Summary
The gaseous plant hormone ethylene plays important roles in plant growth and development. Recent discoveries have expanded our linear view of ethylene signaling by revealing an elaborate signaling network with multiple regulatory circuits. At the membrane, the ethylene receptors form heteromeric and higher order complexes providing enhanced sensitivity and fine-tuning of signaling. Ethylene sensitivity is further enhanced by the rapid degradation of ethylene receptors upon ethylene binding and by dependence on a novel protein REVERSION-TO-ETHYLENE SENSITIVITY1 (RTE1)/GREEN-RIPE (GR). In the nucleus, EIN3-BINDING F-BOX1 and 2 (EBF1/2) coordinately control 26S proteasome degradation of the critical transcription factors EIN3 and EIL1. EBF1/2 expression is repressed by ETHYLENE-INSENSITIVE5 (EIN5), which encodes the exoribonuclease XRN4. Additionally, EIN3 possesses two mitogen-activated protein kinase (MAPK) phosphorylation sites that have opposing effects on EIN3 stability.
Introduction
Ethylene is a gaseous plant hormone that plays a key role in many processes, including seed germination, leaf senescence, fruit ripening, abscission and responses to abiotic and biotic stresses [1]. The molecular dissection of ethylene signal transduction began with genetic screens based on the well-documented triple response phenotype of ethylene-treated etiolated Arabidopsis seedlings [2]. Initial studies uncovered a linear framework for the ethylene-signaling pathway, leading from ethylene perception at the membrane to transcriptional activation in the nucleus.
Briefly, ethylene is perceived by a family of membrane-bound receptors [2,3] having similarity to two-component histidine protein kinase receptors and derived from a cyanobacterial origin [4**,5]. Each receptor has an N-terminal membrane-spanning domain that binds ethylene with a copper cofactor [6] provided by the RAN1 copper transporter [7]. Although the receptors display protein kinase activity in vitro, their biochemical signaling mechanism is unknown [2,3]. Genetically, the receptors are negative regulators of ethylene signaling [8,9*]; in the absence of ethylene, the receptors repress downstream ethylene responses through the Raf-like protein kinase CTR1 [10] and, when ethylene is bound, the receptors no longer repress ethylene responses [2,3]. CTR1 negatively regulates ethylene responses by repressing the positive regulator EIN2 [11], which relays the ethylene signal by an unknown mechanism to the transcription factors EIN3 and EIL, which in turn activate the ERF1 transcription factor [12]. ERF1 activates transcription of ethylene responsive genes such as PDF1.2 [12]. EIN3 and EIL1 are constitutively expressed and controlled by protein degradation through a 26S proteasome-dependent pathway [13–15]. The components and overall mechanisms of ethylene signaling are conserved among dicots and monocots [2, 16–18].
Recent advances have expanded our linear view of the ethylene-signaling pathway into an increasingly complex signaling system that includes multiple pathways of regulation and feedback (Figure 1). In this review, we focus on discoveries in ethylene signaling reported in the last two years. These latest findings have revealed new levels of regulation, particularly with respect to the ethylene receptors and the EIN3/EIL1 transcription factors. Due to space limitations, we do not discuss ethylene crosstalk with other pathways or descriptions of ethylene responses, which can be found in recent reviews on ethylene signaling [2], ethylene biosynthesis [19] and crosstalk [20–23].
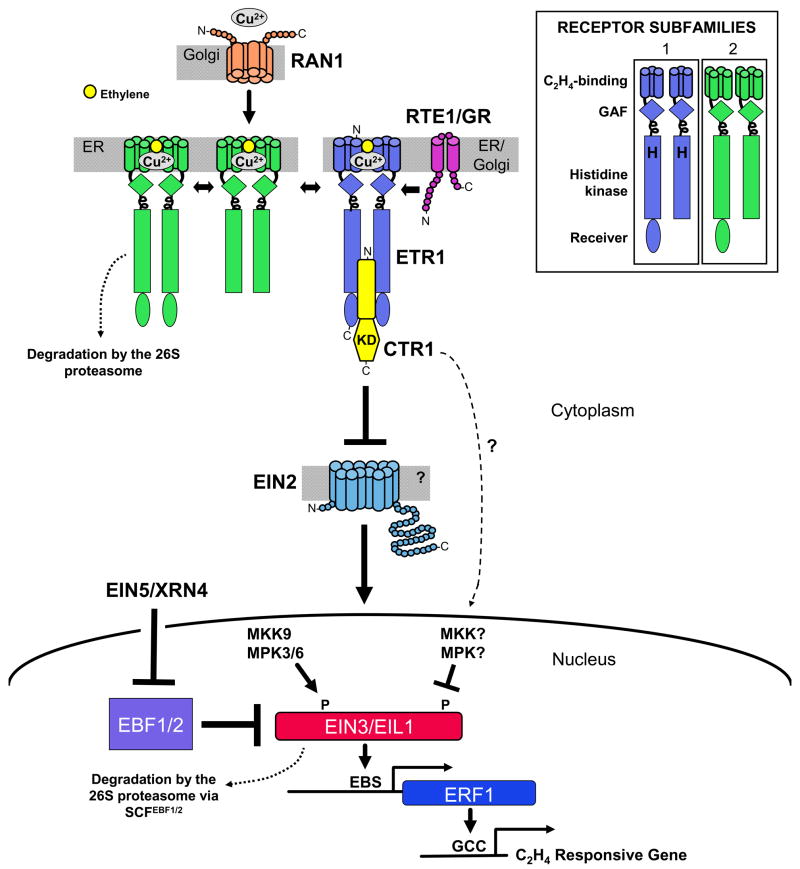
Figure 1. Current model of the ethylene-signaling pathway.
Ethylene is perceived at the endomembranes by a family of receptors (see inset) that share similarity to prokaryotic two-component histidine kinase receptors [3]. The receptors form higher order complexes of homodimers and heterodimers [29*; Gao et al., unpublished]. RAN1 is a P-Type ATPase copper transporter homolog [7] in the Golgi membrane [52] that provides the copper cofactor for ethylene binding. RTE1/GR is a novel protein [32**,34**] in the Golgi and ER [30*] that positively regulates ETR1 receptor signaling in Arabidopsis [32**,33*]. In the absence of ethylene binding, the receptors repress ethylene responses by signaling through CTR1, a Raf-like MAPKK kinase that negatively regulates responses [10]. When ethylene binds to the receptors, receptor signaling is inactivated, causing the CTR1 kinase domain (KD) to be inactivated, allowing downstream signaling to proceed through EIN2, which has similarity to theNramp family of metal ion transporters [11]. EIN2 is a positive regulator of ethylene responses, and loss of EIN2 renders the plant completely insensitive to ethylene [11]. EIN2 regulates a transcriptional cascade initiated by EIN3 and EIL1, two members of a small family of DNA-binding proteins [41]. EIN3 activates ethylene responses by binding to the EIN3-binding site (EBS) in the promoter of ERF1 [12]. ERF1 encodes a transcriptional activator that binds to the GCC-box in the promoters of several ethylene-responsive genes. A key regulatory step in the pathway is the degradation of EIN3 and EIL1 by the 26S proteasome-dependent pathway, mediated by an SCFEBF1/2 E3 ligase complex containing F-Box proteins EBF1 and EBF2 [13–15,42,43**]. Stability of EIN3 is promoted by phosphorylation of T174 through a MAP kinase cascade consisting of MKK9 signaling to MPK3/6, whereas degradation of EIN3 is promoted by phosphorylation on T592, possibly through a separate MAP kinase cascade involving CTR1[44**]. Repression of EBF1 and EBF2 transcription is mediated by an exoribonuclease encoded by EIN5/XRN4 [47**,48**].
Inset: Each receptor has a transmembrane (TM) N-terminal domain, which binds ethylene with a copper cofactor and localizes the receptor to the endomembranes [3]. Subfamily I receptors have three TM domains. Subfamily II receptors have a fourth TM domain, which might serve as a signal sequence. In the cytosol, the receptor contains a GAF domain adjacent to a coiled coil region followed by a histidine kinase (HK)-like domain. In some receptors, the HK domain is fused to a receiver domain, which appears to have a subtle role in signaling [3]. The GAF domain may play a role in transmitting the signal between the receptors [28,29*; Gao et al., unpublished], as well as from the N-terminal domain to the HK and receiver. Although the HK domain is required for proper signaling, protein kinase activity does not play a major role [3]. Subfamily I receptors have autokinase activity on the histidine (H) [3]. Subfamily II receptors have degenerate HK domains and possess serine/threonine protein kinase activity [3]. (Subfamily I receptor ERS1 in Arabidopsis is capable of both activities [3].)
Multiple modes of ethylene receptor regulation
Subfamily I versus Subfamily II receptors
The ethylene receptors fall into two subfamilies as shown in Figure 1. Subfamily I receptors have three amino-terminal transmembrane domains, while subfamily II receptors have four [3]. Subfamily I receptors possess histidine kinase activity, whereas subfamily II receptors possess serine/threonine kinase activity. Arabidopsis has five ethylene receptors, two of which (ETR1 and ERS1) are members of subfamily I. Tomato has six ethylene receptors, three of which belong to subfamily I [24].
Individual receptor isoforms contribute differentially to overall signaling. In Arabidopsis, subfamily I plays a larger role than subfamily II and cannot be replaced by subfamily II members [2,3]. In tomato, subfamily II receptors (LeETR4 and LeETR6) play the largest role in ethylene responses [24,25**]. To clarify the role of Arabidopsis subfamily I receptors, Qu et al. [9*] analyzed new T-DNA insertion alleles of ETR1 and ERS1. The new ers1-3 mutant exhibits hypersensitivity to ethylene not seen previously for ers1-2, and correspondingly, the subfamily I double null mutant carrying ers1-3 has a stronger phenotype. In addition to having a more severe constitutive triple response, the double mutant is dwarfed with reduced fertility, premature leaf senescence, and novel filamentous structures at the base of the flower. The double mutant still shows a response to ethylene, representing signaling from subfamily II.
Repression of ETR1 signaling by the ETR1 N-terminal domain
The first mutations isolated in the ethylene receptor genes were dominant gain-of-function mutations conferring ethylene insensitivity. Single receptor loss-of-function mutants are essentially indistinguishable from the wild type due to a high degree of functional redundancy among the receptors [3], but null mutant combinations, such as the subfamily I mutant described above, exhibit varying degrees of constitutive ethylene responses, demonstrating that the ethylene receptors are negative regulators of ethylene responses. From this, it was deduced that ethylene perception shuts off, rather than activates ethylene receptor signaling [8]. Interestingly, all of the known dominant mutations reside within or immediately after the N-terminal ethylene-binding domain.
Greater insight into the relationship between the N-terminal domain and signaling domain was recently obtained from a structure/function analysis of ETR1 [4**]. Wang et al. [4**] introduced amino acid substitutions for 37 residues of the ETR1 N-terminal domain and measured the effects on both ethylene binding and signal output. Interestingly, only two of these mutations cause a loss of function with respect to ETR1 signaling. Most of the remaining mutations result in gain-of-function (constitutive) ETR1 signaling. A subset of these constitutive signaling mutations disrupt ethylene binding (and are primarily located in the mid-regions of transmembrane helices I and II), but the others (located near the cytoplasmic ends of transmembrane helices I and III) do not. The latter hold the receptor in an intermediate state, in which ethylene is bound to the receptor but receptor signaling remains on. These results suggest that the predominant function of the N-terminal domain is to inhibit the signaling domain.
Ethylene receptor degradation: enhancing sensitivity
Protein degradation plays a key role both in ethylene biosynthesis through the regulation of ACC synthase [26] and in ethylene signaling through the regulation of EIN3 [13–15]. Recently, it was found that one or more ethylene receptors are subject to regulated degradation as well. Upon ethylene binding, the Arabidopsis ETR2 receptor is targeted for endoplasmic reticulum (ER)-associated degradation by a proteasome-dependent pathway, triggered perhaps by a conformational change in the receptor [27**]. Similarly, tomato LeETR4 and LeETR6 receptors are degraded rapidly in the presence of ethylene, most likely by the 26S proteasome [25**]. The degradation of either LeETR4 or LeETR6 results in early fruit ripening, providing a nice demonstration of how receptor levels can control the sensitization of plant tissues to ethylene [25**]. This degradation helps to explain why the dramatic increase in ethylene receptor gene transcript levels during ripening does not result in the inhibition of ripening.
Whether all ethylene receptors are regulated in this manner remains to be seen. As explained by Chen et al. [27**], the receptors exhibit slow ethylene dissociation kinetics when expressed in yeast cells, but in plants, both rapid (< 30 minute half-life) and slow (> 12 hour half life) release kinetics have been observed. The degradation of certain ethylene receptors might account for this rapid dissociation component.
Interactions among the ethylene receptors
New data indicate that the Arabidopsis ethylene receptors are capable of forming heteromeric complexes, which has implications for signal amplification and fine-tuning of ethylene responses. It was shown previously that ETR1 and ERS1 exist as disulfide-linked homodimers [3], although the cysteine residues required for the disulfide bonds do not appear to be essential for receptor signaling [28]. Recently, protein-protein interactions for all possible ethylene receptor combinations have been observed, both in a membrane recruitment assay in tobacco cells and in the yeast split ubiquitin assay [29*]. Similar interactions, as well as higher order complexes, have been detected by co-purification of epitope-tagged ethylene receptors in Arabidopsis [Gao et al., submitted]. Such interactions may explain why a truncated form of ETR1 (residues 1–349 comprising the ethylene-binding domain, GAF domain and coiled coil domain) is capable of signaling in the presence of other ethylene receptors [3,28]. In several studies, the GAF domain has been indicated as a site of interaction between the receptors [28,29*; Gao et al., submitted]. Because the ethylene receptor genes have overlapping but distinct expression patterns, the composition of the receptor complex is likely to vary among different plant tissues, resulting in a broad range of differential responses to ethylene [29*].
Membrane localization and topology of the ethylene receptors
The ethylene receptors might be present at one or more membrane systems. Arabidopsis ETR1 and ETR2 were previously localized to the ER by sucrose density gradient fractionation [3], and recently, all five Arabidopsis ethylene receptors were localized to the ER when expressed in tobacco leaf epidermal cells [29*]. In contrast, immunohistochemistry showed ETR1 to be primarily at the Golgi apparatus in Arabidopsis roots [30*], whereas tobacco NTHK1 (subfamily II) was reported to be at the plasma membrane in protoplasts [3].
The predicted membrane topology of the receptors was confirmed using CmERS1, a melon subfamily I ethylene receptor that localizes to the ER [31]. The N-terminal domain of CmERS1 spans the membrane three times with the N-terminus on the lumenal side and the C-terminus in the cytosolic side of the ER membrane.
RTE1, a membrane protein that represses ethylene responses through ETR1
Arabidopsis REVERSION-TO-ETHYLENE SENSITIVITY1 (RTE1) encodes a novel integral membrane protein involved in regulating ETR1 function [32**]. RTE1 is highly conserved in plants, animals and some protists, but its molecular function is unknown. The RTE1 protein co-localizes with ETR1 in the Golgi and ER [30*]. RTE1 was identified on the basis of loss-of-function rte1 mutants that suppress the ethylene-insensitive receptor mutation etr1-2 [32**]. rte1 mutants have hypersensitivity to ethylene, similar to the etr1 null mutant [32**,33*]. GREEN-RIPE (GR) is a tomato homolog of RTE1. The dominant Green-ripe (Gr) mutant exhibits inhibition of fruit ripening and other ethylene-insensitive phenotypes [34**]. The Gr mutant carries a deletion in the GR promoter region that causes ectopic over-expression of GR [34**]. In Arabidopsis, ethylene insensitivity from RTE1 over-expression is largely dependent on ETR1 but not on the other ethylene receptors, suggesting that RTE1 has specificity for ETR1 [32**,33*]. In addition, rte1 does not suppress ethylene insensitivity conferred by the etr1-1 allele nor ethylene-insensitive mutations in the other ethylene receptor genes [32**]. RTE1 expression is ethylene-inducible, suggesting that RTE1 is involved in negative feedback on ethylene signaling [32**].
Downstream regulation of ethylene signaling
Regulation of the Raf-like kinase CTR1
Acting downstream of the ethylene receptors is a putative MAPK kinase kinase CTR1, a negative regulator of ethylene responses with similarity to Raf protein kinases [10]. CTR1 has a non-catalytic N-terminal region that physically interacts with subfamily I receptors, resulting in association of CTR1 with the ethylene receptor complex [35,36]. Arabidopsis has one copy of CTR1, while tomato has three to four copies. The tomato subfamily I receptor NEVER-RIPE (NR) interacts in vivo with LeCTR1, LeCTR3, and LeCTR4 [37*]. Apart from interaction with the receptors, little is understood about CTR1 regulation. Most models of CTR1 regulation are based on what is known about Raf kinases. The mammalian Raf-1 kinase can bind phosphatidic acid (PA), leading to translocation of Raf-1 from the cytosol to the plasma membrane for activation [38]. In plants, PA levels increase rapidly in response to biotic and abiotic stresses [39]. Recently, it was found that PA is capable of binding to the CTR1 kinase domain and blocking kinase activity in vitro [40], raising the possibility that PA negatively regulates CTR1.
Post-transcriptional regulation of the EIN3 transcription factor
The deactivation/inhibition of CTR1 kinase activity leads to the downstream activation of two transcription factors, EIN3 and EIL1, which are positive regulators of ethylene signaling [41]. In the absence of ethylene perception, two F-box proteins, EBF1 and EBF2, in a Skp-Cullin-F-box (SCF) E3 ligase complex, target EIN3 and EIL1 for degradation via the 26S proteasome [13-15,42]. While ebf1 and ebf2 single mutants are only slightly hypersensitive to ethylene, the ebf1 ebf2 double mutant displays severe growth arrest [15]. Binder et al. [43**] showed that by introducing ein3 and eil1 mutations into the ebf1 ebf2 background, the severity of the ebf1 ebf2 double mutant is not only alleviated, but the resulting quadruple mutant is ethylene insensitive. Thus, EIN3 and EIL1 are the predominant targets of EBF1/2 in ethylene signaling. Kinetic analyses of ethylene response in ebf1 and ebf2 suggest that EBF1 degrades EIN3/EIL1 prior to ethylene signaling, whereas EBF2 plays a larger role in degrading EIN3/EIL1 after ethylene responses have been activated [43**]. This role of EBF2 is consistent with previous suggestions that an ethylene-induced negative feedback loop is involved in EIN3 regulation by the SCFEBF1/2 complex [13–15,43**]. The distinct but overlapping roles of EBF1 and EBF1 provide fine-tuned post-transcriptional regulation of EIN3 and EIL1, permitting, for example, the rapid induction of ethylene responses during biotic stresses such as pathogen attack.
Another significant finding is that the stability of EIN3 appears to be controlled through two MAPK phosphorylation sites, one required for stabilization of EIN3 and the other involved in its degradation [44**]. Yoo et al. obtained in vivo evidence that two MAPKs, MPK3 and MPK6, phosphorylate EIN3 on Threonine174 to stabilize EIN3. They also showed that MPK3/6 can be activated by MKK9, forming a potential MAPK cascade in ethylene signaling. In contrast, a constitutively active form of CTR1 results in EIN3 degradation, but loses this effect when the second EIN3 phosphorylation site (Threonine592) is mutated [44**]. This suggests that CTR1 can activate a separate MAPK, but whether CTR1 has direct control of a MAP kinase cascade remains an open question.
EIN3 seems to be a point of convergence of the ethylene, glucose and possibly light signaling pathways [41,42,45]. Therefore, multiple protein kinases serving multiple functions in different pathways may be involved in regulating EIN3. For example, MPK6 plays a role in ethylene biosynthesis [46] and possibly EIN3 stabilization [44**].
EIN5, an exoribonuclease that affects EBF1/EBF2 levels
EBF1 and EBF2 are regulated by ETHYLENE-INSENSITIVE5 (EIN5), which acts downstream of CTR1 and encodes the 5′→ 3′ exoribonuclease XRN4 [47**,48**]. EIN5/XRN4 is homologous to yeast exoribonucleases Xrn1p and Rat1p, which function in mRNA and rRNA degradation, respectively. Similar to Xrn1p, EIN5 localizes to the cytoplasm [48] and can rescue yeast xrn1 but not rat1 [47**]. Interestingly, EBF1/2 transcripts, which are ethylene-inducible, accumulate in the ein5/xrn4 background [47**,48**]. Consequently, EIN3 protein does not accumulate in the ein5/xrn4 background in the presence of ethylene and, similarly, many other ethylene-inducible transcripts are expressed at much lower levels relative to wild type [47**,48**]. EIN5/XRN4 does not appear to directly degrade EBF1/2 transcripts, because the half-life of EBF1/2 transcripts in the ein5 mutant background is the same as in the wild type [48**]. Therefore, the accumulation of EBF1/2 mRNAs may be due to increased transcription with EIN5/XRN4 promoting a repressor of EBF1/2 transcription.
EER3 and EER4 may be involved in transcription
Arabidopsis ENHANCED ETHYLENE RESPONSE3 (EER3) and 4 (EER4) were identified through mutants exhibiting enhanced ethylene response in seedlings [50*, 51]. EER3 encodes a previously uncharacterized prohibitin, AtPHB3 [50*]. In mammals, some prohibitins are involved in the formation of transcriptional complexes. EER4 encodes a transcription factor containing a putative TATA-binding factor (TFIID)-interacting domain [51]. In yeast, TFIID binds to the TATA box and initiates formation of the RNA polymerase II complex. An eer3 mutant completely suppresses ein3-1 ethylene-insensitivity, whereas an eer4 mutant partially suppresses ein3-1. If EER3 and EER4 are functionally conserved with their mammalian and yeast homologs, respectively, then these two proteins may be involved in transcription leading to particular ethylene responses.
Conclusions
In recent years, our understanding of ethylene signal transduction has advanced significantly, making ethylene signaling one of the best-characterized pathways in plants. The latest discoveries have begun to address the complexities surrounding the interplay of the ethylene receptors and how they function to provide exquisite sensitivity and signaling efficiency, and have revealed multiple mechanisms of feedback and regulation. Interactions between the receptors, receptor turnover, differing signaling strengths and differential expression patterns all contribute to the fine tuning of ethylene perception and signaling. Transcription factor EIN3 has emerged as a key player that is regulated by rapid protein turnover. Intriguing data suggests that two distinct MAPK cascades have opposing effects on EIN3 stability. EIN3 (and EIL1) turnover is blocked by down-regulation of EBF1/2 F-box gene expression involving an exoribonuclease. EBF1/2 have distinct but overlapping functions that provide added fine-tuning of this pivotal signaling step.
Remaining key questions include the biochemical mechanism of ethylene receptor signaling, the biochemical function of RTE1/GR, the mechanism of CTR1 regulation and downstream targets of CTR1, the identification of additional MAPK cascade components, the biochemical function, targets and regulation of EIN2, the targets of XRN4, how ethylene perception regulates EBF1/2 and the precise roles of EER3 and EER4. The continued integration of molecular, genetic, cell biological, kinetic and biochemical approaches will help to elucidate the answers to these and many other questions.
Acknowledgments
We thank Chang lab members for comments on the manuscript. The work in our lab is supported by grants from the National Institutes of Health (1R01GM071855) and the U.S. Department of Energy (DE-FG02-99ER20329). C. Chang is supported in part by the University of Maryland Agricultural Experiment Station, and M.D. Kendrick is supported by a USDA National Needs Graduate Fellowship (20053842015761).
Footnotes
Publisher's Disclaimer: This PDF receipt will only be used as the basis for generating PubMed Central (PMC) documents. PMC documents will be made available for review after conversion (approx. 2-3 weeks time). Any corrections that need to be made will be done at that time. No materials will be released to PMC without the approval of an author. Only the PMC documents will appear on PubMed Central -- this PDF Receipt will not appear on PubMed Central.
Contributor Information
Mandy D. Kendrick, Email: mkendric@umd.edu.
Caren Chang, Email: carenc@umd.edu.
References and Annotations
- 1.Abeles FB, Morgan PW, Saltveit ME., Jr . Ethylene in Plant Biology. 2. Academic Press; 2004. [Google Scholar]
- 2.Chen Y-F, Etheridge N, Schaller GE. Ethylene signal transduction. Annals Bot. 2005;95:901–915. doi: 10.1093/aob/mci100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hall B, Shakeel S, Schaller GE. Ethylene Receptors: Ethylene perception and signal transduction. J Plant Growth Regul. 2007;26:118–130. [Google Scholar]
- 4**.Wang W, Esch JJ, Shiu S-H, Agula H, Binder BM, Chang C, Patterson SE, Bleecker AB. Identification of important regions for ethylene binding and signaling in the transmembrane domain of the ETR1 ethylene receptor of Arabidopsis. Plant Cell. 2006;18:3429–3442. doi: 10.1105/tpc.106.044537. This paper provides a detailed structure/function analysis of the ETR1 receptor in terms of identifying residues important for ethylene binding and those important for controlling the conformation of the signaling domain. The authors first identify conserved residues of the N-terminal transmembrane domain (containing the ethylene-binding domain) of the ethylene receptor family, based on comprehensive sequence alignments including homologs from cyanobacteria. Next, 41 amino acid substitutions in mostly conserved residues of the N-terminal transmembrane domain of the ETR1 ethylene receptor are characterized with respect to both ethylene binding ability (measured in a yeast-based assay) and degree of signaling (measured in terms of ethylene insensitivity conferred in transformed plants). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mount SM, Chang C. Evidence for a plastid origin of plant ethylene receptor genes. Plant Physiol. 2002;130:10–14. doi: 10.1104/pp.005397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodriguez F, Esch J, Hall A, Binder B, Schaller GE, Bleecker AB. A copper cofactor for the ETR1 receptor from Arabidopsis. Science. 1999;283:996–998. doi: 10.1126/science.283.5404.996. [DOI] [PubMed] [Google Scholar]
- 7.Hirayama T, Kieber JJ, Hirayama N, Kogan M, Guzman P, Nourizadeh S, Alonso JM, Dailey WP, Dancis A, Ecker JR. RESPONSIVE-TO-ANTAGONIST1, a Menkes/Wilson disease-related copper transporter, is required for ethylene signaling in Arabidopsis. Cell. 1999;97:383–393. doi: 10.1016/s0092-8674(00)80747-3. [DOI] [PubMed] [Google Scholar]
- 8.Hua J, Meyerowitz E. Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell. 1998;94:261–271. doi: 10.1016/s0092-8674(00)81425-7. [DOI] [PubMed] [Google Scholar]
- 9*.Qu C, Hall BP, Gao Z, Schaller GE. A strong constitutive ethylene-response phenotype conferred on Arabidopsis plants containing null mutations in the ethylene receptors ETR1 and ERS1. BMC Plant Biology. 2007;7:3. doi: 10.1186/1471-2229-7-3. The authors report on phenotypes for newly isolated null mutants of the subfamily I receptors, ETR1 and ERS1. The ers1-3 null mutant displays ethylene hypersensitivity that had not been observed in the previous ers1 mutant, which was a partial loss-of-function allele. In addition, the new etr1 ers1 double null mutant has a strong constitutive ethylene response, exhibits premature leaf senescence and develops novel filamentous structures at the base of the flower. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kieber J, Rothenberg M, Roman G, Feldmann A, Ecker J. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the Raf family of protein kinases. Cell. 1993;72:427–441. doi: 10.1016/0092-8674(93)90119-b. [DOI] [PubMed] [Google Scholar]
- 11.Alonso J, Hirayama T, Roman G, Nourizadeh S, Ecker J. EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science. 1999;284:2148–2152. doi: 10.1126/science.284.5423.2148. [DOI] [PubMed] [Google Scholar]
- 12.Solano R, Stepanova A, Chao Q, Ecker JR. Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE FACTOR1. Genes and Development. 1998;12:3703–3714. doi: 10.1101/gad.12.23.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo H, Ecker J. Plant responses to ethylene gas are mediated by SCFEBF1/EBF2-dependent proteolysis of EIN3 transcription factor. Cell. 2003;115:667–677. doi: 10.1016/s0092-8674(03)00969-3. [DOI] [PubMed] [Google Scholar]
- 14.Potuschak T, Lechner E, Parmentier Y, Yanagisawa S, Grava S, Koncz C, Genschik P. EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F Box proteins: EBF1 and EBF2. Cell. 2003;115:679–689. doi: 10.1016/s0092-8674(03)00968-1. [DOI] [PubMed] [Google Scholar]
- 15.Gagne J, Smalle J, Gingerich D, Walker J, Yoo S, Yanagisawa S, Vierstra R. Arabidopsis EIN3-binding F-box 1 and 2 form ubiquitin-protein ligases that repress ethylene action and promote growth by directing EIN3 degradation. Proc Natl Acad Sci USA. 2004;101:6803–6808. doi: 10.1073/pnas.0401698101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klee HJ. Ethylene signal transduction, Moving beyond Arabidopsis. Plant Physiol. 2004;135:660–667. doi: 10.1104/pp.104.040998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen G, Alexander L, Grierson D. Constitutive expression of EIL-like transcription factor partially restores ripening in the ethylene-insensitive Nr tomato mutant. J Exp Bot. 2004;55:1491–1497. doi: 10.1093/jxb/erh168. [DOI] [PubMed] [Google Scholar]
- 18.Mao C, Wang S, Jia Q, Wu P. OsEIL1, a rice homolog of the Arabidopsis EIN3 regulates the ethylene response as a positive component. Plant Mol Biol. 2006;61:141–152. doi: 10.1007/s11103-005-6184-1. [DOI] [PubMed] [Google Scholar]
- 19.Argueso CT, Hansen M, Kieber JJ. Regulation of ethylene biosynthesis. J Plant Growth Regul. 2007;26:92–105. [Google Scholar]
- 20.Li H, Guo H. Molecular basis of the ethylene signaling and response pathway in Arabidopsis. J Plant Growth Regul. 2007;26:106–117. [Google Scholar]
- 21.Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, Shinozaki K. Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr Opin Plant Biol. 2006;9:436–442. doi: 10.1016/j.pbi.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 22.Vandenbussche F, Van Der Straeten D. One for all and all for one: Cross-talk of multiple signals controlling the plant phenotype. J Plant Growth Regul. 2007;26:178–187. [Google Scholar]
- 23.Adie B, Chico JM, Rubi-Somoza I, Solano R. Modulation of plant defenses by ethylene. J Plant Growth Regul. 2007;26:160–177. [Google Scholar]
- 24.Klee HJ. Ethylene signal transduction. Moving beyond Arabidopsis. Plant Physiol. 2004;135:660–667. doi: 10.1104/pp.104.040998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25**.Kevany BM, Tieman DM, Taylor MG, Dal Cin V, Klee HJ. Ethylene receptor degradation controls the timing of ripening in tomato fruit. Plant J. 2008;51:458–467. doi: 10.1111/j.1365-313X.2007.03170.x. This paper shows that the tomato ethylene receptors LeETR4 and Le ETR6 are rapidly degraded in the presence of ethylene via a proteasome-dependent pathway. Moreover, degradation of either of these two receptors results in early fruit ripening, suggesting that receptor levels in the fruit could be responsible for the timing of the onset of ripening. The authors propose that this might serve as a mechanism in fruits to measure cumulative exposure to ethylene to control the timing of ripening. [DOI] [PubMed] [Google Scholar]
- 26.McClellan C, Chang C. The role of protein turnover in ethylene biosynthesis and response. Plant Sci. 2008;175:24–31. doi: 10.1016/j.plantsci.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27**.Chen Y-F, Shakeel SM, Bowers J, Zhao X-C, Etheridge N, Schaller GE. Ligand-incuded degradation of the ethylene receptor ETR2 through a proteosome-dependent pathway in Arabidopsis. J Biol Chem. 2007;282:24752–24758. doi: 10.1074/jbc.M704419200. This paper provides evidence that ethylene perception by the ETR2 ethylene receptor leads to its degradation by a proteasome-dependent pathway in Arabidopsis, possibly increasing the plant’s sensitivity to ethylene. The apparent mechanism is unlike that of animal growth factor receptors, which are degraded via endocytosis and targeting to the lysozome. ETR2 degradation does not require exit of ETR2 from the endoplasmic reticulum (ER), although ER-associated degradation is not generally known to regulate enzyme turnover,. [DOI] [PubMed] [Google Scholar]
- 28.Xie F, Liu Q, Wen C-K. Receptor signal output mediated by the ETR1 N terminus is primarily subfamily I receptor dependent. Plant Physiol. 2006;142:492–508. doi: 10.1104/pp.106.082628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29*.Grefen C, Städele K, Ruzicka K, Obrdlik P, Harter K, Horák J. Subcellular localization and in vivo interactions of the Arabidopsis thaliana ethylene receptor family members. Molecular Plant. 2007;1:308–320. doi: 10.1093/mp/ssm015. This paper demonstrates in planta protein interactions for pairwise combinations of all five Arabidopsis ethylene receptors in tobacco epidermal cells, reflecting the formation of heteromeric receptor complexes that possibly contain receptor heterodimers as well as homodimers. All five receptors localize to the ER in this system. Gene expression patterns show overlapping but distinct patterns for the five receptor genes, suggesting a complex level of interaction among the receptors. [DOI] [PubMed] [Google Scholar]
- 30*.Dong C-H, Rivarola M, Resnick JS, Maggin BD, Chang C. Subcellular co-localization of Arabidopsis RTE1 and ETR1 supports a regulatory role for RTE1 in ETR1 ethylene signaling. Plant J. 2008;53:275–286. doi: 10.1111/j.1365-313X.2007.03339.x. This paper presents the subcellular localization of the novel RTE1 protein and demonstrates co-localization of RTE1 with the ETR1 ethylene receptor at the Golgi apparatus and ER in intact Arabidopsis root cells. This is the first study to visualize ETR1 at the Golgi apparatus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma B, Cui ML, Sun HJ, Takada K, Mori H, Kamada H, Ezura H. Subcellular localization and membrane topology of the melon ethylene receptor CmERS1. Plant Physiol. 2006;141:587–597. doi: 10.1104/pp.106.080523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32**.Resnick JS, Wen C-K, Shockey JA, Chang C. REVERSION-TO-ETHYLENE SENSITIVITY1, a conserved gene that regulates ethylene receptor function in Arabidopsis. Proc Natl Acad Sci USA. 2006;103:7917–7922. doi: 10.1073/pnas.0602239103. This paper presents the cloning and characterization of the RTE1 gene, which is involved in regulating ETR1 receptor function. RTE1 encodes a novel transmembrane protein required for wild-type ETR1 and etr1-2 signaling, but not for etr1-1. The rte1 loss-of-function is hypersensitive to ethylene, whereas RTE1 over-expression confers reduced ethylene sensitivity that is dependent on ETR1. RTE1 is a homolog of tomato GREEN-RIPE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33*.Zhou X, Liu Q, Xie F, Wen C-K. RTE1 is a Golgi-associated and ETR1-dependent negative regulator of ethylene responses. Plant Physiol. 2007;145:75–86. doi: 10.1104/pp.107.104299. This paper provides further characterization of Arabidopsis RTE1, a positive regulator of ETR1 receptor function. The ethylene-insensitive phenotype of RTE1 over-expression phenotype is shown to be dependent on the N-terminal portion of ETR1 (residues 1–349). In addition, RTE1 remains functional when the first 49 amino acids of RTE1 are removed. The authors also provide evidence that RTE1 is associated with the Golgi apparatus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34**.Barry CS, Giovannoni JJ. Ripening in the tomato Green-ripe mutant is inhibited by ectopic expression of a protein that disrupts ethylene signaling. Proc Natl Acad Sci USA. 2006;103:7923–7928. doi: 10.1073/pnas.0602319103. This paper reports on the map-based cloning of the tomato GREEN-RIPE (GR) gene. The Green-ripe mutant, which has reduced ethylene sensitivity particularly in the fruit, is found to carry a deletion in the GR promoter region resulting in increased GR expression. GR is a homolog of Arabidopsis RTE1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clark KL, Larsen PB, Wang X, Chang C. Association of the Arabidopsis CTR1 Raf-like kinase with the ETR1 and ERS ethylene receptors. Proc Natl Acad Sci USA. 1998;95:5401–5406. doi: 10.1073/pnas.95.9.5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao Z, Chen Y-F, Randlett MD, Zhao X-C, Fendell JL, Kieber JJ, Schaller GE. Localization of the Raf-like kinase CTR1 to the endoplasmic reticulum of Arabidopsis through participation in ethylene receptor signaling complexes. J Biol Chem. 2003;278:34725–34732. doi: 10.1074/jbc.M305548200. [DOI] [PubMed] [Google Scholar]
- 37*.Zhong S, Lin Z, Grierson D. Tomato ethylene receptor-CTR1 interactions: visualization of NEVER-RIPE interactions with multiple CTRs at the endoplasmic reticulum. J Exp Bot. 2008;59:965–972. doi: 10.1093/jxb/ern021. The authors provide further support of the current model of receptor-CTR1 interactions. Using BiFC analysis, they demonstrate in vivo interaction of tomato subfamily 1 receptor NR with LeCTR1, LeCTR3, and LeCTR4. Additionally, using yeast-two-hybrid analysis, the authors show that the tomato subfamily 1 receptors interact with LeCTR1, LeCTR3 and LeCTR4, while the subfamily II receptors do not. [DOI] [PubMed] [Google Scholar]
- 38.Ghosh S, Moore S, Bell R, Dush M. Functional analysis of a phosphatidic acid binding domain in human Raf-1 kinase: mutations in the phosphatidate binding domain lead to tail and trunk abnormalities in developing zebrafish embryos. J Biol Chem. 2003;278:45690–45696. doi: 10.1074/jbc.M302933200. [DOI] [PubMed] [Google Scholar]
- 39.Testerink C, Munnik T. Phosphatidic acid: a multifunctional stress signaling lipid in plants. Trends Plant Sci. 2005;10:368–375. doi: 10.1016/j.tplants.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 40.Testerink C, Larsen P, van der Does D, van Himbergen J, Munnik T. Phosphatidic acid binds to and inhibits the activity of CTR1. J Exp Bot. 2007;58:3905–3914. doi: 10.1093/jxb/erm243. [DOI] [PubMed] [Google Scholar]
- 41.Chao Q, Rothenberg M, Solano R, Roman G, Terzaghi W, Ecker JR. Activation of the ethylene gas response pathway in Arabidopsis by nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell. 1997;89:1133–1144. doi: 10.1016/s0092-8674(00)80300-1. [DOI] [PubMed] [Google Scholar]
- 42.Lee JH, Deng XW, Kim WT. Possible role of light in the maintenance of EIN3/EIL1 stability in Arabidopsis seedlings. Biochem Biophys Res Comm. 2006;350:484–491. doi: 10.1016/j.bbrc.2006.09.074. [DOI] [PubMed] [Google Scholar]
- 43**.Binder B, Walker J, Gagne J, Emborg T, Hemmann G, Bleecker A, Vierstra R. The Arabidopsis EIN3 binding F-box proteins EBF1 and EBF2 have distinct but overlapping roles in ethylene signaling. Plant Cell. 2007;19:509–523. doi: 10.1105/tpc.106.048140. In this paper, the authors elucidate different temporal roles of EBF1 and EBF2 F-box proteins, which coordinately fine-tune the regulation of EIN3/EIL1 through degradation. Kinetic analysis of hypocotyl growth rates, as well as phenotypic analysis of double mutants, revealed that EBF2 most likely acts to degrade EIN3 at a later time point than EBF1. The authors also show that EIN3 and EIL1 are the predominant targets of EBF1/2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44**.Yoo S, Cho Y, Tena G, Xiong Y, Sheen J. Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signaling. Nature. 2008;451:789–795. doi: 10.1038/nature06543. This paper identifies the components of two potential MAP kinase cascades that oppositely regulate EIN3 stability. EIN3 is stabilized when phosphorylated on a particular site by MAP kinases MPK3 and MPK6. Data from protoplasts and stably-transformed plants suggests that the MKK9 MAPK kinase positively regulates MPK3/6. A second MAPK phosphorylation site on EIN3 is involved in EIN3 degradation. The authors show that CTR1, a putative MAPK kinase kinase, may have a role in phosphorylation on this site. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yanagisawa S, Yoo S-D, Sheen J. Differential regulation of EIN3 stability by glucose and ethylene signalling in plants. Nature. 2003;425:521–525. doi: 10.1038/nature01984. [DOI] [PubMed] [Google Scholar]
- 46.Liu Y, Zhang S. Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell. 2004;16:3386–3399. doi: 10.1105/tpc.104.026609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47**.Olmedo G, Guo H, Gregory B, Nourizadeh S, Aguilar-Henonin L, Li H, An F, Guzman P, Ecker J. ETHYLENE-INSENSITIVE5 encodes a 5’ 3’ exoribonuclease required for regulation of the EIN3-targeting F-box proteins EBF1/2. Proc Natl Acad Sci USA. 2006;103:13286–13293. doi: 10.1073/pnas.0605528103. By map-based cloning of the EIN5 locus, the authors find that EIN5 encodes the exoribonuclease XRN4. They show that EIN5 rescues the pleiotropic phenotypes of a yeast xrn mutant and find that EBF2 transcripts, and to a lesser extent EBF1, are upregulated in the ein5 mutant background. Consequently, EIN3 protein does not accumulate in the ein5 mutant in the presence of ethylene, supporting a role for EIN5 in regulating EBF1/2 transcript levels. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48**.Potuschak T, Vansiri A, Binder B, Lechner E, Vierstra R, Genschik P. The exoribonuclease XRN4 is a component of the ethylene response pathway in Arabidopsis. Plant Cell. 2006;18:3047–3057. doi: 10.1105/tpc.106.046508. The authors find that ein5 and xrn4 are allelic based on complementation analysis after observing a similar increase in expression of EBF1/EBF2 in these mutants. Using kinetic analysis of hypocotyl growth rates, the authors show that the ein5-1 mutant displays a response similar to the ein3-1 mutant, suggesting that the abundance of EBF1/EBF2 transcripts in the ein5-1 background leads to more rapid EIN3 degradation. Importantly, loss of the exoribonuclease EIN5/XRN4 does not lead to more stable EBF1/EBF2 transcripts, indicating that EIN5/XRN4 does not act by degrading EBF1/2 transcripts. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kastenmayer J, Green P. Novel features of the XRN-family in Arabidopsis: Evidence that AtXRN4, one of several orthologs of nuclear Xrn2p/Rat1p, functions in the cytoplasm. Proc Natl Acad Sci USA. 2000;97:13985–13990. doi: 10.1073/pnas.97.25.13985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50*.Christians MJ, Larsen PB. Mutational loss of the prohibitin AtPHB3 results in an extreme constitutive ethylene response phenotype coupled with partial loss of ethylene-inducible gene expression in Arabidopsis seedlings. J Exp Bot. 2007;58:2237–2248. doi: 10.1093/jxb/erm086. In this paper, the authors identify EER3 as a possible component of the ethylene signaling pathway. eer3-1 was isolated in a screen for Arabidopsis seedlings with enhanced ethylene response. A second allele (a T-DNA insertion), eer3-2, confers a severe constitutive triple-response phenotype and is epistatic to ein2 and ein3 mutations. While this suggests that EER3 may be a negative regulator of ethylene responses acting downstream of EIN2, analysis of ethylene-responsive gene expression suggests that EER3 instead may act to positively regulate certain ethylene responsive genes and to promote growth in the presence of ethylene. [DOI] [PubMed] [Google Scholar]
- 51.Robles L, Wampole J, Christians M, Larsen P. Arabidopsis enhanced ethylene response 4encodes an EIN3-interacting TFIID transcription factor required for proper ethylene response, including ERF1 induction. J Exp Bot. 2007;58:2627–2639. doi: 10.1093/jxb/erm080. [DOI] [PubMed] [Google Scholar]
- 52.Dunkley TPJ, Hester S, Shadforth IP, Runions J, Weimar T, Hanton SL, Griffin JL, Bessant C, Brandizzi F, Hawes C, et al. Mapping the Arabidopsis organelle proteome. Proc Natl Acad Sci USA. 2000;103:6518–6523. doi: 10.1073/pnas.0506958103. [DOI] [PMC free article] [PubMed] [Google Scholar]