Abstract
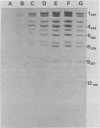
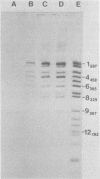
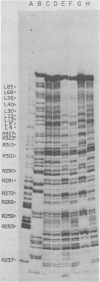
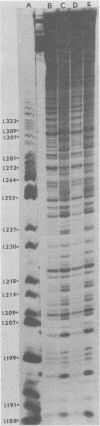
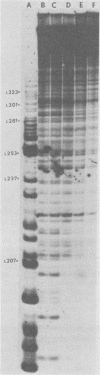
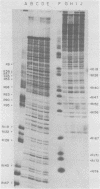
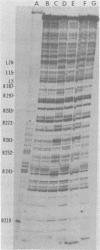
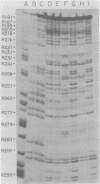
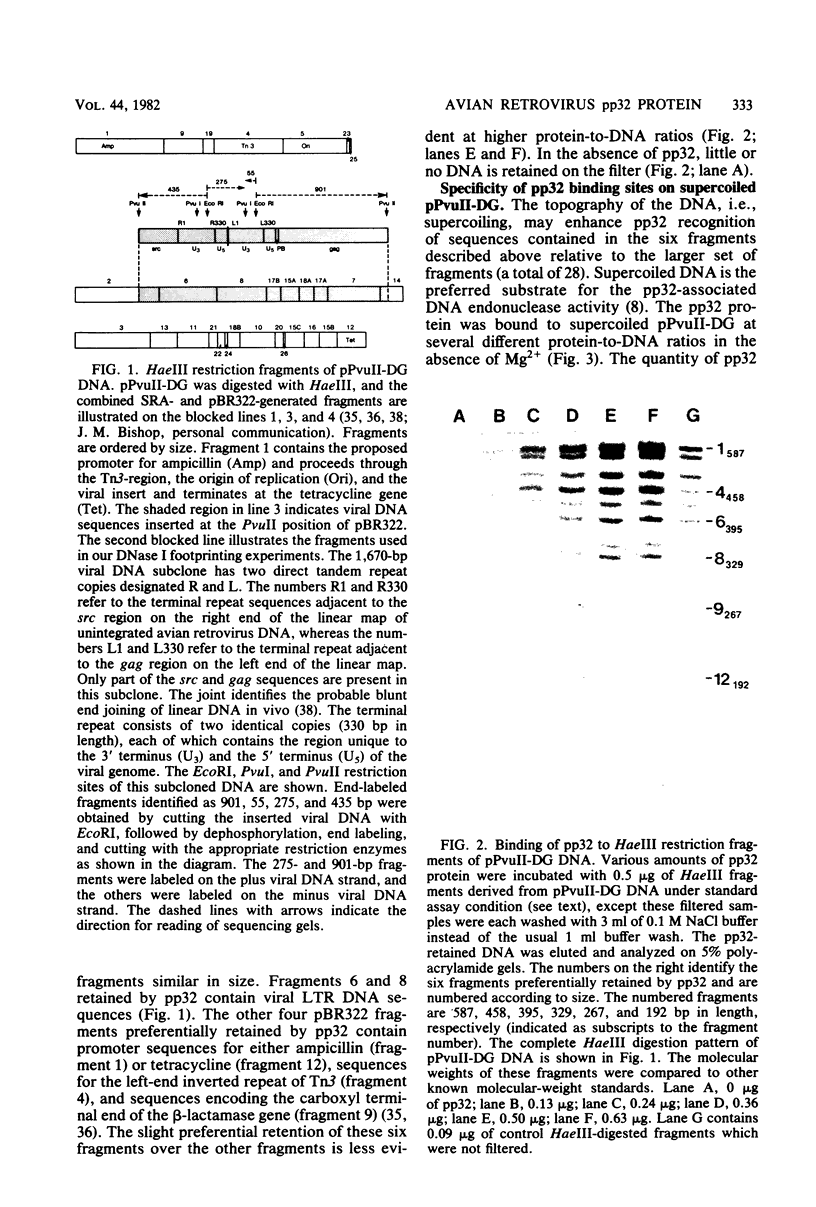
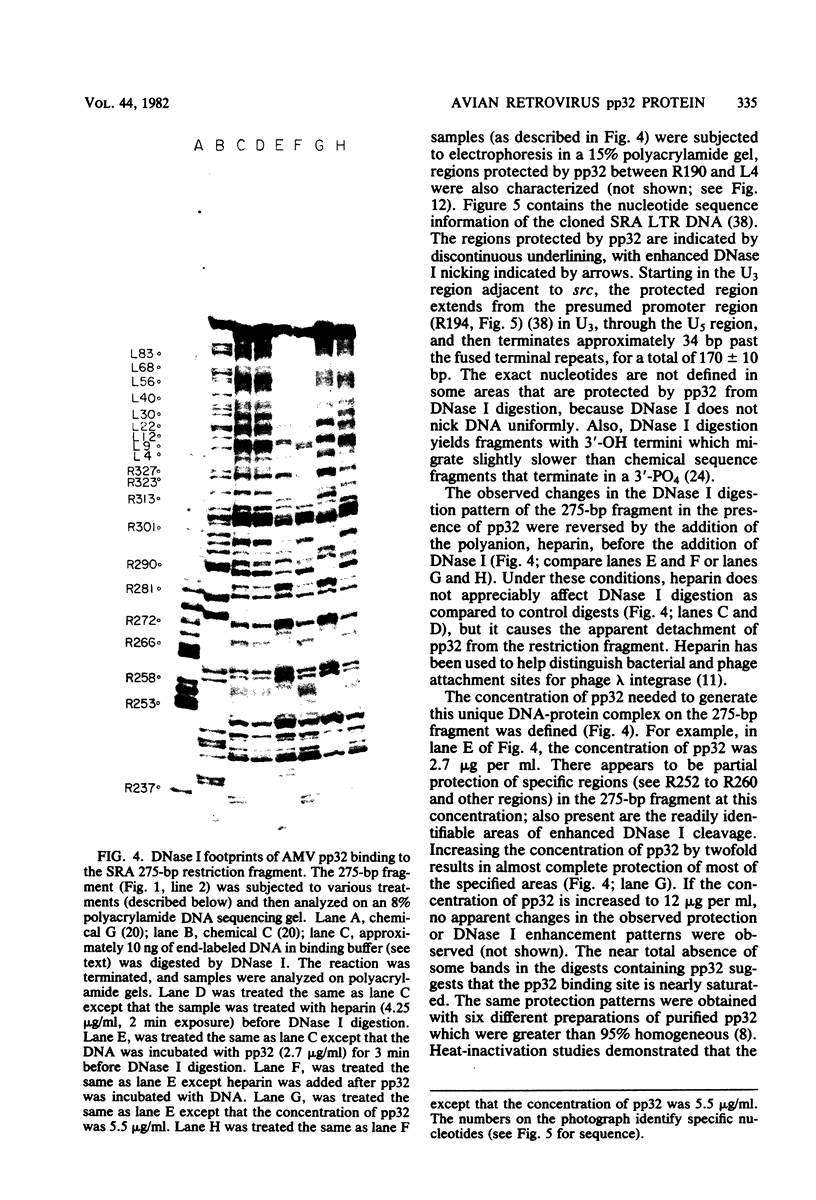
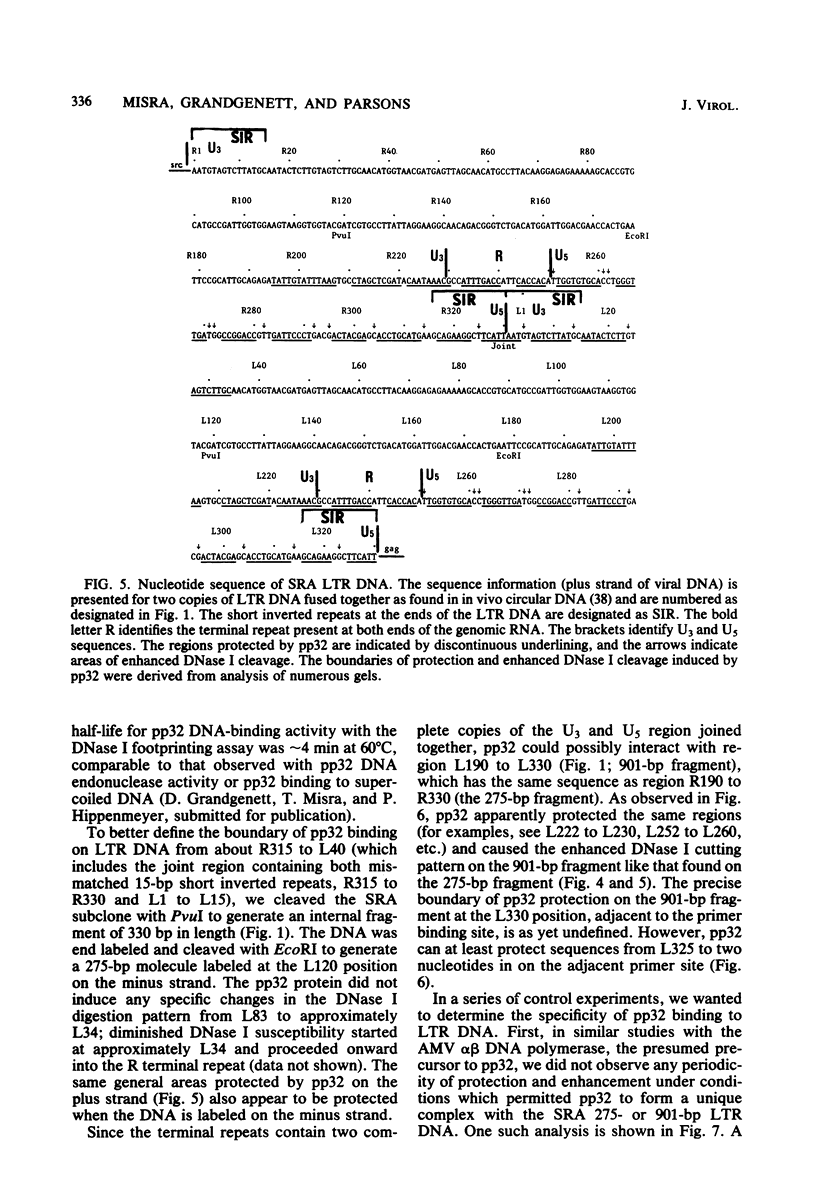
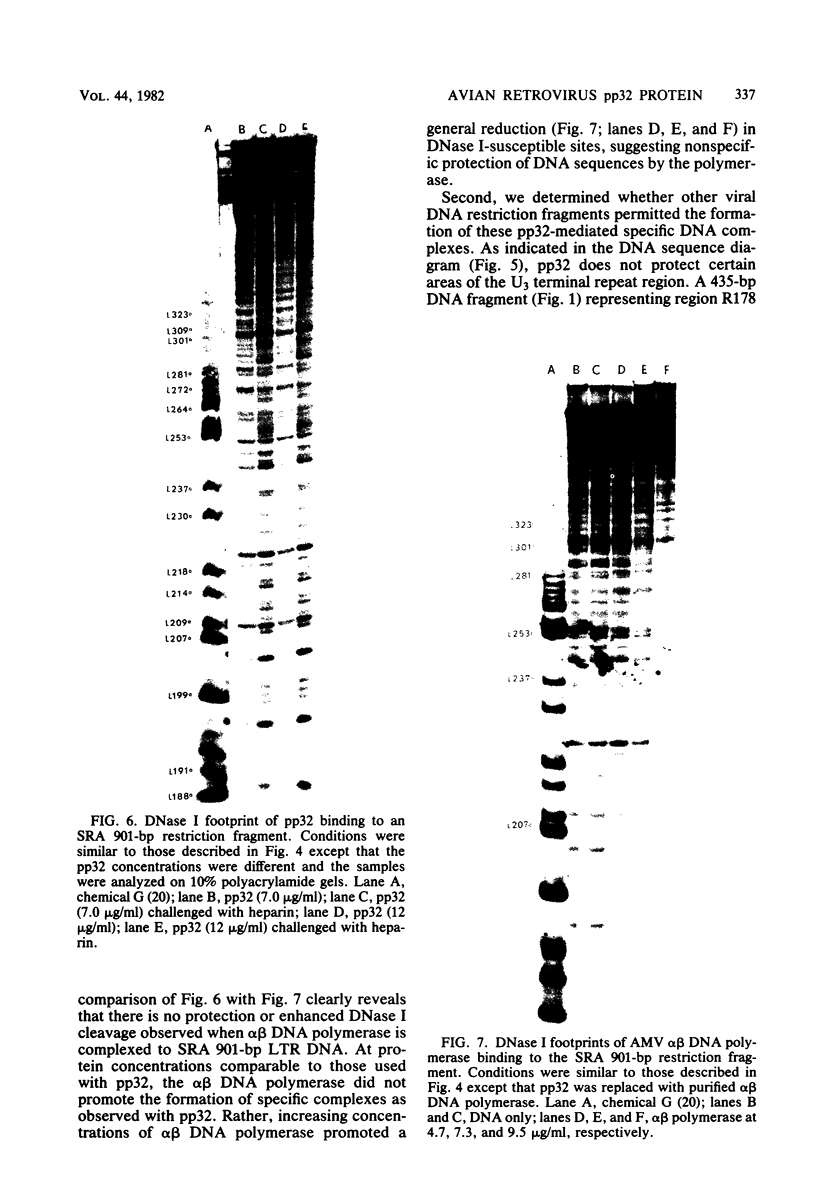
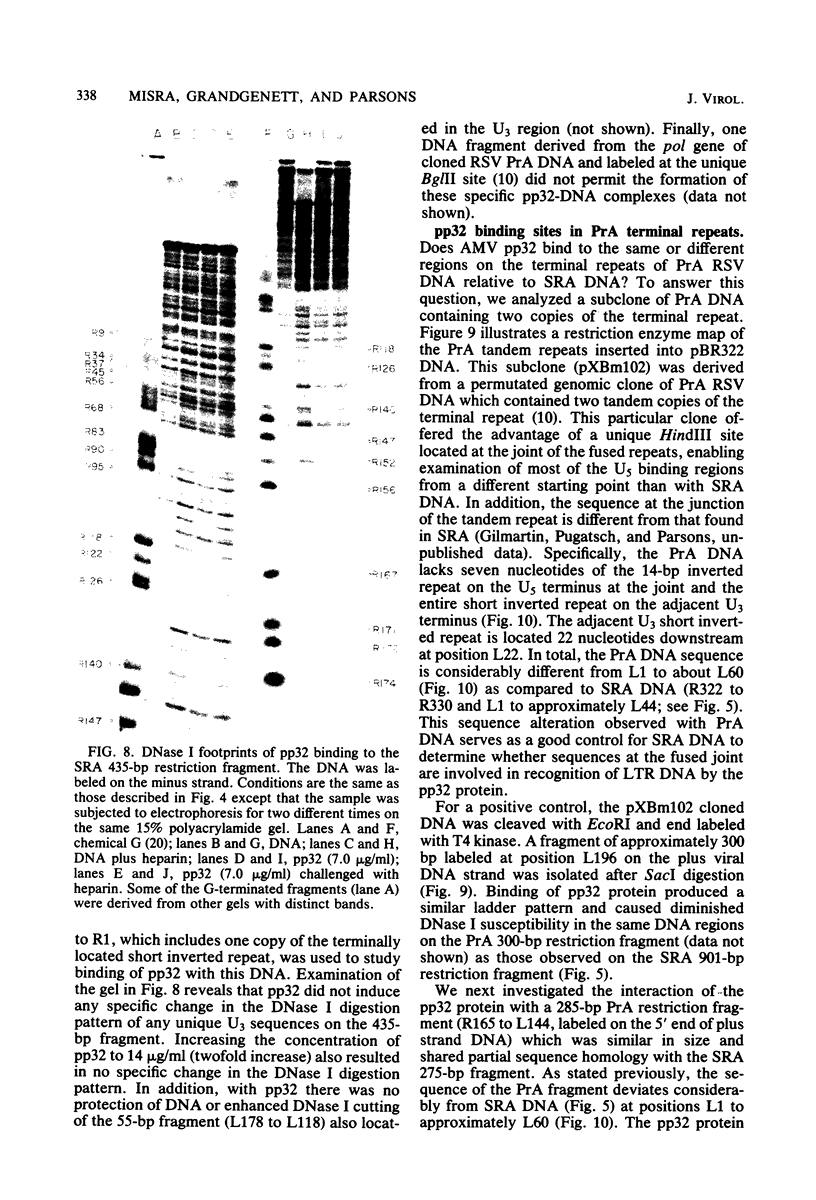
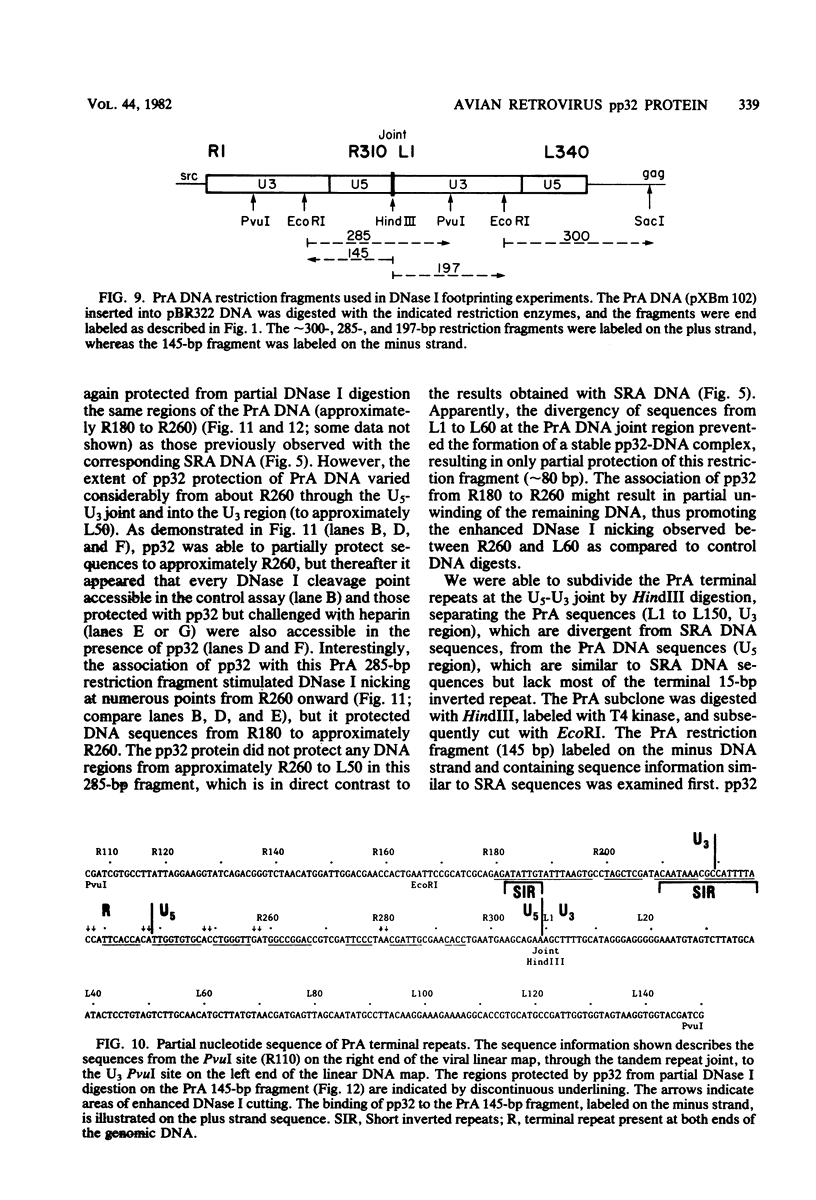
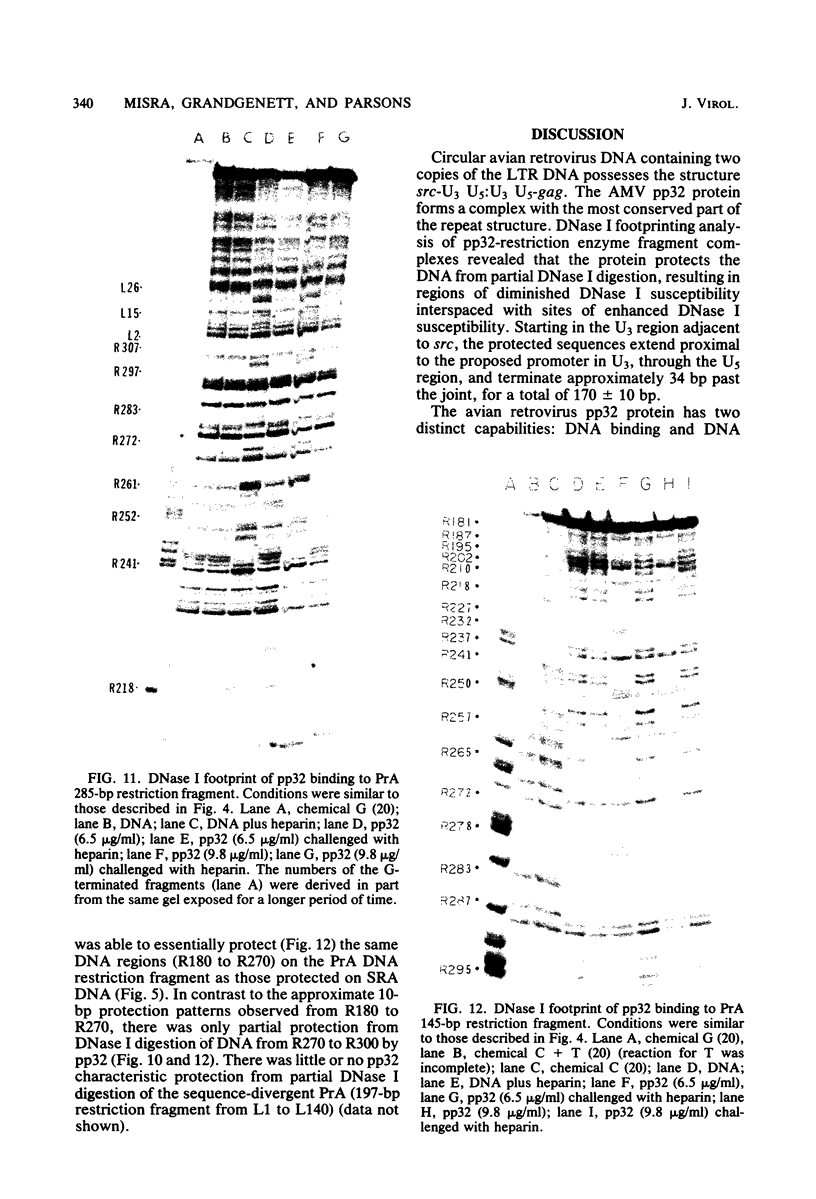
The avian retrovirus pp32 protein possesses a DNA-nicking activity which prefers supercoiled DNA as substrate. We have investigated the binding of pp32 to avian retrovirus long terminal repeat (LTR) DNA present in both supercoiled and linear forms. The cloned viral DNA was derived from unintegrated Schmidt-Ruppin A (SRA) DNA. A subclone of the viral DNA in pBR322 (termed pPvuII-DG) contains some src sequences, tandem copies of LTR sequences, and partial gag sequences in the order src-U3 U5:U3 U5-gag. Binding of pp32 to supercoiled pPvuII-DG DNA followed by digestion of this complex with a multicut restriction enzyme (28 fragments total) permitted pp32 to preferentially retain on nitrocellulose filters two viral DNA fragments containing only LTR DNA sequences. In addition, pp32 also preferentially retained four plasmid DNA fragments containing either potential promoters or Tn3 “left-end” inverted repeat sequences. Mapping of the pp32 binding sites on viral LTR DNA was accomplished by using the DNase I footprinting technique. The pp32 protein, but not the avian retrovirus αβ DNA polymerase, is able to form a unique protein-DNA complex with selected regions of either SRA or Prague A LTR DNAs. Partial DNase I digestion of a 275-base pair SRA DNA fragment complexed with pp32 gives upon electrophoresis in denaturing gels a unique ladder pattern, with regions of diminished DNase I susceptibility from 6 to 10 nucleotides in length, in comparison with control digests in the absence of protein. The binding of pp32 to this fragment also yields enhanced DNase I-susceptible sites that are spaced between the areas protected from DNase I digestion. The protected region of this unique complex was a stretch of 170 ± 10 nucleotides that encompasses the presumed viral promoter site in U3, which is adjacent to the src region, extends through U5, and proceeds past the joint into U3 for about 34 base pairs. No specific protection or DNase I enhancement by pp32 was observed in experiments with a 435-base pair SRA DNA fragment derived from a part of U3 and the adjacent src region or a 55-base pair DNA fragment derived from another part of U3. The DNA sequence of Prague A DNA at the fused LTRs differs from that of SRA DNA. The alteration in the sequence at the juncture of the LTRs prevented pp32 from forming a stable complex in this region of the LTR. Our results are relevant to two aspects of the interaction between pp32 and LTR DNA. First, the pp32 protein in the presence of selected viral DNA restriction fragments possibly forms a higher order oligomer analogous to Escherichia coli DNA gyrase-DNA complexes or eucaryotic nucleosome structures. Second, the specificity of the binding suggests a role for pp32 and the protected DNA sequences in the retrovirus life cycle. The preferred sequences to which pp32 binds include two adjacent 15-base pair inverted terminal repeats at the joint between U5 and U3 in SRA DNA. This region is involved in circularization of linear DNA and is perhaps the site that directs integration into cellular DNA.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Calos M. P., Miller J. H. Transposable elements. Cell. 1980 Jul;20(3):579–595. doi: 10.1016/0092-8674(80)90305-0. [DOI] [PubMed] [Google Scholar]
- Dhar R., McClements W. L., Enquist L. W., Vande Woude G. F. Nucleotide sequences of integrated Moloney sarcoma provirus long terminal repeats and their host and viral junctions. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3937–3941. doi: 10.1073/pnas.77.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilboa E., Mitra S. W., Goff S., Baltimore D. A detailed model of reverse transcription and tests of crucial aspects. Cell. 1979 Sep;18(1):93–100. doi: 10.1016/0092-8674(79)90357-x. [DOI] [PubMed] [Google Scholar]
- Golomb M., Grandgenett D. P. Endonuclease activity of purified RNA-directed DNA polymerase from avian myeloblastosis virus. J Biol Chem. 1979 Mar 10;254(5):1606–1613. [PubMed] [Google Scholar]
- Golomb M., Grandgenett D. P., Mason W. Virus-coded DNA endonuclease from avian retrovirus. J Virol. 1981 May;38(2):548–555. doi: 10.1128/jvi.38.2.548-555.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golomb M., Vora A. C., Grandgenett D. P. Purification of reverse transcriptase from avian retroviruses using affinity chromatography on heparin-sepharose. J Virol Methods. 1980;1(3):157–165. doi: 10.1016/0166-0934(80)90012-9. [DOI] [PubMed] [Google Scholar]
- Grandgenett D. P., Vora A. C., Schiff R. D. A 32,000-dalton nucleic acid-binding protein from avian retravirus cores possesses DNA endonuclease activity. Virology. 1978 Aug;89(1):119–132. doi: 10.1016/0042-6822(78)90046-6. [DOI] [PubMed] [Google Scholar]
- Hamilton D., Yuan R., Kikuchi Y. The nature of the complexes formed between the int protein and DNA. J Mol Biol. 1981 Oct 15;152(1):163–169. doi: 10.1016/0022-2836(81)90100-5. [DOI] [PubMed] [Google Scholar]
- Highfield P. E., Rafield L. F., Gilmer T. M., Parsons J. T. Molecular cloning of avian sarcoma virus closed circular DNA: structural and biological characterization of three recombinant clones. J Virol. 1980 Oct;36(1):271–279. doi: 10.1128/jvi.36.1.271-279.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P. L., Ross W., Landy A. The lambda phage att site: functional limits and interaction with Int protein. Nature. 1980 May 8;285(5760):85–91. doi: 10.1038/285085a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu T. W., Sabran J. L., Mark G. E., Guntaka R. V., Taylor J. M. Analysis of unintegrated avian RNA tumor virus double-stranded DNA intermediates. J Virol. 1978 Dec;28(3):810–818. doi: 10.1128/jvi.28.3.810-818.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. A., Walseth T. F. The enzymatic preparation of [alpha-32P]ATP, [alpha-32P]GTP, [32P]cAMP, and [32P]cGMP, and their use in the assay of adenylate and guanylate cyclases and cyclic nucleotide phosphodiesterases. Adv Cyclic Nucleotide Res. 1979;10:135–167. [PubMed] [Google Scholar]
- Jones O. W., Berg P. Studies on the binding of RNA polymerase to polynucleotides. J Mol Biol. 1966 Dec 28;22(2):199–209. doi: 10.1016/0022-2836(66)90126-4. [DOI] [PubMed] [Google Scholar]
- Kirkegaard K., Wang J. C. Mapping the topography of DNA wrapped around gyrase by nucleolytic and chemical probing of complexes of unique DNA sequences. Cell. 1981 Mar;23(3):721–729. doi: 10.1016/0092-8674(81)90435-9. [DOI] [PubMed] [Google Scholar]
- Kopchick J. J., Harless J., Geisser B. S., Killam R., Hewitt R. R., Arlinghaus R. B. Endodeoxyribonuclease activity associated with Rauscher murine leukemia virus. J Virol. 1981 Jan;37(1):274–283. doi: 10.1128/jvi.37.1.274-283.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S. Y., Riggs A. D. Lac repressor binding to non-operator DNA: detailed studies and a comparison of eequilibrium and rate competition methods. J Mol Biol. 1972 Dec 30;72(3):671–690. doi: 10.1016/0022-2836(72)90184-2. [DOI] [PubMed] [Google Scholar]
- Liu L. F., Wang J. C. DNA-DNA gyrase complex: the wrapping of the DNA duplex outside the enzyme. Cell. 1978 Nov;15(3):979–984. doi: 10.1016/0092-8674(78)90281-7. [DOI] [PubMed] [Google Scholar]
- Lutter L. C. Precise location of DNase I cutting sites in the nucleosome core determined by high resolution gel electrophoresis. Nucleic Acids Res. 1979 Jan;6(1):41–56. doi: 10.1093/nar/6.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Morrison A., Cozzarelli N. R. Contacts between DNA gyrase and its binding site on DNA: features of symmetry and asymmetry revealed by protection from nucleases. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1416–1420. doi: 10.1073/pnas.78.3.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen-Meyer J., Nes I. F. Purification and properties of DNA endonucleases associated with Friend leukemia virus. Nucleic Acids Res. 1980 Nov 11;8(21):5043–5055. doi: 10.1093/nar/8.21.5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes D., Klug A. Helical periodicity of DNA determined by enzyme digestion. Nature. 1980 Aug 7;286(5773):573–578. doi: 10.1038/286573a0. [DOI] [PubMed] [Google Scholar]
- Ross W., Landy A., Kikuchi Y., Nash H. Interaction of int protein with specific sites on lambda att DNA. Cell. 1979 Oct;18(2):297–307. doi: 10.1016/0092-8674(79)90049-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushlow K. E., Lautenberger J. A., Reddy E. P., Souza L. M., Baluda M. A., Chirikjian J. G., Papas T. S. Nucleotide sequence analysis of the long terminal repeat of avian myeloblastosis virus and adjacent host sequences. J Virol. 1982 Jun;42(3):840–846. doi: 10.1128/jvi.42.3.840-846.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabran J. L., Hsu T. W., Yeater C., Kaji A., Mason W. S., Taylor J. M. Analysis of integrated avian RNA tumor virus DNA in transformed chicken, duck and quail fibroblasts. J Virol. 1979 Jan;29(1):170–178. doi: 10.1128/jvi.29.1.170-178.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiff R. D., Grandgenett D. P. Partial phosphorylation in vivo of the avian retrovirus pp32 DNA endonuclease. J Virol. 1980 Dec;36(3):889–893. doi: 10.1128/jvi.36.3.889-893.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz A., Galas D. J. Sequence-specific interactions of the tight-binding I12-X86 lac repressor with non-operator DNA. Nucleic Acids Res. 1980 Feb 11;8(3):487–506. doi: 10.1093/nar/8.3.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shank P. R., Varmus H. E. Virus-specific DNA in the cytoplasm of avian sarcoma virus-infected cells is a precursor to covalently closed circular viral DNA in the nucleus. J Virol. 1978 Jan;25(1):104–104. doi: 10.1128/jvi.25.1.104-104.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro J. A. Molecular model for the transposition and replication of bacteriophage Mu and other transposable elements. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1933–1937. doi: 10.1073/pnas.76.4.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimotohno K., Mizutani S., Temin H. M. Sequence of retrovirus provirus resembles that of bacterial transposable elements. Nature. 1980 Jun 19;285(5766):550–554. doi: 10.1038/285550a0. [DOI] [PubMed] [Google Scholar]
- Shoemaker C., Goff S., Gilboa E., Paskind M., Mitra S. W., Baltimore D. Structure of a cloned circular Moloney murine leukemia virus DNA molecule containing an inverted segment: implications for retrovirus integration. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3932–3936. doi: 10.1073/pnas.77.7.3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G., Shinnick T. M., Verma I. M., Lerner R. A. Nucleotide sequence of Moloney leukemia virus: 3' end reveals details of replications, analogy to bacterial transposons, and an unexpected gene. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3302–3306. doi: 10.1073/pnas.77.6.3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanstrom R., DeLorbe W. J., Bishop J. M., Varmus H. E. Nucleotide sequence of cloned unintegrated avian sarcoma virus DNA: viral DNA contains direct and inverted repeats similar to those in transposable elements. Proc Natl Acad Sci U S A. 1981 Jan;78(1):124–128. doi: 10.1073/pnas.78.1.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Beveren C., Goddard J. G., Berns A., Verma I. M. Structure of Moloney murine leukemia viral DNA: nucleotide sequence of the 5' long terminal repeat and adjacent cellular sequences. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3307–3311. doi: 10.1073/pnas.77.6.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg R. A. Structure of the intermediates leading to the integrated provirus. Biochim Biophys Acta. 1977 Mar 21;473(1):39–55. doi: 10.1016/0304-419x(77)90006-3. [DOI] [PubMed] [Google Scholar]