Abstract
Notch3 gene amplification and pathway activation have been reported in ovarian serous carcinoma. However, the primary Notch3 ligand that initiates signal transduction in ovarian cancer remains unclear. In this report, we identify Jagged-1 as the highest expressed Notch ligand in ovarian tumor cells as well as in peritoneal mesothelial cells that are in direct contact with disseminated ovarian cancer cells. Cell-cell adhesion and cellular proliferation were reduced in Notch3-expressing ovarian cancer cells that were cocultured with Jagged-1 knockdown mesothelial and tumor feeder cells. Interaction of Notch3 expressing ovarian cancer cells with Jagged-1 expressing feeder cells activated the promoter activity of candidate Notch3 target genes and this activity was attenuated by Notch3 siRNA. Constitutive expression of the Notch3 intracellular domain significantly suppressed the Jagged-1 shRNA-mediated growth inhibitory effect. In Notch3-expressing ovarian cancer cells, Jagged-1-stimulating peptides enhanced cellular proliferation, which was suppressed by γ-secretase inhibitor and Notch3 siRNA. Taken together, our results demonstrate that Jagged-1 is the primary Notch3 ligand in ovarian carcinoma and Jagged-1/Notch3 interaction constitutes a juxtacrine loop promoting proliferation and dissemination of ovarian cancer cells within the intraperitoneal cavity.
Keywords: Notch3 receptor, microenvironment, ovarian cancer, Notch receptor ligand, Jagged-1
INTRODUCTION
Ovarian serous carcinoma represents one of the most aggressive neoplastic diseases in women. Although the molecular etiology of ovarian serous carcinoma remains mostly unknown, the majority of high-grade serous carcinomas harbor TP53 mutations. They also exhibit high levels of chromosomal instability as reflected by frequent changes in DNA copy number including allelic loss and gain involving almost all chromosomes (1-2). Genome-wide analysis has demonstrated amplification in genes with oncogenic potential at several loci. Based on digital karyotyping and SNP array analyses, our research group found that cyclin E1, AKT2, Notch3, Rsf-1 and PIK3CA loci were among the most frequently amplified genomic regions (2). One of the genes we have characterized is Notch3 because of its well-established role in a variety of physiological and pathological processes including cancer development. Copy number gain in Notch3 at chromosome 19p13.12 occurred in approximately 20% of high-grade serous carcinomas, and overexpression of Notch3 was observed in nearly 50% of the cases examined (3), suggesting that Notch3 signaling contributes to tumor progression in ovarian cancer.
The Notch signaling pathway is evolutionally conserved. Members of this pathway include Notch ligands (Delta and Jagged), Notch receptors, the nuclear transcription factors such as CSL (also known as RBP-J and CBF1) that bind to Notch intracellular fragment, as well as the target genes that are controlled by Notch3/CSL co-activators. The mammalian Notch family is comprised of four Notch receptors encoded by Notch1, 2, 3, 4. Five Notch ligands including Jagged-1, Jagged-2, Delta-like-1 (DLL1), Delta-like-3 (DLL3) and Delta-like-4 (DLL4) have been reported in mammals. Notch signaling is initiated by receptor-ligand interaction, which leads to proteolytic cleavages that liberate the Notch intracellular cytoplasmic domain (NICD) from the membrane. NICD then translocates to the nucleus where it binds to the transcription factor CSL complex, and converts CSL into a transcriptional activator that promotes the transcription of genes downstream in the Notch pathway (4). We have previously demonstrated that inactivation of the Notch3 pathway by γ-secretase inhibitor, or by Notch3-specific siRNA resulted in suppression of proliferation and induction of apoptosis in ovarian cancer cells, suggesting that targeting of Notch3 may offer a therapeutic intervention in ovarian cancer with Notch3 amplification and overexpression (3, 5).
As the first step in elucidating the molecular mechanisms underlying the role of the Notch signaling pathway in the progression of ovarian serous carcinomas, we analyzed all known Notch ligands for their expression levels in ovarian tumor cell lines and found that Jagged-1 was expressed at the highest level among all the ligands. In ovarian cancer tissues, expression of Jagged-1 and nuclear localization of Notch3 was highly correlated. This suggests that in ovarian carcinoma Jagged-1 interacted with Notch3 in juxtacrine fashion. We then characterized the functional role of Jagged-1 and Notch3 interaction in promoting cellular binding and proliferation. Because mesothelial cells constitute the tumor microenvironment in advanced stage ovarian cancer, we also analyzed the expression profile of Notch ligands in mesothelial cells. Our result demonstrated that Jagged-1 was the predominant form of Notch ligand expressed by mesothelial cells. Its biological role in supporting adhesion and growth of ovarian cancer cells was further investigated in this study.
MATERIALS AND METHODS
Tissue samples
A total of 77 high-grade serous carcinomas and 12 low-grade serous carcinomas of the ovary were retrieved from the Ovarian Cancer Tissue Bank in the Johns Hopkins Medical Institutions. In addition, mesothelial cells from benign ascites or primary cultures were harvested from fresh specimens. Acquisition of tissue specimens and clinical information was approved by an institutional review board.
Reagents and cell lines
Ovarian cancer cells including OVCAR3, A2780, ES2, SKOV3 and TOV21G cells were purchased from ATCC (Rockville, MD). Immortalized ovarian surface epithelial cells (OSE) by SV40 large T antigen were used in this study (6). Parental L cell and J cells stably expressing the hemagglutinin (HA)-tagged Jagged-1 protein were generously provided by Dr. G Weinmaster (Department of Biological Chemistry, UCLA). Type 1 γ-secretase inhibitor was purchased from Calbiochem (San Diego, CA) and was dissolved in DMSO. Jagged-1 peptide (CDDYYYGFGCNKFCRPR) and scrambled peptide (RCGPDCFDNY GRYKYCF) were synthesized by GenScript Corporation (Piscataway, NJ). Disuccinimidyl glutarate (DSG), a crosslinking reagent, was purchased from Pierce (Rockford, IL).
Primary Mesothelial Cell Culture
Mesothelial cells were derived from either benign peritoneal effusions or from primary tumor tissues containing benign mesothelium. The enrichment of mesothelial cells was performed by dissociating tissues with collagenase A followed by incubation with magnetic beads conjugated with Ber-EP4 (EpCAM) antibody to immunosort epithelial cells. The negative cellular fraction was short-term cultured to expand the number of mesothelial cells. The mesothelial cells used in this study were found to be positive for calretinin (a mesothelial cell marker) in more than 99% of cells and negative for Ber-EP4 and mucin 4 (carcinoma markers) as determined by immunocytochemistry. Representative stains for mucin 4 and calretinin in mesothelial cell and carcinoma cell cultures were shown in Fig. S1. The procedure to isolate epithelial cells has been previously described (7) and the protocol is available upon request.
Real-time PCR
Relative mRNA expression was measured by quantitative real-time RT-PCR using an iCycler (Bio-Rad, Hercules, CA). Threshold cycle numbers (Ct) were obtained using the iCycler Optical system interface software. PCR primers were designed using the Primer 3 program and the nucleotide sequences of the primers for determining transcript expression were listed in Table S1. Mean Ct of the gene of interest was calculated from duplicate or triplicate measurements and normalized with the mean Ct of a control gene, beta-amyloid precursor gene (APP), for which mRNA expression is relatively constant among the SAGE libraries (8). Data was further normalized to the result obtained from ovarian surface epithelial cell, OSE7.
Gene knockdown using siRNA and shRNA
Jagged-1 small hairpin RNA (shRNA) vectors were purchased from Sigma-Aldrich (St. Louis, MO). Jagged-1 shRNA sequence templates (CCGGCCGAATGGAGTACATCG TATACTCGAGTATACGATGTACTCCATTCGGTTTTTG) and (CCGGCCAGGAT AACTGTGCGAACATCTCGAGATGTTCGCACAGTTATCCTGGTTTTTG) were inserted into lentiviral plasmids (pLKO.1-puro). Notch3-specific small interfering RNA (siRNA) (GUCAAUGUUCACUUCGCAGUU) and (GCGUGGAUUCGGACCAGUCUGAGAGGG) and control siRNA that targets the Luciferase gene (GAUUAAAUCUUCUAGCGACUGCUUCGC) were synthesized by Integrated DNA Technologies (Coralville, IA). Cells were transfected with siRNA or shRNA at a final concentration of 200 nM or 2 μg, respectively, using lipofectamine method (Invitrogen, Carlsbad, CA). Six hours after transfection, cells were replaced with fresh medium. On the following day, the treated cells were harvested and used for cell growth, binding and coculture assays.
Retrovirus transduction
The NICD3 retrovirus expressing the active intracellular domain of Notch3 was kindly provided by Dr. Michael Wang (University of Michigan, Ann Arbor, MI) (9). Packaging cells (Phoenix cells) were transiently transfected with the NICD3 or empty vector using lipofectamine method (Invitrogen). On the following day, the supernatant was harvested and passed through a 0.45 μM syringe filter. The filtered viral supernatant was resuspended in 4 μg/ml polybrene and added to cancer cell culture. Twenty-four hours after infection, cells were harvested and used for assays.
Immunohistochemistry
Anti-Notch3 rabbit polyclonal antibody and anti-Jagged-1 goat polyclonal antibody were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). An EnVision+System peroxidase kit (DAKO, Carpinteria, CA) was used for detection. Tissue microarrays (triplicate 1.5 mm cores from each specimen) including 60 high-grade serous carcinomas were used to facilitate immunohistochemistry. Immunointensity for Jagged-1 was scored as negative/low and high; nuclear staining for Notch3 was scored as negative (-) and positive (+) by two investigators.
Cell growth and colony formation assays
Cells were grown in 96-well plates at a density of 3,000 per well. Cell number was measured by the incorporation of SYBR green I nucleic acid gel stain (Molecular Probes, Eugene, OR) using a fluorescence microplate reader (Fluostar from BMG, Durham, NC). Data was determined from five replicates and was expressed as the fold increase of control group. For colony formation assay, cells were seeded into 25-cm2 flasks at a cell density of 1,500 or 4,500 cells per flask. The colonies were counted after staining with crystal violet dye (Sigma) at day 10.
Cell-cell binding and coculture assay
For binding assay, the feeder cells were transfected with empty or Jagged-1 shRNA vector. One day after, the cells were seeded in 12-well plates at a density of 1.0×106 cells per well and were allowed to grow to confluence. A2780 cells (1.0×105) expressing green fluorescence protein (A2780-GFP) were laid on top of the feeder cells. Fifteen or forty-five minutes after coculture, A2780-GFP cells which did not anchor to the bottom layer were harvested and classified as non-adherent cells. The A2780-GFP cells adhering to the bottom monolayer cells were dissociated by PBS containing 2 mM EGTA and determined as adherent cells. For cell proliferation assay, 1.0×104 A2780-GFP cells were laid on top of the monolayer cells, which were previously irradiated after transfection with Jagged-1 shRNA. OVCAR3 cells were irradiated for 30 minutes with 68 cGy/min while L cell, J cell, and mesothelial cells were irradiated at the same intensity for 8 minutes. The number of A2780-GFP cells was counted under a fluorescent microscope at day 2 of coculture.
Coprecipitation of Jagged-1 and Notch3
1.0×107 A2780 cells were cocultured over the monolayer of J cells and incubated with a buffer containing 20m M HEPES (pH 7.5), 150 mM NaCl, and 0.9 mM CaCl2 at room temperature for 45 min. Disuccinimidyl glutarate (DSG), a crosslinking reagent, was added to the culture at a final concentration of 20 μM and incubated for 30 min. Following crosslinking reaction, the cells were resuspended in TNE buffer (20mMTris-Cl, pH7.4,150mM NaCl,1% NP-40, 5 g/ml aprotinin, and 1mM EDTA) (10), and the lysate was immunoprecipitated with a Notch3 antibody or a control rabbit serum. To reverse the crosslink, a portion of the pull-down immunocomplex was treated with 50 mM DTT (dithiothreitol) and boiled for 10 minutes before electrophoresis.
Promoter activity assay
Hes1-luc and Hes5-luc reporter constructs were gifts from Dr. R Kageyama (Kyoto University, Japan). Pbx1-luc containing the 3Kb region of Pbx1 promoter was gift by Dr. S Higashiyama (Ehime University, Japan). Promoter reporter constructs and pRL-Renilla control plasmid (Promega) were transiently transfected into OVCAR3 cells by lipofectamine (Invitrogen). Twenty-four hours after transfection, the cells were seeded onto 48 well plates pre-layered with J cells or L cells. Luciferase activity was determined by the Dual-Glo luciferase reagent (Promega). The reporter luciferase was normalized to Renilla luciferase and the ratio of luminescence from the experimental reporter to luminescence from the control reporter was calculated.
RESULTS
Jagged-1 is the primary Notch ligand expressed in ovarian cancer cells
Gene expression levels among the known Notch ligands (Jagged-1, Jagged-2, DLL1, DLL3, and DLL4) were analyzed in a panel of ovarian normal and cancer cell lines by quantitative RT-PCR (Fig. 1A). Jagged-1 was found to be highly expressed in ovarian cancer cell lines but not in ovarian surface epithelial cells (OSE). Since the expression level of other Notch ligands was relatively low in ovarian cancer cells, a different scale was used to present the relative expression levels among cell lines (Fig. S2). To extrapolate the findings from ovarian cell lines to ovarian cancer tissues, we performed two additional experiments. First, quantitative RT-PCR was conducted to determine if Jagged-1 was over-expressed in ovarian carcinoma tissues. The data demonstrated that Jagged-1 was highly expressed in ovarian carcinoma tissues as compared to either low-grade ovarian carcinomas or ovarian surface epithelial cells (p<0.0001, Mann-Whitney test, Fig. 1B). Second, immunohistochemistry was performed on a larger panel of ovarian high-grade carcinomas (N=60) to assess the expression of Jagged-1 and Notch3. The specificity of the Jagged-1 antibody was demonstrated by Western blot analysis. A single protein band corresponding to Jagged-1 protein was observed in OVCAR3, an ovarian cancer cell line showing high Jagged-1 mRNA expression, and in J cells, a mouse fibroblast cell line engineered to express full-length Jagged-1, but not in non-transfected parental L cells (Fig. 2A). The Notch3 antibody used for immunohistochemistry has been previously reported (3) and its specificity was further validated by Western blot. In this analysis, HeLa cells were transduced with retrovirus expressing NICD and a single band corresponding to the molecular weight of NICD was detected (Fig. S3). In addition, this band was absent in either non-transduced or empty vector-transduced group. We used prominent nuclear Notch3 immunoreactivity as a surrogate marker for Notch3 signaling activation in immunohistochemistry. By parallel comparison of Jagged-1 and Notch3 immunoreactivity in the same tissue samples, the data demonstrated a significant correlation between intense Jagged-1 immunoreactivity and Notch3 nuclear immunoreactivity (Fig. 2B) (p< 0.0001, Fisher’s exact test).
Figure 1. Expression of Notch ligands in ovarian cancer cells.

A. mRNA expression levels of known Notch ligands including Jagged-1, Jagged-2, DLL1, DLL3, and DLL4 were measured by quantitative RT-PCR in six immortalized ovarian surface epithelial (OSE) cell lines and six ovarian cancer cell lines. B. mRNA expression levels of Jagged-1 in ovarian carcinoma tissues were measured by quantitative RT-PCR. Each symbol represents an individual specimen. OSE; ovarian surface epithelial cells, LGCA; low-grade ovarian carcinoma, HGCA; high-grade ovarian carcinoma.
Figure 2. Correlation of Jagged-1 immunoreactivity with nuclear localization of Notch3 in ovarian carcinomas.
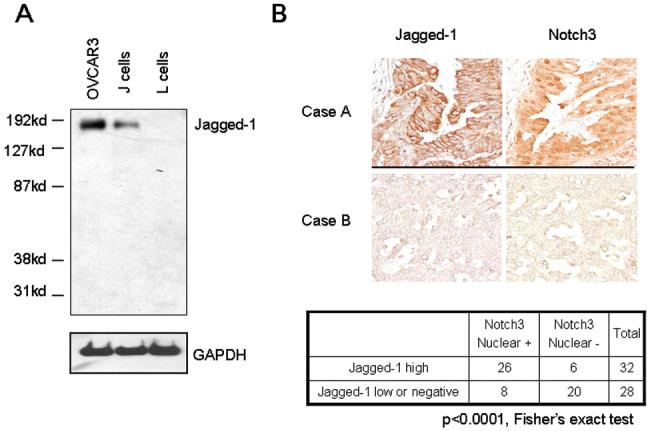
A. Jagged-1 protein expression was determined by Western blot in OVCAR3, J, and L cells to validate the antibody specificity. J cell is an Ltk- mouse fibroblast cell line that was engineered to express the full-length Jagged-1. L cell is the parental Ltk- cell line. Jagged-1 protein bands (∼175 kD) were detected in both OVCAR3 and J cells but not in L cells. B. Upper panel: Immunoreactivity of Jagged-1 and Notch3 in two representative ovarian high-grade carcinoma tissues. Case A shows intense Jagged-1 membrane staining and prominent Notch3 nuclear immunoreactivity. In contrast, case B shows very weak Jagged-1 and Notch3 staining. Lower panel: Summary table of Jagged-1 and Notch3 immunohistochemistry for 60 high-grade carcinomas. Jagged-1 protein expression and nuclear localization of Notch3 is significantly correlated (p<0.0001, Fisher’s exact test).
Biological effects of Jagged-1 expression on cell adhesion and proliferation
The finding that Jagged-1 and Notch3 were co-expressed in ovarian carcinoma tissues and cell lines suggested that this ligand-receptor interaction could play an important functional role in ovarian cancer development. In this regard, we performed a cocultivation experiment to determine the ability of Jagged-1-expressing cells to enhance cell-cell adhesion and to stimulate cell growth of Notch-expressing tumor cells. OVCAR3, which expresses abundant Jagged-1, was selected to represent Jagged-1-expressing cells. Aliquots of OVCAR3 cells were transfected with Jagged-1 shRNAs to knock down Jagged-1 protein expression (Fig. 3A, left panel). Notch3 receptor-expressing cells, A2780, were pre-engineered to stably express green fluorescent protein (GFP) to facilitate quantification of cell numbers (designated as A2780-GFP). The A2780-GFP cells were overlaid onto a sublethally irradiated OVCAR3 monolayer and at indicated time points, the number of adherent and non-adherent A2780-GFP cells was counted. Our results showed that a significant fraction of the overlaid A2780-GFP cells bound to the vector control-transfected OVCAR3 cells at both 15 min and 45 min of coculture (middle panel, Fig. 3A). In contrast, knockdown of Jagged-1 by either shRNA1 or shRNA2 significantly reduced the number of A2780 cells that adhered to the OVCAR3 monolayer at 45 min. The binding of A2780-GFP to the OVCAR3 monolayer increased gradually and the vast majority of cells bound to the OVCAR3 monolayer (with or without Jagged-1 shRNA treatment) after two hours of coculture.
Figure 3. Coculture experiments demonstrate Jagged-1 expressed in feeder cells is essential in promoting cell adhesion and growth of Notch3 expressing cancer cells.
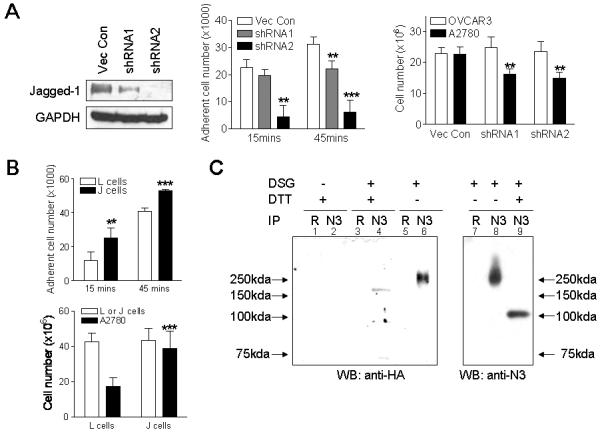
A. Left panel: Western blot analysis shows both Jagged-1 shRNAs, shRNA1 and shRNA2, significantly reduce Jagged-1 protein expression in OVCAR3 cells as compared to the empty vector control. When compared to shRNA1, shRNA2 shows a more potent inhibitory effect. Middle panel: Binding of A2780-GFP cells to Jagged-1 shRNA transfected OVCAR3 cells was examined in a cell-cell association assay. The cell number of Notch3 expressing A2780-GFP cells that adhered to Jagged-1 shRNA-transfected OVCAR3 cells (feeder monolayer) was reduced when compared to the vector-transfected cells at both15 min and 45 min of cocultivation. Right panel: The growth of A2780-GFP cells is significantly reduced when cocultured with Jagged-1 shRNA-transfected feeder cells (OVCAR3) as compared to vector control transfected cells. Data was measured on the 2nd day of coculture. Data also shows similar number of feeder cells (OVCAR3) was present in each experimental group. ** p<0.01; *** p<0.001, Student’s t test. B. Upper panel: Coculture of A2780-GFP cells and J or L feeder monolayer. The cell number of A2780-GFP bound to L cells (without Jagged-1 expression) was lower than that of J cells (with Jagged-1 expression) at both 15 min and 45 min coculture incubation. Lower panel: The growth of A2780-GFP cells is promoted when cocultured with J cells comparing to L cells. Similar number of L and J feeder cells was present in each experiment. ** p<0.01; *** p<0.001, Student’s t test. C. Binding of Jagged-1 to Notch3 was verified by immunoprecipitation/Western experiment. The J cell and A2780-GFP cell coculture lysates were immunoprecipitated with an anti-Notch3 rabbit polyclonal antibody (N3) and were blotted with an HA antibody or a Notch3 antibody. HA antibody was used to detect expression of Jagged-1 in J cells because the expression construct contained an HA epitope tag. Rabbit anti-serum (R) was used as the control in immunoprecipitation. DSG: disuccinimidyl glutarate; DTT: dithiothreitol
Cell growth assay was performed by seeding the same number of A2780-GFP cells and measuring A2780-GFP cell numbers at 48 hours after coculture. The data demonstrated that Jagged-1 knockdown in OVCAR3 cells reduced the ability of OVCAR3 cells to stimulate the growth of A2780-GFP cells (Fig. 3A, right panel).
To further demonstrate that biological effects of cell-cell adhesion and cell growth were due to interactions between Jagged-1 and Notch3, we performed an additional coculture assay using the J cell, which is an Ltk-mouse fibroblast cell line engineered to express Jagged-1 (11), as the feeder monolayer. The parental Ltk- cell (L cell) was used as a control. Similarly, in the cell-binding assay, the A2780-GFP cells were overlaid on top of sublethally irradiated J cells or L cells and the number of adherent A2780-GFP cells was counted. The data showed that a higher number of A2780-GFP cells adhered to J-cell monolayer than to L-cell monolayer at 15 min and 45 min after coculture (Fig. 3B, upper panel). The number of A2780-GFP cells that adhered to J cells and L cells gradually increased and almost all A2780 cells bound to J cells and L cells after two hours after coculture. In the cell growth assay, same number of A2780-GFP cells was laid on top of irradiated J cells or L cells and total number of A2780-GFP cells was determined 48 hours after coculture. The results showed a higher A2780-GFP cell number when cocultured with J cells as compared to L cells (Fig. 3B, lower panel). The above results imply that the presence of Jagged-1 in feeder cells is important in mediating cell binding and growth.
To determine if Jagged-1 directly bound to Notch3, a co-immunoprecipitation experiment was performed using a Notch3 antibody to pull down the receptor/ligand complex from a coculture of J cells and A2780-GFP cells (Fig. 3C). The coculture was first treated with a crosslinking reagent, disuccinimidyl glutarate (DSG), before immunoprecipitation. Western blot was then performed in the presence or absence of DTT (to reverse crosslink). Our results showed that in the absence of DTT, high molecular weight immunoprecipitate was detected by both HA antibody (to detect Jagged-1-HA fusion protein expressed in J cells, lane 6) and Notch3 antibody (lane 8). When the same cell lysates were treated with DTT, the higher molecular weight band disappeared and lower molecular weight bands corresponding to monovalent Jagged-1 (lane 4) and Notch3 (lane 9) were detected. The Notch3-Jagged-1 immunocomplex was not detectable in the control experiment using rabbit control serum in the immunoprecipitation step. The Notch3 protein under the crosslink/DTT denaturing condition migrated at a molecular weight higher than native NICD (∼86 kD), probably because it contained the N-terminal transmembrane domain, a subunit that was not yet cleaved by secretases.
Interaction of Notch3-expressing tumor cells and Jagged-1-expressing mesothelial cells
Mesothelial cells are the main cell type in direct contact with ovarian cancer cells in the peritoneal cavity. It is possible that expression of Notch ligands in mesothelial cells creates a microenvironment suitable for ovarian cancer cells to survive and disseminate. Therefore, expression of Notch ligands in mesothelial cells derived from benign effusions or purified from tumor tissues was analyzed. The results demonstrated that Jagged-1 was expressed in the majority of mesothelial cell samples, and was the primary Notch ligand expressed by mesothelial cells (Fig. 4A, p<0.01 Mann-Whitney test). To determine if mesothelial cells would support tumor adhesion and growth, we performed coculture experiments using mesothelial cells as the feeder layer. Primary mesothelial cell cultures were treated with Jagged-1 shRNA to reduce Jagged-1 expression. Although expression of Jagged-1 was significantly reduced, growth of mesothelial cells was not significantly affected (Fig. S4). However, the cell-cell binding activity measured at 45 minutes was significantly reduced in A2780-GFP cells cocultured with mesothelial cells pretreated with Jagged-1 shRNA as compared to those cocultured with control shRNA-treated mesothelial cells (Fig. 4B). Similar to previous coculture systems, almost all A2780-GFP eventually adhered to mesothelial feeder cells 2 hours after coculture. In cell growth assay, knockdown of Jagged-1 expression in mesothelial feeder cells significantly suppressed cell growth (Fig. 4C). The data indicated that the Jagged-1 expression level in mesothelial cells is important for binding and growth of adjacent tumor cells.
Figure 4. Expression of Jagged-1 in human peritoneal mesothelial cells is important in supporting adhesion and growth of ovarian tumor cells.
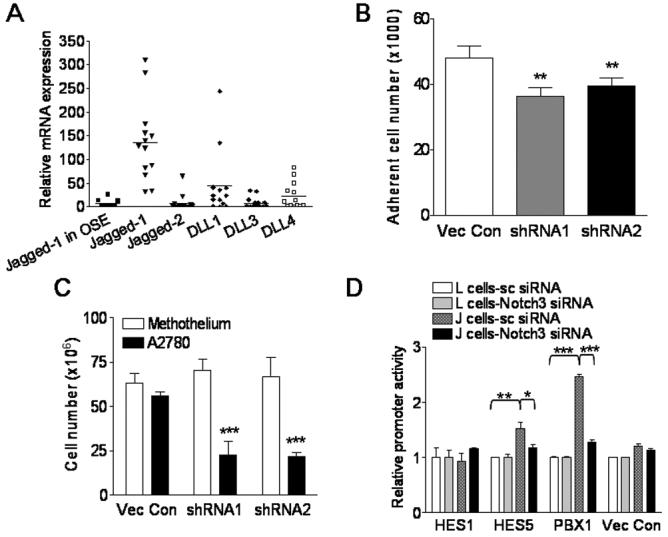
A. Quantitative RT-PCR measured Notch ligand expression levels in peritoneal mesothelial cells. B-C Coculture experiments. Human mesothelial cells expressing Jagged-1 were transfected with Jagged-1 shRNA and served as feeder monolayer. They were cocultured with Notch3-expressing A2780-GFP cells. B. Compared to vector-transfected control, Jagged-1 shRNA treatment in mesothelial cells significantly reduced the binding capability of A2780-GFP cells. C. Mesothelial cells pretreated with Jagged-1 shRNA significantly lost the ability to support growth of A2780-GFP cells. Similar number of mesothelial feeder cells was present in each experiment. Data was measured at 48 hours after coculture. D. Coculture of J cells with ovarian cancer cells stimulates the promoter activity of candidate Notch3 target genes. Promoter reporter constructs for Notch3 candidate target genes including Hes1, Hes5, and Pbx1 were transfected into Notch3-expressing OVCAR3 cells. The transfected cells were then cocultivated with J cells or L cells. High luciferase activity was detected in Pbx1 and Hes5 promoter construct-transfected groups when cocultured with J cell. This luciferase activity could be potently suppressed by Notch3-specific siRNA. * p<0.1; ** p<0.01; *** p<0.001, Student’s t test.
To further determine if Jagged-1 expressed by the feeder cells could stimulate Notch3 signaling in tumor cells, we performed promoter reporter assays in which A2780 cell was transiently transfected with reporter plasmids containing promoter regions of candidate Notch3 downstream target genes including Hes1, Hes5 (12), and Pbx1 (Park et al, unpublished observation), and the transfected cells were cocultured with J cells or L cells. The data demonstrated that Hes5 and Pbx1 promoter activities in A2780 cell were significantly induced by co-culturing with J cells but not with L cells. Furthermore, these promoter activities were significantly suppressed by Notch3-siRNA (Fig. 4D) or γ-secretase inhibitor (data not shown). In contrast, we did not detect luciferase activity from Hes1 promoter reporter or vector control plasmid in any of the experimental conditions.
Jagged-1 gene knockdown reduces cellular proliferation in ovarian cancer cells overexpressing Jagged-1
To determine if Jagged-1 expression in cancer cells was essential for tumor cell growth, we knocked down Jagged-1 in TOV21G and OVCAR3 ovarian cancer cell lines, both expressed relatively high levels of Jagged-1 (Fig. 5A). In addition, two immortalized ovarian surface epithelial cell lines (IOSE7 and IOSE10), neither of which expressed robust level of Jagged-1, were used as negative controls. The results showed that both of the Jagged-1-negative cell lines continued proliferating after shRNA transfection (Fig. 5B). There was no statistically significant difference between Jagged-1 shRNA-treated versus vector control-treated groups (Student’s t test). In contrast, both Jagged-1-high cell lines showed a significant growth inhibitory effect following transfection with Jagged-1 shRNA as revealed by growth curves (Fig. 5C) and colony formation assay (Fig. 5D).
Figure 5. Effect of Jagged-1 knockdown on cell proliferation in ovarian cancer cells.
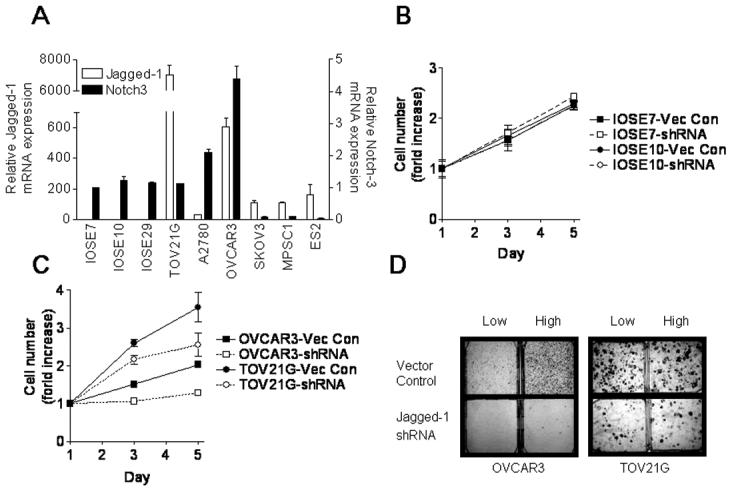
A. Expression of Jagged-1 and Notch3 mRNA in IOSE cells (IOSE7, IOSE10 and IOSE29) and ovarian cancer cell lines (TOV21G, A2780, OVCAR3, SKOV3, MPSC1 and ES2) was measured by quantitative RT-PCR. TOV21G and OVCAR3 showed highest levels of Jagged-1 expression. B. Jagged-1 shRNA does not have a significant effect in cell growth at both day 3 and day 5 in IOSE 7 and IOSE10 cells. C. Jagged-1 shRNA significantly inhibits cell growth in ovarian cancer cell lines which over-expresses Jagged-1. p<0.01 (day 3); p<0.001 (day 5), Student’s t test. D. Colony formation is significantly reduced in both OVCAR3 and TOV21G cells transfection with Jagged-1 shRNA, comparing to vector-transfected group (p<0.01, Student’s t test). Cells were seeded at both low and high density and cell colonies were measure at 2 weeks after plating.
The effects of Jagged-1 are mediated by the Notch3 signaling pathway
To determine if the observed biological effects of Jagged-1 involved the Notch3 signaling pathway, we applied two independent but complementary approaches. First, we asked if constitutive expression of the NICD could reverse the growth inhibitory effect of Jagged-1 shRNA. As shown in Fig. 6A, ectopic expression of NICD had minor effect on cell growth; however it significantly abrogated the growth inhibitory effect of Jagged-1 shRNA. Second, a Jagged-1 peptide known to activate Notch signaling comparably to full-length Jagged-1 was employed. When cultured in medium containing Jagged-1 peptide, the number of ovarian cancer cell OVCAR3 was significantly increased as compared to the number of cells cultured in medium containing control peptide (Fig. 6B). The increase in cell number was likely due to increased proliferation activity because BrdU incorporation was enhanced in OVCAR3 cells incubated with Jagged-1 peptides (Fig. S5). This proliferation-stimulating effect of Jagged-1 peptide could be inhibited by inactivating Notch3 signaling using either γ-secretase inhibitor (Fig. 6C) or Notch3-specific siRNA (Fig. 6D).
Figure 6. Involvement of Notch3 in Jagged-1-induced cell growth in ovarian cancer cells.
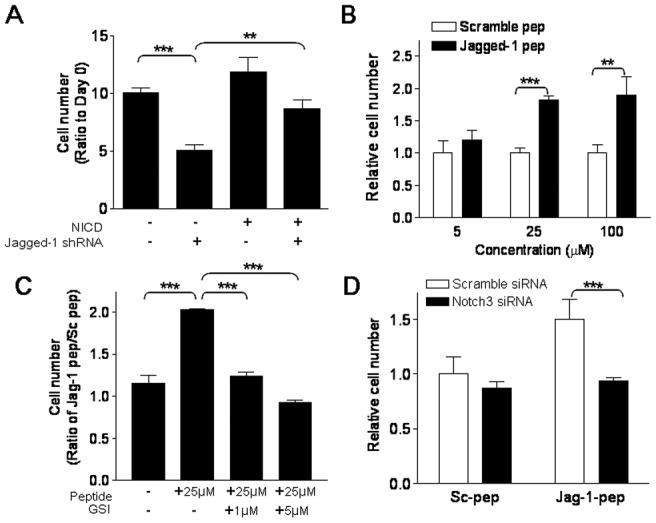
A. OVCAR3 cells were transduced with retrovirus expressing active form of Notch3 (NICD) or with retrovirus prepared from empty vector. Jagged-1 shRNA-inhibited cell proliferation in OVCAR3 cells and this growth inhibitory effect can be rescued by ectopic NICD expression. B. Incubation of Jagged-1 stimulatory peptide in A2780 cells significantly increases cell number in a dose dependent manner. Conversely, scramble peptide did not show growth stimulatory effect. C. The growth stimulatory effect of Jagged-1 peptide is blocked in a dose-dependent manner by treatment of Υ-secretase inhibitor (GSI). D. Similar to GSI, attenuation of Notch3 pathway using Notch3-specific siRNA suppressed Jagged-1 peptide-induced cell growth. ** p<0.01; *** p<0.001, Student’s t test.
To determine if Notch3 could regulate Jagged-1 expression and thus initiate a positive-feedback loop in Notch signaling, we applied Notch3-siRNA in A2780, TOV21G, and OVCAR3 and measured the amount of Jagged-1 mRNA using quantitative real-time PCR. As shown in Fig. S6, the results demonstrated that Notch3 siRNA did not significantly affect Jagged-1 mRNA expression, suggesting a lack of such feedback loop in ovarian cancer cells. Taken together, the above data indicated that the biological effects of Jagged-1 observed in this study were mediated at least in part through Notch3 signaling.
DISCUSSION
Growth and survival of tumor cells, as well as their ability to metastasize depend on intricate interactions with their microenvironment. Despite accumulation of a variety of genetic lesions, human ovarian cancer cells remain dependent on their microenvironment during the progression of the disease. Our previous studies have demonstrated Notch3 gene amplification and overexpression in a significant fraction of ovarian carcinomas (2, 3). In the current study, we provided evidence that Jagged-1 expressed by mesothelial and ovarian cancer cells forms a juxtacrine loop with Notch receptor expressed on the surface of ovarian cancer cells. This promotes adhesion and proliferation of cancer cells within the peritoneal cavity.
Although Jagged-1 has been shown to be one of the Notch ligands, its role in initiating Notch signaling has not been well-established in ovarian cancer. In this study we have demonstrated the following pieces of evidence to suggest an important role of the Notch3/Jagged1 axis in promoting adhesion and growth in ovarian cancer cells. First, Jagged-1 was co-expressed with Notch3 in a significant number of ovarian cancers. Second, Jagged-1 expression in feeder cells is responsible for the binding and growth of cocultured ovarian cancer cells. Third, Jagged-1 activating peptide increased cell number in Notch3-expressing ovarian cancer cell line, in which the effect could be reduced by γ-secretase inhibitor or by Notch3 specific siRNA. Forth, the growth suppression effect of Jagged-1 shRNA in ovarian cancer cells can be rescued by ectopic expression of NICD. Finally, in the coculture system, Jagged-1 expressed by the feeder cells induced promoter activation of candidate Notch3 target genes, Hes5 and Pbx1. In aggregate, these findings suggest that Jagged-1 and Notch3 form a functional signaling network. Our results are consistent with a previous report that expression of Notch1 and its ligands, Jagged-1 and Delta-like-1, is critical for cell survival and proliferation in glioma (13).
Expression of Jagged-1 in ovarian cancer cells and peritoneal mesothelial cells has significant biological implications. First, in primary ovarian tumors, reciprocal binding of Jagged-1 to Notch3 between adjacent tumor cells acts as a juxtacrine mediator that initiates and sustains Notch3 pathway activation, which is responsible for ovarian tumor development (Fig. S7). Second, during tumor cell dissemination in the peritoneal cavity, Jagged-1 expressed by mesothelial cells may enhance tumor cell binding to and growth on the peritoneal surface, thus facilitating intra-peritoneal tumor dissemination, a cardinal feature in ovarian serous carcinoma (Fig. S7). Ovarian serous carcinoma is associated with a devastating clinical outcome because most patients present at advanced stages when the tumor has widely spread in the peritoneal cavity (14). This “transcoelomic” dissemination involves multiple processes including tumor cell detachment, migration, and implantation on mesothelial cells carpeting the peritoneal cavity and the surface of abdominal organs. Transcoelomic dissemination is a major factor contributing to morbidity and mortality in women with ovarian carcinomas. It is plausible that ovarian cancer cells detached from the primary site directly contact with peritoneal mesothelial cells which provide abundant Jagged-1 to facilitate the attachment of cancer cells to mesothelial cells and to enhanced proliferation of ovarian cancer cells. This process would contribute establishment of cancer cell colonies on the peritoneal surface, where the tumor cells might invade the underlying stromal tissue and establish implanted tumors.
While the above represents our preferred view how Jagged-1 contributes to tumor progression in ovarian cancer, it should be noted that other mechanisms may exist. For example, Jagged-1 may have its own signaling function that is independent of the canonical Notch pathway. It has been demonstrated that Jagged-1 and Delta-like-1, upon binding to Notch receptors, are sequentially processed by α- and γ-secretase, which leads to the release of nuclear signaling fragments (15, 16). The soluble Jagged-1 intracellular fragment translocates into the nucleus and activates gene expression via the transcription factor AP1 (15). Furthermore, ectopic expression of Jagged-1 was found to transform kidney epithelial cells and this ability depends on the PDZ ligand domain at the C terminus of Jagged-1 (17). Therefore, in ovarian cancer cells that coexpress Notch3 and Jagged-1, Jagged-1 may play two functional roles. First, Jagged-1 serves as a membrane ligand to stimulate adjacent tumor cells in a juxtacrine manner through Notch3 receptor. Second, the intracellular domain of Jagged-1 may trigger signaling pathway distinct from Notch3 and promote tumor cell growth. Although the biological effects of Notch activation are well-known in human cells, it would be of interest to determine the Notch-independent role of Jagged-1 in human neoplasms.
In summary, we provided new evidence that Jagged-1 is the primary Notch3 ligand expressed by ovarian cancer cells and mesothelial cells. Interaction of Jagged-1 and Notch3 which activates intracellular Notch3 signaling may provide growth advantage of ovarian cancer cells in the peritoneal microenvironment. The above results demonstrate the dependence of ovarian cancer cells on a single Notch ligand and suggest that antagonizing Jagged-1 or disrupting the interaction between Jagged-1 and Notch3 can be potential therapeutic strategies for ovarian cancer.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Kevin Lee for the help with the manuscript preparation. Grant support: Individual Investigator Grant, Ovarian Cancer Research Fund (TLW); Institutional Research Grant, American Cancer Society (TLW); Department of Defense Ovarian Cancer Research Program OC040060 (TLW); National Cancer Institute RO1 CA103937 and RO1 CA129080 (IMS).
REFERENCES
- 1.Kohler MF, Marks JR, Wiseman RW, et al. Spectrum of mutation and frequency of allelic deletion of the p53 gene in ovarian cancer. J Natl Cancer Inst. 1993;85:1513–9. doi: 10.1093/jnci/85.18.1513. [DOI] [PubMed] [Google Scholar]
- 2.Nakayama K, Nakayama N, Jinawath N, et al. Amplicon profiles in ovarian serous carcinomas. Int J Cancer. 2007;120:2613–7. doi: 10.1002/ijc.22609. [DOI] [PubMed] [Google Scholar]
- 3.Park JT, Li M, Nakayama N, et al. Notch-3 gene amplification in ovarian cancer. Cancer Res. 2006;66:6312–8. doi: 10.1158/0008-5472.CAN-05-3610. [DOI] [PubMed] [Google Scholar]
- 4.Weng AP, Aster JC. Multiple niches for Notch in cancer: context is everything. Curr Opin Genet Dev. 2004;14:48–54. doi: 10.1016/j.gde.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 5.Shih Ie M, Wang TL. Notch signaling, gamma-secretase inhibitors, and cancer therapy. Cancer Res. 2007;67:1879–82. doi: 10.1158/0008-5472.CAN-06-3958. [DOI] [PubMed] [Google Scholar]
- 6.Nitta M, Katabuchi H, Ohtake H, Tashiro H, Yamaizumi M, Okamura H. Characterization and tumorigenicity of human ovarian surface epithelial cells immortalized by SV40 large T antigen. Gynecol Oncol. 2001;81:10–7. doi: 10.1006/gyno.2000.6084. [DOI] [PubMed] [Google Scholar]
- 7.Nakayama K, Nakayama N, Kurman RJ, et al. Sequence mutations and amplification of PIK3CA and AKT2 genes in purified ovarian serous neoplasms. Cancer Biol Ther. 2006;5:779–85. doi: 10.4161/cbt.5.7.2751. [DOI] [PubMed] [Google Scholar]
- 8.Buckhaults P, Zhang Z, Chen YC, et al. Identifying tumor origin using a gene expression-based classification map. Cancer Res. 2003;63:4144–9. [PubMed] [Google Scholar]
- 9.Dang L, Yoon K, Wang M, Gaiano N. Notch3 signaling promotes radial glial/progenitor character in the mammalian telencephalon. Dev Neurosci. 2006;28:58–69. doi: 10.1159/000090753. [DOI] [PubMed] [Google Scholar]
- 10.Shimizu K, Chiba S, Hosoya N, et al. Binding of Delta1, Jagged1, and Jagged2 to Notch2 rapidly induces cleavage, nuclear translocation, and hyperphosphorylation of Notch2. Mol Cell Biol. 2000;20:6913–22. doi: 10.1128/mcb.20.18.6913-6922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindsell CE, Shawber CJ, Boulter J, Weinmaster G. Jagged: a mammalian ligand that activates Notch1. Cell. 1995;80:909–17. doi: 10.1016/0092-8674(95)90294-5. [DOI] [PubMed] [Google Scholar]
- 12.Ohtsuka T, Ishibashi M, Gradwohl G, Nakanishi S, Guillemot F, Kageyama R. Hes1 and Hes5 as notch effectors in mammalian neuronal differentiation. Embo J. 1999;18:2196–207. doi: 10.1093/emboj/18.8.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Purow BW, Haque RM, Noel MW, et al. Expression of Notch-1 and its ligands, Delta-like-1 and Jagged-1, is critical for glioma cell survival and proliferation. Cancer Res. 2005;65:2353–63. doi: 10.1158/0008-5472.CAN-04-1890. [DOI] [PubMed] [Google Scholar]
- 14.Tan DS, Agarwal R, Kaye SB. Mechanisms of transcoelomic metastasis in ovarian cancer. Lancet Oncol. 2006;7:925–34. doi: 10.1016/S1470-2045(06)70939-1. [DOI] [PubMed] [Google Scholar]
- 15.LaVoie MJ, Selkoe DJ. The Notch ligands, Jagged and Delta, are sequentially processed by alpha-secretase and presenilin/gamma-secretase and release signaling fragments. J Biol Chem. 2003;278:34427–37. doi: 10.1074/jbc.M302659200. [DOI] [PubMed] [Google Scholar]
- 16.Bland CE, Kimberly P, Rand MD. Notch-induced proteolysis and nuclear localization of the Delta ligand. J Biol Chem. 2003;278:13607–10. doi: 10.1074/jbc.C300016200. [DOI] [PubMed] [Google Scholar]
- 17.Ascano JM, Beverly LJ, Capobianco AJ. The C-terminal PDZ-ligand of JAGGED1 is essential for cellular transformation. J Biol Chem. 2003;278:8771–9. doi: 10.1074/jbc.M211427200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


