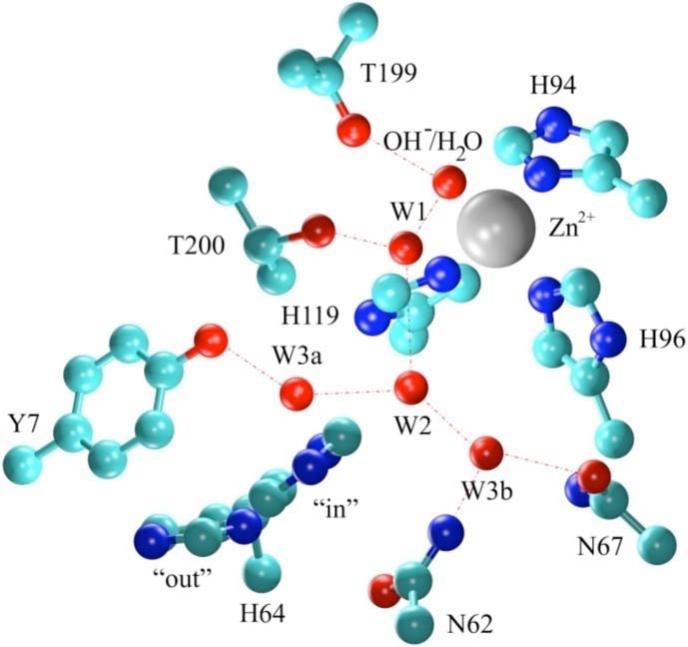
Representation of the WT HCA II active site. Coordinates were taken from the X-ray crystallographic structure (2CBA).9 Red dotted lines indicate proposed hydrogen bonding.
2. Methodology
Molecular Dynamics Simulations (MD)
To evaluate the effect of the zinc-bound water/hydroxide on the active site of HCA II several systems representing the hydration of CO2 and the dehydration of were created. The first system contains a zinc-bound water and an unprotonated His64 (ZnH2O2+-His) and was modeled with His64 in both the “in” and “out” orientations while the second system contains a zinc-bound hydroxide and a protonated His64 (ZnOH+-HisH+) and was modeled with His64 in the “out” position only. The “in” and “out” positions were selected for the ZnH2O2+-His system based on the dual occupancy of the histidine residue as seen in X-ray experiments and MD simulations.9,12,41 The ZnOH+-HisH+ system was modeled with His64 in the “out” orientation only, based on recent computational evidence that the ZnOH+-HisH+ system predominantly adopts the “out” orientation and due to several independent “in” oriented simulations spontaneously converting to the “out” orientation.12,15,43 To evaluate the effect of residue 7 on the stabilization of the intramolecular water wire, the ZnH2O2+-His and ZnOH+-HisH+ systems were created for the WT and the Y7F mutant of HCA II. The 1.54 Å resolution X-ray structure9 (Protein Data Bank reference 2CBA) was used for the initial coordinates of the WT simulations. The Y7F mutant was created by using the Swiss-PDB Viewer44 to mutate Tyr7 to Phe7 and to subsequently adjust Phe7 to the most appropriate rotamer. The WT and Y7F mutant of HCA II along with the 220 X-ray waters were then solvated in a cubic box (L ≈ 75 Å) of modified TIP3P water.45 The parm9946 force field was used to describe the enzyme while the zinc active site was described by the parameters used in an earlier publication.12 All systems were equilibrated for 250 ps in the constant NVE ensemble followed by 1.25 ns in the constant NPT ensemble.47 The “out” orientations were created in the initial 250 ps of the constant NPT simulations by placing a harmonic restraint on the distance between the zinc and the Nδ of His64 using a spring force constant of 40 kcal mole−1 Å−2. After the equilibration phase was completed, a data collection phase consisting of 2.0 ns was conducted in the constant NVT ensemble which was used to determine water occupancy in the active site as well as correlated movements of the WT and Y7F mutant.47 All simulations used periodic boundary conditions with long-range Coulombic interactions calculated by particle mesh Ewald summation, while short-ranged non-bonded interactions and forces were subjected to a 10 Å cutoff.48 Simulations were conducted at 298.15 K and used an MD time integration step within the leap-frog Verlet integrator of 0.5 fs. During the constant NPT ensemble the simulations were run at 1 atm, while all simulations used Langevin dynamics for the thermostat.
Correlation Analysis
To determine which water positions in the active site are most stable over the course of the trajectories, the trajectories were first centered and rms fitted to the average configuration and then the volmap tool in VMD49 was used to generate the average occupancy of the water oxygens in the active site. The water positions that were stable for over 75% of the trajectories were used to determine which waters would subsequently be used for correlation analysis.50 The water in each trajectory frame that was closest to the conserved water position (75% occupancy) was selected. Each residue in the protein was mapped to one site, at its center of mass, and the mass-weighted correlation of the movement of these points with the movement of the water oxygens, selected above, was calculated using the AMBER 8 program, yielding a mass weighted correlation matrix51
Water Wire Analysis
In addition to the previous simulations an additional independent set of simulations were created which were used to analyze water wire properties such as water wire size and lifetime in the WT and Y7F mutant. The independent simulations were created by using the final structures from the data collection period from the first set of simulations. The structures were then re-equilibrated using the same protocol described above followed by a data collection phase of 2.5 ns conducted in the constant NVT ensemble. The data collection phase was then analyzed for continuous water wires connecting either the zinc-bound water to His64 or connecting His64H+ to the zinc-bound hydroxide. A water wire is defined as a continuously hydrogen bonded network of waters that has the donating and accepting oxygens within 3.5 Å, the acceptor oxygen and donating oxygen's hydrogen within 2.5 Å, and an angle of less than 120° between the donating oxygen-donating hydrogen vector and the donating oxygen-accepting oxygen vector.
Umbrella Sampling Simulation
The use of biased sampling procedures such as umbrella sampling allow for the analysis of fundamental processes that have barriers larger than kBT. The rotation of His64 between the “in” to “out” orientation, which is believed to be critical to the overall proton transport process of HCA II, is one such process.12 To sample the rotation of His64 about the χ1 dihedral (N-Cα-Cβ-Cγ) 30 umbrella windows of the form
| (6) |
were used to restrain sampling of the dihedral angle. A dihedral force constant of 40 kcal mol−1 rad−2, kn, was placed equidistantly over the range of = −65° to 80° to enhance sampling. The biased sampling data was then recombined using the weighted histogram analysis method (WHAM) to generate a continuous free energy plot of the rotation of His64 about the χ1 dihedral.52,53 Each umbrella window was equilibrated for 500 ps followed by a data collection period of 2 ns in the constant NVT ensemble. The MD time integration step within the leap-frog Verlet integrator was set to 1 fs for all umbrella windows. Convergence of the resulting potential of mean force (PMF) was achieved when the a PMF calculated with the first half and second half of the data possessed differences ≤ 0.25 kcal/mole, and the error determined by the Monte Carlo bootstrap error analysis was relatively small, ≤ 0.1 kcal/mole.
3. Results and Discussion
The effect of mutating residue 7, Y7F, from an amino acid that participates in the hydrogen bonding intramolecular water wire to an amino acid that does not participate has significant effects on the catalytic ability of HCA II as seen by experimental data.1 To understand the effect of the Y7F mutation on catalysis an extensive analysis of the intramolecular water wire and the orientation of His64 was conducted for both the WT HCA II and the Y7F mutant. The distribution, probability of formation, and the lifetime of the intramolecular water wire were analyzed to understanding how HCA II utilizes the intramolecular water wire for the PT event. In addition to these properties the spatial occupancy of water wire oxygens and their correlated movements with respect to each other and with the enzyme were investigated to understand how the WT and Y7F mutant stabilize their respective intramolecular water wires. Theoretical studies on PT through water wires of different sizes indicate that smaller water wires transport protons more efficiently.36 Therefore, focusing analysis on relatively small water clusters should prove insightful when evaluating the impact of mutations on the intramolecular water wire.
Probability and Lifetime of the Intramolecular Water Wire
Non-biased MD simulations were analyzed for continuous hydrogen bonding water wires that would allow for the transport of an excess proton from the zinc-bound water to the unprotonated Nδ of His64 for the ZnH2O2+-His system or the transport of an excess proton from the protonated Nδ of His64 to the zinc-bound hydroxide in the ZnOH+-HisH+ system. The water wire analysis was conducted for both the WT and Y7F simulations. In all cases the cluster size includes the zinc-bound water/hydroxide. After the average probability of forming a continuous hydrogen bonded water wire was evaluated (Supplemental Figure 1) the data was further analyzed for the distribution of the smallest water cluster size given that a continuous hydrogen bonded water wire existed. The smallest water cluster distribution was chosen for evaluation because it is believed that the smaller water wires are more conducive to PT. In addition this analysis eliminates the counting of multiple water wires of the same size at a given point in time.
The average probability for the smallest continuous hydrogen bonded water wires in the active site for the ZnH2O2+-His and ZnOH2+-HisH+ systems for both the WT and Y7F mutant are found in Figure 2. When evaluating water cluster sizes there is an important distinction between water clusters of size 3 and those that are larger than size 3. The water cluster size of 3 is unique in that it represents an un-branched water wire connecting the donating zinc-bound water to the accepting Nδ of His64. It is possible that a water cluster of size 3 will proceed in a concerted fashion that is almost barrier-less. Water wires of sizes greater than 3 represent clusters that may possess branching and therefore could exhibit an Eigen cation, H9O4+, in the water cluster, which has been shown to elevate the proton transport (PT) barrier when compared to smaller un-branched water wires.36 In the Y7F's ZnH2O2+-His system with His64 in the “in” orientation the dominant cluster size is 3 (72 ± 7%), while the WT's dominant cluster size is 4 (41 ± 4%). The WT ZnH2O2+-His system does form water clusters of size 3 but at a significantly reduced probability when compared to the Y7F mutant. The probabilities for forming water clusters of sizes 5 and larger are typically less than 25% and show only moderate differences between WT and the Y7F mutant. These results indicate that the Y7F mutant, because it possesses a significantly elevated probability of forming water clusters of size 3, may favor a possible concerted proton transfer event that is likely to account for the elevated turnover number reported by kinetic studies.1 The WT system, while possessing a moderate number of size 3 water clusters (12 ± 5%) favors water clusters of size 4. This would indicate that concerted PT in WT is possible but that PT through a water cluster of size 4 is more likely, which may involve an Eigen cation and hence a higher overall barrier to PT.
Figure 2.
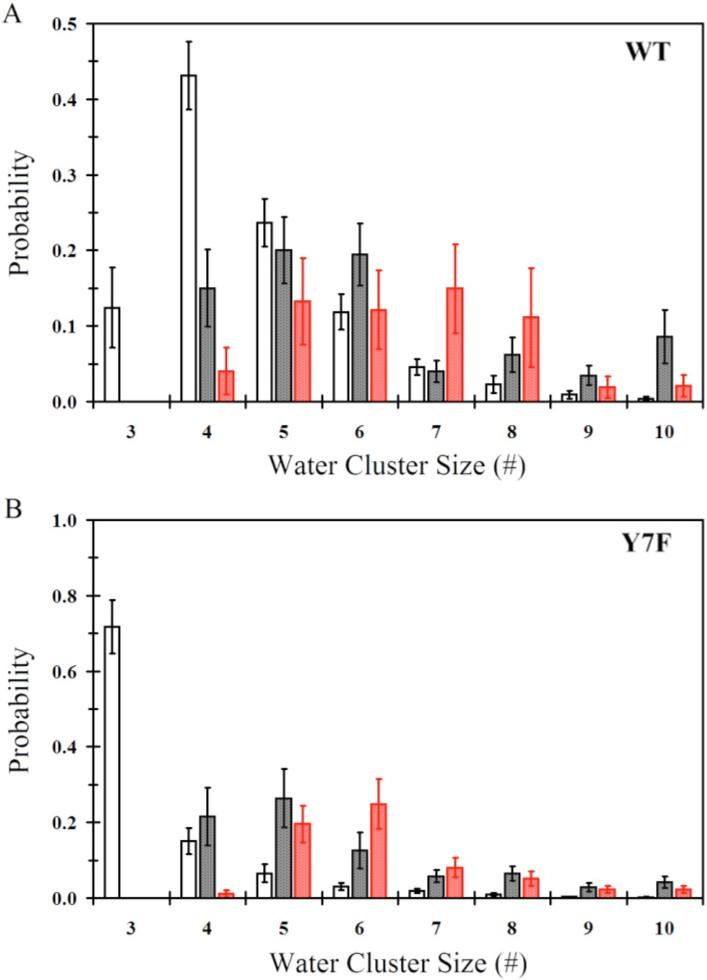
Smallest continuous hydrogen bonded water wire that connects the zinc-bound water/hydroxide and the protonated/unprotonated Nδ of His64. (A) Water wire distribution for WT-ZnH2O2+-His in the “in” orientation (black no fill), WT-ZnH2O2+-His in the “out” orientation (black with black fill), WT-ZnOH+-HisH+ in the “out” orientation (red with red fill). (B) Water wire distribution for Y7F-ZnH2O2+-His in the “in” orientation (black no fill), Y7F-ZnH2O2+-His in the “out” orientation (black with black fill), Y7F-ZnOH+-HisH+ in the “out” orientation (red with red fill).
When comparing the WT and Y7F systems for both ZnH2O2+-His and ZnOH+-HisH+ with His64 in the “out” orientation, larger water cluster sizes are expected because His64 is farther away from the catalytic zinc-bound solvent. The WT and the Y7F mutant systems contain water wires of size 5 and greater with probabilities of less than or equal to 15% and 25%, respectively. There appears to be little significant difference between the probability of forming these larger cluster sizes (size 5 or greater) for the WT and Y7F mutant systems and, therefore, due to the similarities between the WT and the Y7F mutant systems for forming these clusters in the His64 “out” orientation it can be reasoned that these systems in the “out” position do not account for the differences seen in the enzyme turnover numbers.
Another important aspect of the water wires is their respective lifetimes as seen in Figure 3. For the WT system water wire lifetimes are typically less than 3 ps with water wires of size 4 having the longest lifetime (2.8 ± 0.5 ps). The water wire lifetimes for the WT system are in agreement with previously reported lifetimes (∼1 ps)33,34 and are significantly shorter than the Y7F mutant's water wires lifetimes. The Y7F mutant exhibits lifetimes greater than twice that of the WT water wires with lifetimes of 5.0 ± 0.4 ps and 4.4 ± 0.3 ps for cluster sizes 3 and 4, respectively. In contrast to the hydrogen bonded water wire lifetimes found in the ZnH2O2+-His systems the water wire lifetimes found in the ZnOH+-HisH+ systems for WT and the Y7F mutant are similar, with lifetimes of around 0.5 ps. The similarities in the lifetime of the water wires for the “out” orientations of His64 is another indication that these particular water wires are not the main determining factor in the increased turnover number seen in the Y7F mutant. Instead, the data indicates that it is the behavior of the ZnH2O2+-His system and in particular its water wires of sizes 3 and 4 that are the main contributor to the experimentally observed increase in the turnover numbers.
Figure 3.
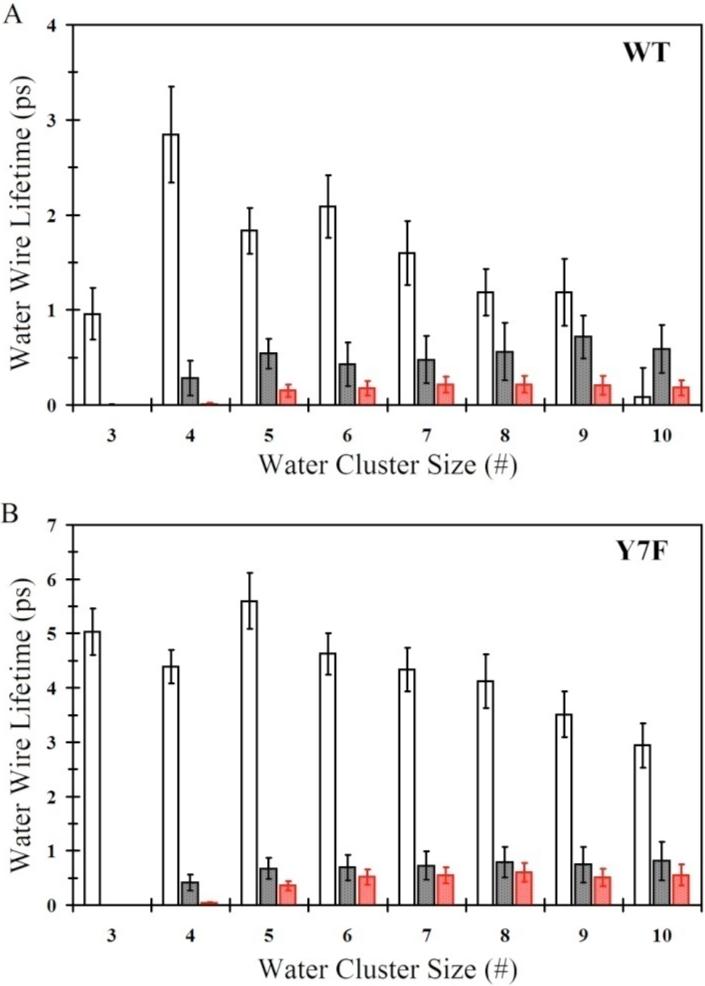
Water wire lifetimes for the continuous hydrogen bonded water wire that connects the zinc-bound water/hydroxide and the protonated/unprotonated Nδ of His64. (A) Water wire lifetime distribution for WT-ZnH2O2+-His in the “in” orientation (black no fill), WT-ZnH2O2+-His in the “out” orientation (black with black fill), WT-ZnOH+-HisH+ in the “out” orientation (red with red fill). (B) Water wire lifetime distribution for Y7F-ZnH2O2+-His in the “in” orientation (black no fill), Y7F-ZnH2O2+-His in the “out” orientation (black with black fill), Y7F-ZnOH+-HisH+ in the “out” orientation (red with red fill).
Occupancy, Radial Distribution Function, and Correlation Analysis of the Intramolecular Water Wire
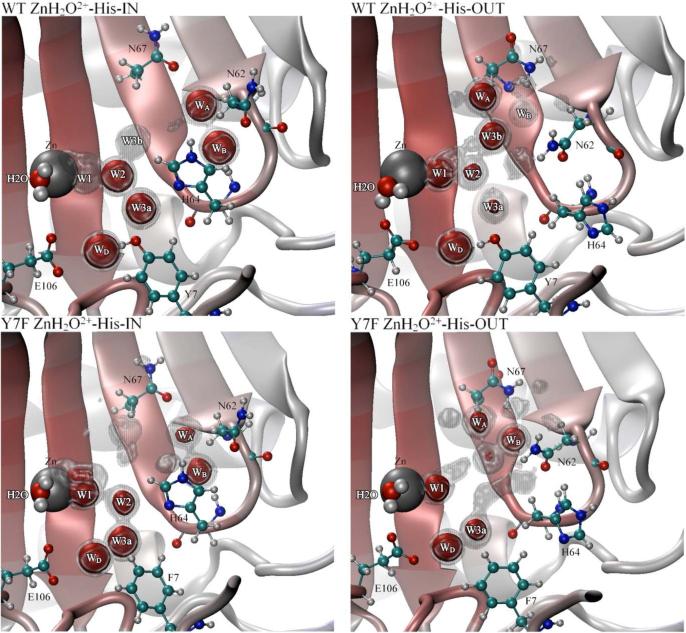
To understand why there is such a significant difference in water cluster sizes and lifetimes between the WT and Y7F enzymes, the average occupancy of water oxygens was evaluated. When analyzing the average occupancy it is convenient to look at different isosurfaces described by the occupancy cutoff. The occupancy isosurface describes the occupancies equal to the cutoff value. In Figure 4 the ZnH2O2+-His “in” orientation clearly shows a difference in the stabilized water oxygen locations between the WT and Y7F enzymes. In the WT enzyme, at the 50% occupancy cutoff, a branched water wire can be seen (Zn-H2O-W1-W2-W3a or W3b) which agrees well with experimental crystal structure results (PDB accession # 2CBA and the recent 1.05 Å resolution crystal structure PDB accession # 2ILI)9,41 However, when the stringency of the cutoff is increased to 75%, only the zinc-bound water, W2 and W3a are still present. This indicates that the W1 water is not as spatially stable as the W2 or W3a waters, suggesting that the position of W1 is the limiting factor in formation of a continuous water wire between the zinc-bound water and N of His64. By contrast, in the Y7F mutant a clear un-branched 3-water wire structure (Zn-H2O-W1-W2) is present that is stable when evaluated at both 50% and 75% occupancy cutoffs. The occupancy data for the Y7F mutant indicates that the intramolecular water wire is more stable than the corresponding water wire found in the WT enzyme. These results agree with the water wire formation and lifetime data reported earlier since, in the WT, W1 is more mobile thereby disrupting the water wire and leading to a reduced probability of water wire formation and shorter lifetimes. In the Y7F enzyme, the water oxygens are less mobile (more stable) and therefore facilitate an increase in water wire formation and longer water wire lifetimes. The water occupancy plot for the “out” orientation of the WT enzyme (Figure 4) is very similar to the “in” orientation. The major differences are a slight change in the position and increased stabilization of W1 and a loss of stabilization of the W3a, W3b and W2 waters, consistent with the idea that W3a is stabilized in part by His64 and that W2 is stabilized primarily by W3a. For the Y7F enzyme, the “in” and “out” occupancy plots were also very similar with the major difference being a reduced stabilization of W2 in the “out” orientation which supports the idea that W2 is partially stabilized by His64 when in the “in” orientation.
Figure 4.
Occupancy data for water oxygens inside the active site of HCA II. Gray regions represent 50% occupancy and the red regions represent 75% occupancy for the WT ZnH2O2+−His-IN system, WT ZnH2O2+-His-OUT system (Top left and right panel, respectively), Y7F ZnH2O2+-His-IN system and the Y7F-ZnH2O2+-His-OUT system (Bottom left and right panel, respectively).
One possible explanation for the destabilization of W1 in the WT enzyme's “in” orientation is found by examining the radial distribution functions (RDF) from the catalytic zinc to the surrounding waters, g(RZn-OW), (Figure 5). Inspection of WT's g(RZn-OW) reveals that there are changes in the active site solvation structure between the ZnH2O2+-His “in” and “out” systems. The main difference is in the location of the first solvation peak. In the “out” orientation, as well as in the Y7F enzymes “in” and “out” systems, the first solvation peak is seen to lie further from the zinc than is seen in the “in” orientation of the WT enzyme. In the WT “in” orientation it appears that the water wire is compressed, as reflected by the shortening of the first solvation shell distance. The compression of the water wire may occur in order to accommodate a continuous hydrogen bonded water wire. This compression, which destabilizes W1 (evident by the occupancy plots), is relieved when His64 is in the “out” orientation as indicated by the shift and increase in occupancy of W1 in the water occupancy plots, as well as the shift in the position of the first solvation shell peak seen in the g(RZn-OW). The Y7F enzyme does not show this compression since the position of W2 and W3a have shifted toward Phe7, which has reduced the orientational strain on W1. In the Y7F mutant W1 does not need to move closer to the zinc-bound water in order to accommodate a hydrogen bond with W2, therefore a hydrogen bonded network can form (ZnH2O2+-W1-W2-His64) without any orientational strain on W1. Comparing the g(RZn-OW) for the ZnH2O2+-His “in” and “out” systems of the WT with those for the Y7F enzyme reveals differences in the second and third solvation shells. In the mutant there is no clear second or third shell, which can be explained by looking at the water occupancy plots. In the WT enzyme the stabilized waters are oriented approximately in radial shells around the zinc (W1, W2 and WD, W3a and W3b) as seen by the distinct peaks in the g(RZn-OW). In the Y7F enzyme W1 occupies a discrete shell but W2, W3a, and WD do not reside at distinct radial distances from the zinc, which has the effect of merging peaks in the g(RZn-OW). This result though should not be interpreted as disorder in the active site. In fact inspection of the occupancy plot reveals that the Y7F mutant possesses a much more ordered and localized water wire than that seen in WT.
Figure 5.
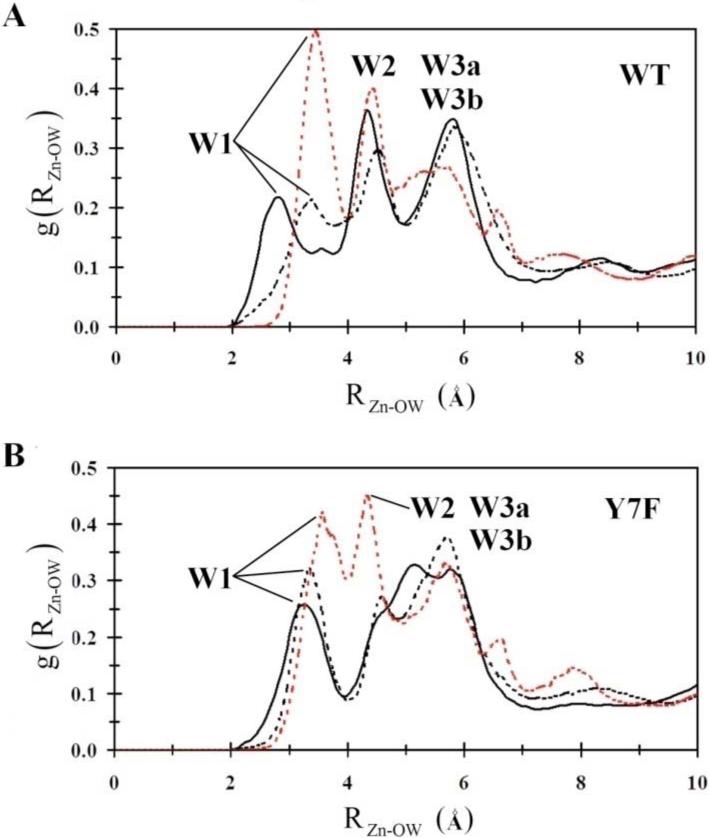
Intramolecular water wire radial distribution functions. (A) WT-ZnH2O2+-His in the “in” orientation (black), WT-ZnH2O2+-His in the “out” orientation (black dotted), WT-ZnOH+-HisH+ in the “out” orientation (red dotted). (B) Y7F-ZnH2O2+-His in the “in” orientation (black), Y7F-ZnH2O2+-His in the “out” orientation (black dotted), Y7F-ZnOH+-HisH+ in the “out” orientation (red dotted).
While the g(RZn-OW) and water occupancy data reveal the orientation and stability of active site waters, correlated movements of the stable waters and the enzyme indicate recurring hydrogen bonding patterns and long range interactions. Therefore, residue-residue and residue-water position-position correlation functions were calculated. In looking at the correlations between protein residues, there is significantly more correlated movement of the protein in the mutant compared to the WT, Figure 6A. All of the regions involved in this difference are outside the active site, thus the increase in correlated motion indicates increased coupling between distant regions of the enzyme. These differences cannot be directly related to changes in proton transport, but do indicate an overall change in protein dynamics and long range interactions caused by the Y7F mutation.
Figure 6.
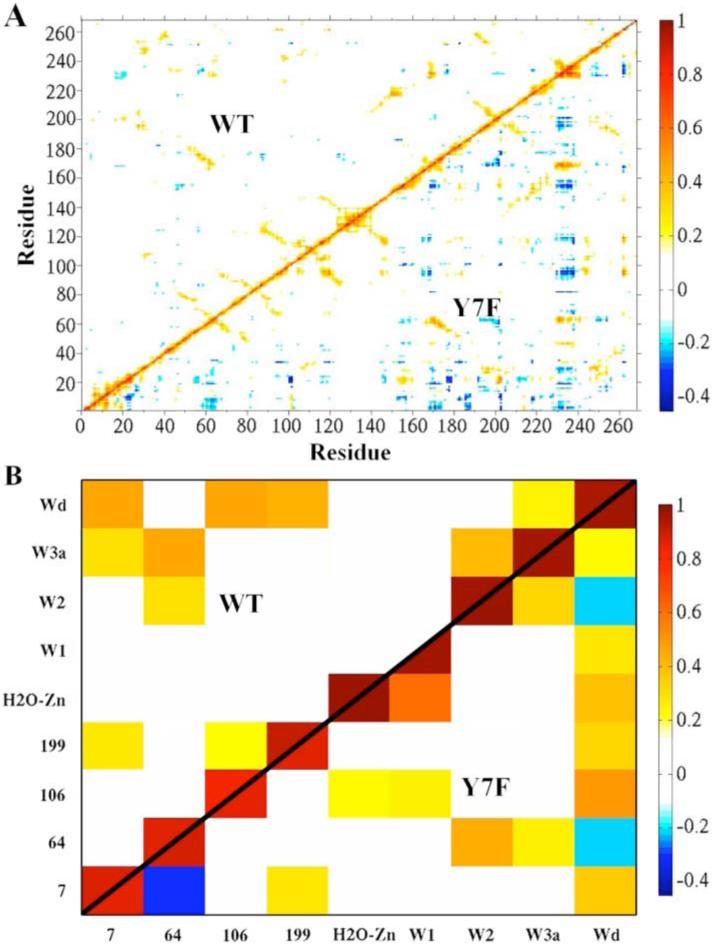
Correlation plots. (A) Residue-residue correlation plot of WT (upper diagonal) and the Y7F mutant (lower diagonal) of the HCA II enzyme. (B) The active site water-water and active site water-residue correlation plot of WT (upper diagonal) and the Y7F (lower diagonal).
Correlations involving the waters at positions showing at least 75% occupancy and residues of the enzyme are shown in Figure 6B. In the WT enzyme, the water wire can be viewed as different components of correlated or uncorrelated waters. When analyzing the water wire in this fashion three distinct components are found to comprise the water wire. The first component contains the zinc-bound water which is not correlated to the rest of the intramolecular water wire (W1, W2, W3a or W3b). The second component contains W1 which is also not correlated to the other waters of the intramolecular water wire, and the third component which contains W2, W3a, WD, His64, and Tyr7. In the third correlated component W2 is correlated to W3a which in turn is correlated to Tyr7 and His64. The strong correlation of W3a to His64 suggests that this water is primarily the final water in the water wire while the smaller correlation between His64 and W2 suggests it may also act as the final water which mediates the terminal step in proton transport to His64. These conclusions are supported by the earlier water wire formation data that shows a low probability of forming a water wire of size 3 which would use W2 as the final proton donator, and a larger probability of proceeding through a water wire of size 4, which would use W3a as the final proton donator. This correlation analysis creates a picture where three separate components of the water wire must come together to form a continuous hydrogen bonded network. The connection of these components is dependent on W1 (component 2) which is not correlated with the components on either side of it, namely the zinc-bound water and the correlated cluster around His64. The lack of correlation between the three components helps explain the reduced probability of forming water wires (when compared to the Y7F mutant) which is further supported by the water occupancy, water wire formation and water wire lifetime data reported earlier.
In contrast, the Y7F enzyme possesses two components of water molecule correlation instead of the three seen in the WT. The first component consists of the zinc-bound water and W1 while the second component consists of W2, W3a, WD and His64. In the second correlated component W2 is correlated to His64 and W3a while W3a is correlated to WD. The strong correlation between W2 and His64 suggests that it is the final water in the water wire and mediates the final step in proton transport to His64 in Y7F. While there is a weak correlation between W3a and His64 in Y7F, the spatial orientation revealed by the occupancy data precludes W3a from acting as the final proton transfer water, which may account for the relatively small probability of forming water wires of size 4. The ability of the Y7F mutant to form a greater number of smaller water wires and for a longer time than WT can be attributed to the Y7F mutant having two components of correlation instead of three. Due to the fact that W1 is now correlated with the zinc-bound water, completing the continuous hydrogen bonded water wire (in Y7F) only relies on the two components hydrogen bonding to one another. That is to say that the Y7F mutant has eliminated W1 as being the uncorrelated connection between the zinc-bound water and the correlated component around His64. In addition to the elimination of W1 as the connector, the underlying correlation of the component surrounding His64 has changed from that of the WT. The correlation between the W2, W3a, WD and His64 component indicates that W3a stabilizes the water wire (W2) in the Y7F mutant instead of the hydroxyl group of Tyr7 in WT. The movement of Phe7 towards the active site cavity wall (in order to interact with other hydrophobic residues) has allowed W3a to occupy this new space and interact via hydrogen bonding with WD and W2 (Figure 4). Due to the flexibility of the interactions in this hydrogen bonded network W3a has an increased ability to adjust to the fluctuations of W2 and His64, thereby allowing for greater accommodation of the inward orientation of His64. This flexibility also has the effect of relieving the compression of the W1 water (seen in the WT) which is believed to be the cause of its mobility.
In the WT enzyme, WD stabilizes W3a, allowing it to act as the final member of the water wire. WD's correlation with both residue 106 and residue 7 is indicative of hydrogen bonding linking these two parts of the active site. This association orients and stabilizes residue 7, which in turn hydrogen bonds with W3a. By contrast, in the Y7F system, W2 is the final member of the water wire in the Y7F mutant and the stability of the water wire is reflected in its strong correlation with His64. The bottom part of the active site is not directly involved in the stabilization of the water wire and as a result, when the enzyme undergoes isotropic expansion and contraction, the top and bottom of the active site show anti-correlated movement, observed between His64 and Tyr7. WD is most highly correlated with residue 106, suggesting that it will tend to move with the bottom part of the active site. By contrast, W2 is most highly correlated to His64, suggesting that it will tend to move with the top part of the active site. While W2 and WD both have weak correlations with W3a, they are only indirectly linked with each other. For this reason, even though WD and W2 are considered part of the same water wire component based on their hydrogen bonding with W3a, they show weakly anti-correlated movements because their correlations to distinct regions of the enzyme are larger than their indirect correlation via W3a. Part of the explanation for longer water wire lifetimes in the Y7F mutant may be that the enzyme can expand and contract without placing strain on the water wire responsible for proton transport.
The water analysis of the intramolecular water wire suggests that the Y7F's ZnH2O2+-His system with His64 in the “in” orientation increases the lifetime of all cluster sizes and substantially increases the probability of forming water clusters of size 3. Given that clusters of size 3 in the PT event of HCA II are believed to be almost barrier-less and proceed in a concerted fashion, one would expect Y7F to be more conducive to PT than the WT enzyme. In addition to the Y7F forming more water wires overall than the WT (Supplemental Figure 1), the Y7F mutant possesses longer water wire lifetimes. Occupancy and correlation analysis have given insight into how the Y7F mutant increases the formation and stabilization of smaller water wires. It is evident that increased correlation of His64 with the water wire and the stabilization of the water wire by W3a allow the Y7F mutant to both form smaller water wires more frequently and for a longer time. It is this ability of the Y7F mutant to form and subsequently use a concerted 3 water wire cluster in its rate limiting PT event that is hypothesized here to elevate its turnover. Indeed, experimental data does show an increase in the turnover number of Y7F.1
Orientation of His64
The size and lifetime of the intramolecular water wire is intimately linked to the orientation of His64. As indicated previously an “in” orientation of His64 favors smaller water wires and therefore, presumably, a faster turnover number. In an effort to understand the complete effect of the Y7F mutation on catalysis, the orientation of His64 was also investigated. The 1 dihedral angle describes the “in” and “out” orientations of His64 in HCA II while the χ2 dihedral angle describes the orientation of the imidazole ring. Both dihedral angles are important in describing the spatial orientation of His64 and therefore its ability to participate in the intramolecular water wire. Comparing the χ1 and χ2 dihedral angle observed in the WT and the Y7F mutant at different stages in the catalytic cycle of HCA II reveals how different enzyme environments stabilize the orientation of His64. To evaluate the stability of His64 at various χ1 dihedral angles a PMF was evaluated, F(î÷1). The free energy profile F(î÷1) is capable of indicating the relative stability of His64 as the χ1 dihedral angle changes, and indicates the barrier to rotation, Fb, between the “in” and “out” orientations. The F(î÷1) may be integrated according to the following equation (Equation 7) to yield “in” and “out” probabilities
| (7) |
where P is the probability, ξ is the reaction coordinate, a and b represent the subset range of configurations, F(î) is the free energy, and where kB is Boltzmann's constant and T is temperature. The subset range for the “out” and “in” orientation are −180° ≤ ÷1 ≤ Transition State (TS) and TS ≤ ÷1 ≤ 180°, respectively. The χ2 dihedral angles dependence on the χ1 dihedral angle is best visualized in a two-dimensional (2-D) PMF, F(î÷1,î÷2), which is constructed using the method described by Alen et al.54
The free energy profile for the rotation of His64 about the χ1 dihedral for the Y7F's ZnH2O2+-His and Y7F's ZnOH+-HisH+ systems is shown in Figure 7, while the 2-D PMF for the χ2 dihedral angles is given in Figure 8. From Figure 7 it is evident that the Y7F mutation significantly changes the “in” and “out” probabilities of His64 for both the ZnH2O2+-His and the ZnOH+-HisH+ systems when compared to the WT HCA II.12 While the WT system predicted His64 to occupy the “in” orientation (86%) for the ZnH2O2+-His systems, which is in close agreement with the crystal structure (80%),12,41 the Y7F system indicates an almost equal distribution of “in” and “out” (41% and 59%, respectively), which is in contrast to recent crystal structure data.1 This discrepancy may be due to the MD simulations sampling a greater region of conformational space that includes the conformational space sampled by the X-ray data (for further discussion see Supplemental Material).
Figure 7.
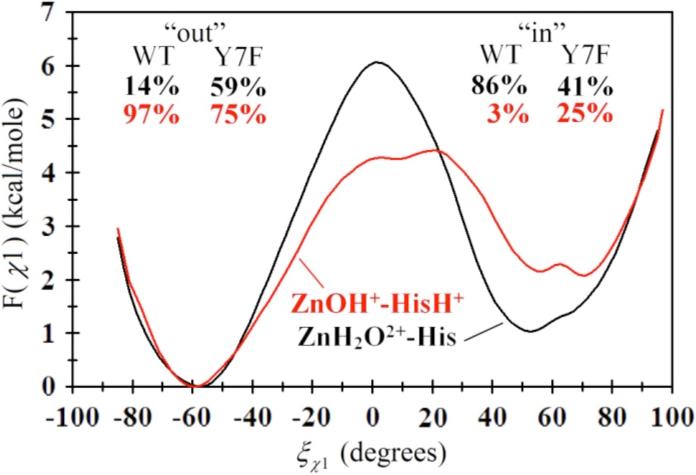
Free energy profile (PMF) for the rotation of His64 about the χ1 dihedral angle in the ZnH2O2+-His system (black) and the ZnOH+-HisH+ system (red) for the Y7F mutant of HCA II. The subset percentages correspond to the “in” and “out” percentages as determined by integrating F(î÷1) for both the WT (curves not shown, from Ref. 12) and the Y7F mutant.
Figure 8.
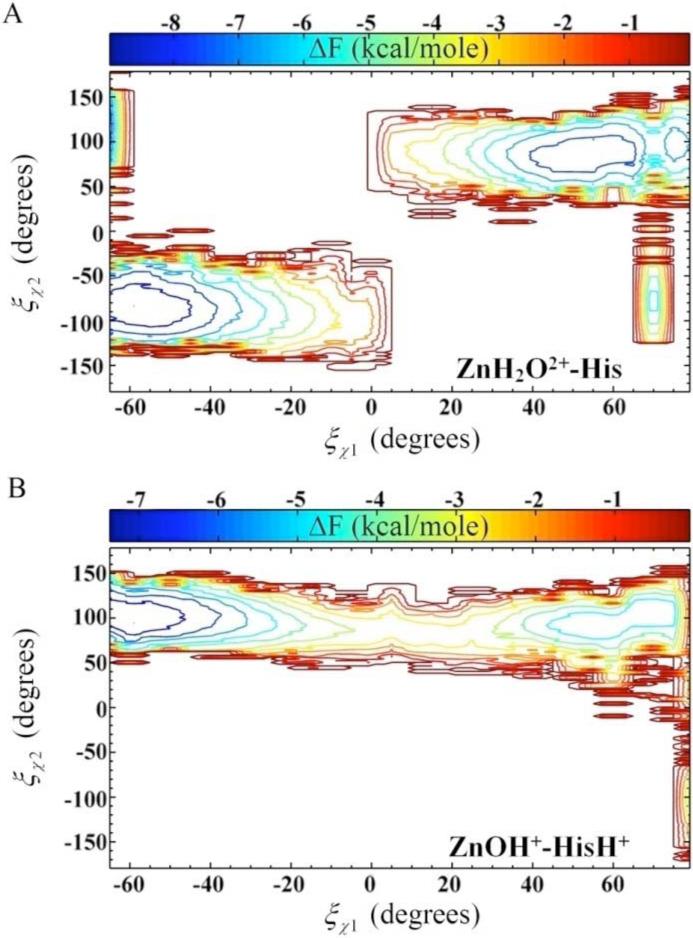
Two-dimensional free energy profile (PMF) for the rotation about the dihedral angles as a function of χ1 and χ2. (A) F(î÷1,î÷2) for the ZnH2O2+-His system of the Y7F mutant of HCA II. (B) F(î÷1,î÷2) for the ZnOH+-HisH+ system of the Y7F mutant of HCA II.
To understand how the HCA II enzyme stabilizes the orientation of His64, occupancy data for solvating waters (Figure 4) and the χ2 dihedral angle of His64 (Figure 7) were used to analyze the primary factors that influence the orientation and stabilization of His64. From the occupancy data it appears that the stabilization of His64 in the ZnH2O2+-His system is due to several competing affects (the ZnOH+-HisH+ systems are not reported here due to the instability of the “in” orientation and the similarities between the “out” orientations). One common effect between WT and the Y7F mutant is the charge rearrangement between the ZnH2O2+-His and the ZnOH+-HisH+ systems. Due to the similarity of this charge rearrangement between the systems of interest its direct effect will be assumed to cancel and therefore will not be discussed further. The indirect effect of the charge rearrangement however is seen in the solvation environment and the enzyme fluctuations which accounts for the other effects believed to be crucial in the stabilization of His64. The solvation environment stabilizing His64 is composed of the intramolecular water wire and a hydrogen bonded network of waters near N67 and N62 that also help to stabilize His64, Figure 4. Waters WA and WB are the two waters near Asn62 and Asn67 which are likely involved in stabilizing the fluctuations of Asn62 and Asn67 in addition to creating a hydrogen bonding network with His64. It is the ability of these waters to stabilize/destabilize the loop region between Asn62, His64 and Ans67 that may play a major role in the observed orientation of His64. In addition to WA and WB the orientation of Asn62 is also thought to be a factor in determining the observed orientation of His64.15 From all the simulation data it was observed that when His64 occupied the “in” orientation Asn62's functional group was pointing “out” of the active site, and when His64 occupied the “out” orientation Asn62's functional group was pointing “in” toward the active site (Figure 4). While direct interconversion was not observed during the simulation, these alternate conformational states indicate that in addition to the hydrogen bonding network a steric effect is at play between Asn62 and His64 that determines their respective orientations. The interplay of these factors may account for the orientational preference of His64 in different enzyme environments.
In the WT ZnH2O2+-His system, His64 prefers to occupy the “in” orientation, due to favorable solvation and loop region affects. The hydrogen bonded network between WD, W3a and Tyr7 create a stable solvation environment for His64 when in the “in” orientation. In addition to the intramolecular water wire, WA hydrogen bonds to N67 and WB while WB is hydrogen bonding to His64 and N62. This hydrogen bonding network creates a stable solvation environment for His64 in the “in” orientation in addition to stabilizing the loop region. The loop region between Asn62, His64 and Asn67 is now connected via a hydrogen bonded network through the WA and WB waters. This hydrogen bonded network has the effect of reducing the loop fluctuation and in particular the fluctuation of Asn62, thereby stabilizing Asn62's outward orientation (occupancy data not shown). In contrast when viewing the “out” orientation of His64 many of these favorable hydrogen bonds are eliminated. When His64 occupies the “out” orientation W2 and W3a of the intramolecular water wire are no longer as stabilized (Figure 4). Waters WA and WB are shifted as Asn62 now points toward the active site. While WA appears to retain most of its occupancy WB has a noticeable reduced occupancy. The elimination of the stable WA and WB hydrogen bonded network present when His64 was in the “in” orientation, now allows for a greater mobility of Asn67 and Asn62. This enhanced mobility creates a greater fluctuation in the loop region and an overall destabilization of the “out” orientation for His64. This increased mobility of Asn62 may increase its probability of reorienting to face out of the active site and subsequently cause His64 flip to the “in” orientation. The χ2 dihedral angle for the WT ZnH2O2+-His system reveals a limited sampling of orientational space when in the “in” orientation and a greater sampling of orientational space when in the “out” orientation.12 This mobility of the imidazole ring complements the solvation and fluctuation analysis which together indicate a stabilized His64 when in the “in” orientation and a destabilized (more mobile) His64 when in the “out” orientation. These analyses help to explain why the WT ZnH2O2+-His system prefers the “in” orientation over the “out” orientation. Similar occupancy analysis for the ZnOH+-HisH+ system is not possible due to inability to achieve a stable “in” orientation for His64; this inability to achieve a stable “in” trajectory is not unreasonable given that the enzyme only samples the “in” orientation 3% of the time.
In the Y7F ZnH2O2+-His system, His64 has an almost 50/50 distribution between the “in” and “out” orientation which, like the WT system, can be explained by the solvating water and the flexibility of the loop region. When His64 is in the “in” orientation the intramolecular water wire (W1 and W2) are very stable and therefore have a strong stabilizing effect on His64. Waters WA and WB behave very similar to the WT systems WA and WB with the exception that these waters in the Y7F mutant are not as stable as they are in the WT. The reduced stability of WA and WB is believed to destabilize the loop region when compared to the WT system. When His64 is in the “out” orientation there are a number of significant differences as compared to the WT system. In the “out” orientation WB is more localized than in the WT, WA has slightly reduced stability, and there appears to be more water density around the loop region (50% iso-surface). This increased water density and the increased localization of WB is believed to stabilize the loop region when compared to the WT. Asn62 and Asn67 are also less mobile in the Y7F “out” orientation than in the WT “out” orientation (data not shown). Inspection of the χ2 dihedral angle also shows significant differences compared to the WT. When His64 is in the “in” orientation a limited orientational space is sampled by the imidazole ring of His64, similar to the WT. Yet, when His64 is in the “out” orientation the orientational freedom seen in the WT is not present. Instead a reduced sampling of orientational space is seen. This would indicate that the destabilizing effects in the WT for the “out” orientation are eliminated in the Y7F mutant. These combined affects have the effect of increasing the “out” orientation of His64 in the Y7F mutant as seen by the integration of F(î÷1).
Taking the “in” and “out’ probabilities of His64 in addition to the water wire formation probabilities it is possible to compare the overall water wire formation for WT and Y7F. Using a linear combination of the probabilities for His64 “in|out” and the probability of forming water wires of different sizes for “in|out” a total probability of water wire formation can be estimated as
| (8) |
where is the total probability of forming a water wire of size N, is the probability of His64 occupying the “in” or “out” orientation, is the probability of forming a water wire of size N when His64 is in the “in” or “out” orientation. For the WT ZnH2O2+-His system there is a total probability of 11 ± 5% and 39 ± 5% for forming size 3 and 4 water wires respectively. For the Y7F ZnH2O2+-His system there is a total probability of 29 ± 3% and 19 ± 6% for forming size 3 and 4 water wires respectively. Once again analysis of the ZnOH+-His+ systems yielded very similar probabilities for both WT and Y7F. Even though the Y7F His64 orientation is predicted to occupy around a 50/50 distribution of “in” and “out” the enzyme still forms significantly more water wires of size 3. In conjunction with the total probability of water wire formation the increased lifetime of water wires in the Y7F mutant increases the probability of the PT event utilizing the water wires present. Combining the information from F(î÷1), the water cluster size data, and the water cluster lifetime data indicates that the Y7F mutant will have an elevated turnover number due to the increased probability of forming water clusters of size 3. This prediction will be verified in the future using the multi-state empirical valence bond method for including explicit proton solvation and transport in the MD simulations.14,24
4. Conclusions
From the current investigation it is clear that Tyr7 has an important role in stabilizing the orientations of His64 and scaffolding the intramolecular water wire in the WT enzyme. Interestingly, the mutant Y7F, by altering the stability of His64 and the active site waters, has the overall effect of creating a more efficient enzyme. The Y7F mutant creates an environment that changes the relative distribution of His64 such that while in the ZnH2O2+-His protonation state the His64 in the “in” to “out” orientation is almost equally probable. Evaluation of the water wire distribution and water wire lifetime data indicates that the Y7F mutant also has an increased ability to form water wires of size 3 and an increased water wire lifetime when compared to the WT enzyme. This stabilization of the water wires is due to an increased occupancy of critical waters in forming a water wire of size 3. Correlation analysis further indicates the Y7F mutant sets up correlated movements that are more conducive to forming and stabilizing smaller water wires than those seen in WT. Taking into account the “in/out” probabilities and the water wire distributions and lifetimes it is evident that the Y7F mutant creates more size 3 water wires, which can conduct protons in with a lower barrier to reaction, than the WT enzyme. Therefore, the Y7F mutant is an enzyme that is expected to have an elevated catalytic turnover when compared to the WT enzyme, as observed experimentally.1
The ZnH2O2+-His “out” and the ZnOH+-HisH+ “out” configurations for the Y7F mutant, while different in some features from the analogous WT systems, do not differ to the same degree as the corresponding ZnH2O2+-His “in” systems. Therefore it is reasoned that the “out” orientations do not significantly contribute to the experimentally observed catalytic turnover difference between the WT and the Y7F HCA II enzyme.
In addition to His64's orientation and the water wire formation data there are also differences in the enzyme active site solvation environment as seen by water occupancy and g(RZn-OW) data. This data shows that in the WT's ZnH2O2+-His “in” orientation the first solvation shell is compressed toward the catalytic zinc, while in the “out” orientation for both the ZnH2O2+-His and the ZnOH+-HisH+ systems this compression is relieved. The compression is likely due to the solvation interaction of His64, Tyr7, and the W3a water. In the Y7F mutant none of the g(RZn-OW) plots exhibit this compression of the solvent environment. This relieving of solvent compression may also play a role in the elevated turnover number of the Y7F mutant.
Mutations such as Y7F that can alter water structure for enhanced proton transport have broad implications for protein engineering. The concept of employing an amino acid change to modulate water structure (and thus catalytic activity) via indirect, long-range coupling is an area that has not been widely explored in the context of enzyme catalyzed reactions. Given that most enzymatic reactions occur in aqueous solvent, this approach may well find applications in other enzyme systems, offering protein engineers with yet another route by which the properties of enzymes may be tuned for specific purposes. Moreover, modulating PT by altering water structure could also be investigated in the context of synthetic PT constructs. These and other implications of the present work will be explored in future research.
Acknowledgement
This work was supported by the National Institutes of Health (R01-GM53148). The computational resources for this project have been provided in part by the National Institutes of Health (Grant # NCRR 1 S10 RR17214-01) on the Arches Metacluster, administered by the University of Utah Center for High Performance Computing.
Footnotes
Publisher's Disclaimer: This PDF receipt will only be used as the basis for generating PubMed Central (PMC) documents. PMC documents will be made available for review after conversion (approx. 2−3 weeks time). Any corrections that need to be made will be done at that time. No materials will be released to PMC without the approval of an author. Only the PMC documents will appear on PubMed Central -- this PDF Receipt will not appear on PubMed Central.
Supplemental Data Available. Data on the total continuous hydrogen bonded water wire probabilities. Discussion of the His64 orientational discrepancies between X-ray data and MD simulations. Complete Ref. 51. This material is available free of charge via the Internet at http://pubs.acs.org.
Supplementary Material
References
- 1.Fisher SZ, Tu C, Bhatt D, Govindasamy L, Agbandje-McKenna M, McKenna R, Silverman DN. Biochemistry. 2007;46:3803–3813. doi: 10.1021/bi602620k. [DOI] [PubMed] [Google Scholar]
- 2.Lindskog S, Behravan G, Engstrand C, Forsman C, Jonsson B, Liang Z, Ren X, Xue Y. In: Carbonic Anhydrase--From Biochemistry and Genetics to Physiology and Clinical Medicine. Botre F, Gros G, Storey BT, editors. VCH; Weinheim, Germany: 1991. pp. 1–13. [Google Scholar]
- 3.Nair SK, Christianson DW. J. Am. Chem. Soc. 1991;113:9455–9458. [Google Scholar]
- 4.Christianson DW. Adv. Protein Chem. 1991;42:281–335. doi: 10.1016/s0065-3233(08)60538-0. [DOI] [PubMed] [Google Scholar]
- 5.Lesburg CA, Christianson DW. J. Am. Chem. Soc. 1995;117:6838–6844. [Google Scholar]
- 6.Erikson AE, Jones AT, Liljas A. Proteins. 1988;4:274–282. doi: 10.1002/prot.340040406. [DOI] [PubMed] [Google Scholar]
- 7.Liljas A, Kannan KK, Bergsten PC, Waara I. Nature. 1972;235:131–137. doi: 10.1038/newbio235131a0. [DOI] [PubMed] [Google Scholar]
- 8.Kannan KK, Ramanadham M, Jones TA, Ann NY. Acad. Sci. 1984;429:49–60. doi: 10.1111/j.1749-6632.1984.tb12314.x. [DOI] [PubMed] [Google Scholar]
- 9.Håkansson K, Carlsson M, Svensson LA, Liljas A. J. Mol. Biol. 1992;227:1192–1204. doi: 10.1016/0022-2836(92)90531-n. [DOI] [PubMed] [Google Scholar]
- 10.Duda D, Tu C, Qian M, Laipis P, Agvandje-McKenna A, Silverman DN, McKenna R. Biochemistry. 2001;40:1741–1748. doi: 10.1021/bi002295z. [DOI] [PubMed] [Google Scholar]
- 11.Duda D, Govindasamy L, Agbandje-McKenna, Tu C, Silverman DN, McKenna R. Acta. Crystallogr. Sect. D. 2003;59:93–104. doi: 10.1107/s0907444902019455. [DOI] [PubMed] [Google Scholar]
- 12.Maupin CM, Voth GA. Biochemistry. 2007;46:2938–2947. doi: 10.1021/bi062170f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fisher Z, Hernandez, Prada JA, Tu C, Duda D, Yoshioka C, An H, Govindasamy L, Silverman DN, McKenna R. Biochemistry. 2005;44:1097–1105. doi: 10.1021/bi0480279. [DOI] [PubMed] [Google Scholar]
- 14.Swanson JMJ, Maupin CM, Chen H, Petersen MK, Xu J, Wu Y, Voth GA. J. Phys. Chem. B. 2007;111:4300–4314. doi: 10.1021/jp070104x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roy A, Taraphder S. J. Phys. Chem. B. 2007;111:10563–10576. doi: 10.1021/jp073499t. [DOI] [PubMed] [Google Scholar]
- 16.Silverman DN, McKenna R. Acc. Chem. Res. 2007;40 doi: 10.1021/ar7000588. [DOI] [PubMed] [Google Scholar]
- 17.Marino S, Hayakawa K, Hatada K, Benfatto M, Rizzello A, Maffia M, Bubacco L. Biophysical Journal. 2007;93:2781–2790. doi: 10.1529/biophysj.107.107540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schimara H, Yoshida T, Shibata Y, Shimizu M, Kyogoku Y, Sakiyama F, Nakazawa T, Tate S, Ohki S, Kato T, Moriyama H, Kishida K, Tano Y, Ohkubo T, Kobayashi Y. The Journal of Biological Chemistry. 2007;282:9646–9656. doi: 10.1074/jbc.M609679200. [DOI] [PubMed] [Google Scholar]
- 19.Silverman DN, Lindskog S. Acc. Chem. Res. 1988;21:30–36. [Google Scholar]
- 20.Davies DR. Ann. Rev. Biophys. Biophys. Chem. 1990;19:189–215. doi: 10.1146/annurev.bb.19.060190.001201. [DOI] [PubMed] [Google Scholar]
- 21.Fersht A. Enzyme Structure and Mechanism. W. H. Freeman and Sons; New York: 1985. [Google Scholar]
- 22.Grotthuss C. J. T. d. Ann. Chim. (Paris) 1806;58:54. [Google Scholar]
- 23.Lapid H, Agmon N, Petersen MK, Voth GA. J. Chem. Phys. 2004;122:041506. doi: 10.1063/1.1814973. [DOI] [PubMed] [Google Scholar]
- 24.Voth GA. Acc. Chem. Res. 2006;39:143–150. doi: 10.1021/ar0402098. [DOI] [PubMed] [Google Scholar]
- 25.Maren TH. Physiol. Rev. 1967;47:595–781. doi: 10.1152/physrev.1967.47.4.595. [DOI] [PubMed] [Google Scholar]
- 26.Tashian RE. BioEssays. 1989;10:186–192. doi: 10.1002/bies.950100603. [DOI] [PubMed] [Google Scholar]
- 27.Erikson AE, Kylsten PM, Jones TA, Liljas A. Proteins: Structure, Function, and Genetics. 1988;4:283–293. doi: 10.1002/prot.340040407. [DOI] [PubMed] [Google Scholar]
- 28.Lindskog S, Coleman JE. Proc. Natl. Acad. Sci. U.S.A. 1973;70:2505–2508. doi: 10.1073/pnas.70.9.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lindskog S. In: Zinc enzymes. Spiro TG, editor. John Wiley & Sons; New York: 1983. pp. 78–121. [Google Scholar]
- 30.Silverman DN, Vincent SH. CRC Crit. Rev. Biochem. 1983;14:207–255. doi: 10.3109/10409238309102794. [DOI] [PubMed] [Google Scholar]
- 31.Tu C, Silverman DN, Forsman C, Jonsson B-H, Lindskog S. Biochemistry. 1989;28:7913–7918. doi: 10.1021/bi00445a054. [DOI] [PubMed] [Google Scholar]
- 32.Qian M, Tu C, Earnhardt N, Laipis PJ, Silverman DN. Biochemistry. 1997;36:15758–15764. doi: 10.1021/bi972081q. [DOI] [PubMed] [Google Scholar]
- 33.Toba S, Colombo G, Merz KM., Jr. J. Am. Chem. Soc. 1999;121:2290–2302. [Google Scholar]
- 34.Lu D, Voth GA. Proteins. 1998;33:119–134. [PubMed] [Google Scholar]
- 35.Lu D, Voth GA. J. Am. Chem. Soc. 1998;120:4006–4014. [Google Scholar]
- 36.Cui Q, Karplus K. J. Phys. Chem. B. 2003;107:1071–1078. [Google Scholar]
- 37.Riccardi D, Schaefer P, Yang Y, Yu H, Ghosh N, Prat-Resina X, Konig P, Li G, Xu D, Guo H, Elstner M, Cui Q. J. Phys. Chem. B. 2006;110:6458–6469. doi: 10.1021/jp056361o. [DOI] [PubMed] [Google Scholar]
- 38.König PH, Ghosh N, Hoffmann M, Elstner M, Tajkhorshid E, Frauenheim Th., Cui Q. J. Phys. Chem. A. 2006;110:548–563. doi: 10.1021/jp052328q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Riccardi D, Konig P, Prat-Resina X, Yu H, Elstner M, Frauehneim T, Cui Q. J. Am. Chem. Soc. 2006;128:16302–16311. doi: 10.1021/ja065451j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Riccardi D, Konig P, Guo H, Cui Q. Biochemistry. 2007;47:2369–2378. doi: 10.1021/bi701950j. [DOI] [PubMed] [Google Scholar]
- 41.Fisher SZ, Maupin CM, Govindasamy L, Budayova-Spano M, Govindasamy L, Tu C, Agbandje-McKenna M, Silverman DN, Voth GA, McKenna R. Biochemistry. 2007;46:2930–2937. doi: 10.1021/bi062066y. [DOI] [PubMed] [Google Scholar]
- 42.Rod TH, Radkiewicz JL, Brooks CL., III PNAS. 2003;100:6980–6985. doi: 10.1073/pnas.1230801100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schaefer P, Riccardi D, Cui Q. J. Chem. Phys. 2005;123:014905. doi: 10.1063/1.1940047. [DOI] [PubMed] [Google Scholar]
- 44.Guex N, Peitsch MC. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 45.Day TJ, Soudackov AV, Cuma M, Schmitt UW, Voth GA. J. Chem. Phys. 2002;117:5839–5849. [Google Scholar]
- 46.Wang J, Cieplak P, Kollman PA. J. Comput. Chem. 2000;21:1049–1074. [Google Scholar]
- 47.Kast SM, Nicklas K, Bär H-J, Brickmann J. J. Chem. Phys. 1994;100:566–576. [Google Scholar]
- 48.Hummer G, Pratt LR, Garcia AE. J. Phys. Chem. 1996;100:1206–1215. [Google Scholar]
- 49.Humphrey W, Dalke A, Schulten K. J. Molec. Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 50.Reinhard F, Grubmüller H. J. Chem. Phys. 2007;126:014102. doi: 10.1063/1.2400220. [DOI] [PubMed] [Google Scholar]
- 51.Case DA, et al. 7 ed. University of California; San Francisco.: 2002. [Google Scholar]
- 52.Kumar S, Bouzida D, Swendsen RH, Kollman PA, Rosenberg JM. J. Comput. Chem. 1992;13:1011–1021. [Google Scholar]
- 53.Roux B. Comput. Phys. Commun. 1995;91:275–282. [Google Scholar]
- 54.Allen TW, Andersen OS, Roux B. Proc. Natl. Acad. Sci. 2004;101:117–122. doi: 10.1073/pnas.2635314100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.