Abstract
An intracellular signaling from the endoplasmic reticulum (ER) to the nucleus, called the unfolded protein response (UPR), is activated when unfolded proteins are accumulated in the ER under a variety of stress conditions (“ER stress”). We and others recently identified Hac1p/Ern4p as a transcription factor responsible for the UPR in Saccharomyces cerevisiae. It was further reported that Hac1p (238 aa) is detected only in ER-stressed cells, and its expression is mediated by unconventional splicing of HAC1 precursor mRNA. The splicing replaces the C-terminal portion of Hac1p; it was proposed that precursor mRNA is also translated but the putative product of 230 aa is rapidly degraded by the ubiquitin–proteasome pathway. We have identified and characterized the same regulated splicing and confirmed its essential features. Contrary to the above proposal, however, we find that the 238-aa product of mature mRNA and the 230-aa-type protein tested are highly unstable with little or no difference in stability. Furthermore, we demonstrate that the absence of Hac1p in unstressed cells is due to the lack of translation of precursor mRNA. We conclude that Hac1p is synthesized as the result of ER stress-induced mRNA splicing, leading to activation of the UPR.
INTRODUCTION
Newly synthesized secretory and transmembrane proteins traverse the endoplasmic reticulum (ER), where they fold into correct tertiary and quaternary structures. The productive folding process of these proteins is assisted by molecular chaperones and folding enzymes localized in the ER (reviewed by Gething and Sambrook, 1992; Helenius et al., 1992). When unfolded proteins are accumulated in the ER by a variety of physiological or environmental stress conditions (“ER stress”), synthesis of these chaperones and enzymes is known to be induced at the level of transcription (Lee, 1987; Kozutsumi et al., 1988). This means that eukaryotic cells possess an intracellular signaling pathway from the ER to the nucleus, called the unfolded protein response (UPR) pathway (reviewed by McMillan et al., 1994; Shamu et al., 1994; Pahl and Baeuerle, 1997).
In Saccharomyces cerevisiae, several components of the UPR have been identified. Genetic approach identified the ERN1/IRE1 gene that encodes a transmembrane protein kinase Ern1p localized in the ER (Cox et al., 1993; Mori et al., 1993). Ern1p spans the ER membrane (or possibly the nuclear membrane, which is continuous to the ER membrane) once, with its N-terminal half located inside microsomes and with its C-terminal half carrying essential protein kinase activity on the cytoplasmic side (Mori et al., 1993). The N-terminal domain of Ern1p is likely to sense the accumulation of unfolded proteins and activates the protein kinase domain through its own oligomerization and autophosphorylation (Shamu and Walter, 1996; Welihinda and Kaufman, 1996), although the mechanism remains unknown. Thus, the information about changes in the ER lumen is transmitted to the cytosolic compartment by Ern1p and then to the nucleus. In the nucleus, each of the target genes of the UPR is thought to contain a cis-acting element, termed the unfolded protein-response element (UPRE) as an upstream activator sequence (Mori et al., 1992; Kohno et al., 1993; Partaledis and Berlin, 1993). UPRE was proposed to be a binding site of putative unfolded protein-response factor (UPRF; Mori et al., 1992). So far six ER lumenal proteins have been shown to be regulated by the UPR, including two essential proteins: Kar2p, a member of the heat shock protein 70 family (Normington et al., 1989; Rose et al., 1989), and Pdi1p, protein disulfide isomerase (LaMantia et al., 1991; Tachikawa et al., 1991).
UPRE was originally identified as a 22-bp sequence that is necessary and sufficient for the induction of Kar2p by ER stress (Mori et al., 1992). We conducted extensive mutational analysis and showed that UPRE contains a partial palindrome with a spacer of 1 nucleotide (nt; CAGCGTG) that is essential for its function (Mori et al., 1996). This sequence resembles the E-box consensus (CANNTG) to which trans-acting factors containing a basic region as a DNA-binding domain are known to bind as a homo- or heterodimer (Hurst, 1995; Littlewood and Evan, 1995). However, the presence of a single C residue between CAG and GTG is characteristic of UPRE and this one-base spacing is critical for the response to ER stress. In addition, the sequence GAA located upstream of the partial palindrome was found to be important for the activity of UPRE. Thus, we proposed that yeast UPRF is composed of total three trans-acting polypeptides (a dimer of a basic region-containing protein[s] plus a protein recognizing the sequence GAA; Mori et al., 1996).
We and others recently identified the HAC1/ERN4 gene as an essential component of the UPR (Cox and Walter, 1996; Mori et al., 1996; Nikawa et al., 1996). The nonessential HAC1 gene encodes a basic-leucine zipper (bZIP) protein (Hac1p) of 230 aa. Haploid cells lacking Hac1p (hac1Δ) are unable to induce transcription of any of the target genes tested and exhibit sensitivity to ER stress. By using electrophoretic mobility shift assays, Hac1p was shown to bind specifically to UPRE (Cox and Walter, 1996; Mori et al., 1996). In addition, we demonstrated that Hac1p recognizes the palindromic sequence separated by 1 nt in UPRE both in vivo and in vitro. These results led us to conclude that Hac1p represents a major component of the putative transcription factor UPRF responsible for the UPR (Mori et al., 1996).
Cox and Walter (1996) further analyzed the mechanism of activation of Hac1p and reported that the UPR is regulated by an unconventional type of mRNA splicing that is induced by Ern1p-mediated signaling from the ER: internal 252 nt were removed by the splicing from constitutively expressed HAC1 precursor mRNA (pre-mRNA) to produce mature mRNA in response to ER stress in the wild-type but not in the ern1Δ strain. Moreover, the expression of the intron-less HAC1 cDNA constitutively activated the UPR even in the ern1Δ strain. This splicing is unique in that the sequences around the splice sites do not match the consensus for conventional splicing (Kreivi and Lamond, 1996). Another interesting feature is that the open reading frame switches: the splicing causes replacement of the C-terminal portion of Hac1p due to the location of the 5′ splice site inside the coding region. HAC1 pre-mRNA encodes a protein of 230 aa, whereas mature mRNA encodes a protein of 238 aa, although these two proteins are supposed to share an identical N-terminal 220 aa.
Importantly, only ER-stressed cells produced detectable amounts of Hac1p, which was shown by immunoblotting to contain 18 aa encoded by the second exon and thus translated from mature mRNA. Three lines of evidence led to the proposal (Cox and Walter, 1996) that although 230aa-Hac1p was constitutively translated from pre-mRNA, it was highly unstable and rapidly degraded by the ubiquitin–proteasome pathway, whereas 238aa-Hac1p produced from mature mRNA was stable. First, a majority of pre-mRNA and mature mRNA was shown to be associated with polysomes, suggesting that they both could be translated. Second, β-galactosidase expressed from a reporter gene monitoring the UPR was constitutively elevated in mutants defective in some of the ubiquitin conjugating enzymes. Third, when the last 10 aa present in 230aa-Hac1p was deleted, the resulting 220aa-Hac1p constitutively activated the UPR as effectively as 238aa-Hac1p produced from the intron-less HAC1 cDNA. They inferred from these results that the combination of the N-terminal portion of 220 aa and the 10 aa, but not the last 18 aa present in 238aa-Hac1p, made Hac1p highly unstable.
We independently identified the regulated splicing of HAC1 pre-mRNA in response to ER stress, as described in the initial sections of this article. We then present data focusing on the question of why only Hac1p translated from mature mRNA is detected in cell extracts. Contrary to the previous proposal cited above, the differential protein stability is unlikely to be the main mechanism regulating the expression of Hac1p. The evidence presented below clearly indicates that the presence of intron prevents pre-mRNA from being translated: namely, Hac1p is synthesized only after the mRNA splicing takes place. Accordingly, we propose an alternative model for the regulated expression of Hac1p leading to activation of the UPR in S. cerevisiae.
MATERIALS AND METHODS
Strains and Microbiological Techniques
The yeast strains used in this study are listed in Table 1. The compositions of rich broth medium (YPD) and synthetic complete medium used for selection of transformants such as SC(−Ura, Leu) have been described (Sherman et al., 1986). Tunicamycin was obtained from Sigma (catalogue no. T-7765) and used at a concentration of 5 μg/ml throughout the experiments. Yeast cells were transformed by the lithium acetate method (Ito et al., 1983).
Table 1.
Yeast strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| KMY1005 | MATα leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 | Mori et al. (1996) |
| KMY1015 | KMY1005 ern1Δ::TRP1 | Mori et al. (1996) |
| KMY1045 | KMY1005 hac1Δ::TRP1 | Mori et al. (1996) |
| KMY1105 | KMY1005 | This study |
| ura3-52::URA3-UPRE(Y)-CYC1-lacZ | ||
| KMY1115 | KMY1105 ern1Δ::TRP1 | This study |
| KMY1145 | KMY1105 hac1Δ::TRP1 | This study |
| KMY2005 | MATα leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 sec53-6 | Mori et al. (1996) |
| KMY2105 | KMY2005 | This study |
| ura3-52::URA3-UPRE(Y)-CYC1-lacZ | ||
| KMY2115 | KMY2105 ern1Δ::TRP1 | This study |
| MHY501 | MATα leu2-3,112 ura3-52 his3-Δ200 trp1-1 lys2-801 | Chen et al. (1993) |
| MHY498 | MHY501 ubc4-Δ1::HIS3 | Chen et al. (1993) |
| MHY499 | MHY501 ubc5-Δ1::LEU2 | Chen et al. (1993) |
| MHY508 | MHY501 ubc4-Δ1::HIS3 ubc5-Δ1::LEU2 | Chen et al. (1993) |
| MHY507 | MHY501 ubc7Δ::LEU2 | Chen et al. (1993) |
| MHY500 | MATa leu2-3,112 ura3-52 his3-Δ200 trp1-1 lys2-801 | Chen et al. (1993) |
| MHY513 | MHY500 ubc4-Δ1::HIS3 | Chen et al. (1993) |
Construction of Plasmids
Recombinant DNA techniques were carried out as described (Sambrook et al., 1989). Reporter plasmids, such as pSCZ-Y and pMCZ-Y (the UPRE-CYC1-lacZ gene carried on a single-copy vector and a multicopy vector, respectively), and expression plasmids for the ERN1 gene, such as YCp-ERN1 and YEp-ERN1 (Cen and 2 μm, respectively, in Figure 3), were described previously (Mori et al., 1996). YCp-HAC1 used herein (also referred as YCp-HAC1WT) was previously described as YCp-ERN4, which contains the LEU2-selectable marker (Mori et al., 1996). YCp-HAC1ΔHN or ΔHA in Figure 2 was constructed by deleting the 0.67-kb HindIII818–EcoNI1492 fragment or 0.33-kb HindIII818–AscI1144 fragment from YCp-HAC1WT, respectively. YCp-HAC1Δintron (also referred as 238-type) was constructed by replacing the 0.87-kb MluI272–AscI1144 fragment in YCp-HAC1WT with the corresponding fragment of the reverse transcriptase (RT)- coupled polymerase chain reaction (PCR) product obtained from tunicamycin-treated cells (Figure 4B), thus lacking the intron of the 252 nt. The intron-less HAC1 gene was also inserted into a single-copy vector YCp50 containing the URA3 selectable marker (Rose and Broach, 1991). YCp-HAC1–220-type and 230-type in Figures 6, 7, and 10 were constructed by site-directed mutagenesis (Kunkel, 1985). A XbaI site was introduced at nt 640 in YCp-HAC1WT or YCp-HAC1Δintron [designated WT(XbaI) or Δintron(XbaI), respectively, in Figure 9] by replacing the MfeI fragment (residues 632–645) with a double-stranded oligonucleotide (AATTGATTCTAGA, XbaI site underlined). Three mutant versions of the HAC1 gene described in Figure 9 (Ala221Stop, Ala221Stop′, and Asn216Stop) were constructed by replacing the 0.18-kb XbaI640–HindIII818 fragment with the corresponding fragment of the product obtained by PCR-mediated mutagenesis after its sequence had been confirmed.
Figure 3.
Induction of 1.2-kb HAC1 mRNA correlates with the UPR. (A) The ERN+ (KMY1105) and ern1Δ (KMY1115) strains were transformed with a vector alone (V) or a Ern1p expression plasmid (a single-copy or multicopy plasmid indicated by Cen or 2 μm, respectively). Transformants were grown at 30°C in SC(−Ura, Leu) medium to midlogarithmic phase, and aliquots were incubated for 1 h in the presence (+) or absence (−) of tunicamycin (TM). Total RNAs were extracted and analyzed by Northern blot hybridization using DNA probes specific for HAC1, KAR2, or yeast actin ACT1. Positions of the 1.4-kb and 1.2-kb HAC1 mRNAs are indicated. In addition to these two mRNA species, tunicamycin treatment produced a faint band of 0.7 kb, that was not analyzed in this report. (B) The ERN+ and ern1Δ strains with the sec53 background in which the UPRE-CYC1-lacZ reporter gene had been integrated into the respective chromosome (KMY2105 and KMY2115, respectively) were grown at the permissive temperature of 23°C in SC(−Ura) medium to midlogarithmic phase, and aliquots were incubated for an additional hour at 23°C or at the seminonpermissive temperature of 30°C. Total RNAs were extracted and analyzed as in A. (C) The ERN+, SEC+ strain (KMY1105) was treated with tunicamycin at 30°C for the times indicated as in A, and RNA was similarly analyzed by Northern blot hybridization. Relative radioactivities of each band (HAC1, KAR2, and PDI1) were determined using BioImaging Analyzer BAS-2000 (Fuji Photo Film), corrected for ACT1 values, and plotted after normalization to the 0 time values.
Figure 2.
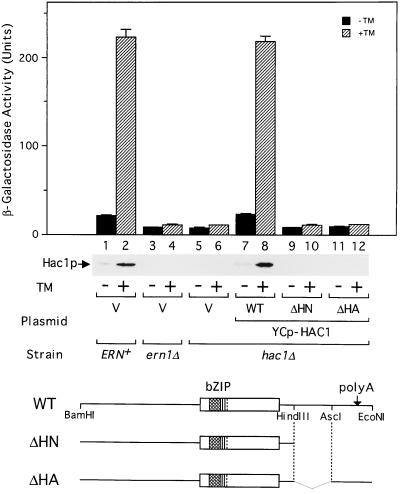
Induction of Hac1p requires 3′UTR of HAC1 mRNA. The ERN+, ern1Δ, and hac1Δ strains with the UPRE-CYC1-lacZ reporter gene integrated into the respective chromosome (KMY1105, 1115, and 1145, respectively) were transformed with a vector alone (V) or a single-copy expression plasmid carrying the wild-type (WT) or a mutant (ΔHN or ΔHA) HAC1 gene, whose structures are schematically drawn at the bottom. bZIP and polyA denote the bZIP region and the polyadenylation site, respectively. Transformants were grown at 30°C in SC(−Ura, Leu) medium to a midlogarithmic phase, and aliquots were incubated in the presence (hatched bars) or absence (solid bars) of tunicamycin (TM). Samples taken after 3 h were used for β-galactosidase assays, and the activities are presented as the mean ± SD, based on duplicate determinations with three independent transformants. Separate samples taken after 1 h were used for extracting total proteins that were analyzed by immunoblotting with purified anti-Hac1p antibodies.
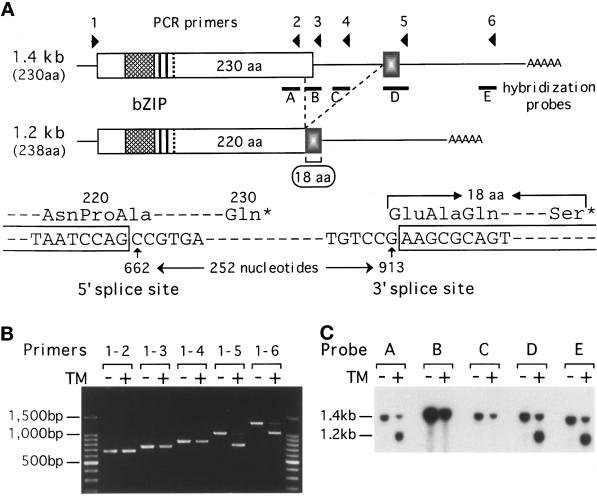
Figure 4.
ER stress-induced mRNA splicing replaces the C-terminal portion of Hac1p. (A) Structures of the 1.4-kb and 1.2-kb HAC1 mRNAs are schematically presented. Locations of primers 1–6 for RT-PCR and DNA probes A-E for Northern blot hybridization are also indicated. The 1.4-kb mRNA can encode a bZIP protein of 230 aa, whereas the 1.2-kb mRNA resulting from ER-stress–induced splicing can encode a bZIP protein of 238 aa. Shown below are the nucleotide sequences around the 5′ and 3′ splice sites. The splicing deletes the internal 252 nt from nt 662 to 913 (the A of the first ATG codon is set as +1), resulting in the replacement of the C-terminal 10 aa with a stretch of 18 aa encoded by the second exon. (B) Total RNAs were extracted from the ERN+ strain grown for 1 h in the presence (+) or absence (−) of tunicamycin (TM). HAC1 cDNA was prepared by RT-PCR using a fixed 5′ primer 1 and various 3′ primers (primers 2–6), and the products were analyzed by 1% agarose gel electrophoresis. The amounts of PCR products did not reflect the amounts of mRNA well due to excessive cycles used. Positions of the 100-bp ladder DNA size markers are indicated. (C) Total RNAs prepared as in B were analyzed by Northern blot hybridization using the various DNA probes indicated in A, which had been labeled with [32P]dCTP by PCR. Positions of the 1.4-kb and 1.2-kb HAC1 mRNAs are indicated.
Figure 6.
Constitutive expression of Hac1p of 238, 220, and 230 aa. (A) Structures of the wild-type (WT) and mutant versions of the HAC1 gene are schematically presented. Thin lines denote nucleotide segments deleted from the HAC1 gene. (B and C) The ern1Δ (KMY1115) and hac1Δ (KMY1145) strains were transformed with a vector alone (V) or a single-copy expression plasmid carrying a mutant version (238, 220, or 230) of the HAC1 gene. Transformants were grown at 30°C in SC(−Ura, Leu) medium to midlogarithmic phase, and aliquots were incubated in the presence (hatched bars and lanes of even numbers) or absence (solid bars and lanes of odd numbers) of tunicamycin (TM). Samples taken after 3 h were used for β-galactosidase assays, and the activities are presented as the mean ± SD, based on duplicate determinations with three independent transformants. Separate samples taken after 1 h were used for extracting total proteins or total RNAs that were analyzed by immunoblotting using anti-Hac1p, anti-Kar2p, and anti-Pdi1p antibodies or by Northern blot hybridization using DNA probes specific for HAC1 and ACT1. Hac1p of 238, 220, and 230 aa were also translated in vitro as described in MATERIALS AND METHODS. The positions of molecular mass markers and Hac1p of 238, 220, and 230 aa are indicated. * denotes a nonspecific band. (B, inset) The levels of Kar2p and Pdi1p in total proteins extracted from tunicamycin-untreated hac1Δ cells.
Figure 7.
Stability of Hac1p of 238, 220, and 230 aa. (A) Transformants of the hac1Δ strain in which each of the mutant versions of the HAC1 gene described in Figure 6A had been introduced were grown at 30°C in SC(−Ura, Leu) medium to midlogarithmic phase, and aliquots were treated with cycloheximide (20 μg/ml). Immediately after the times indicated, the cultures were poured over ice and the cells were collected by centrifugation. Total proteins were extracted and analyzed by immunoblotting using anti-Hac1p and anti-Kar2p antibodies. For Hac1p of 220 aa, a shorter exposure of the film is also shown below. * denotes a nonspecific band. (B) The hac1Δ strain (KMY1045) was cotransformed with a plasmid carrying the mutant (230-type) HAC1 gene and the LEU2-selectable marker and a plasmid carrying the intron-less (238-type) HAC1 gene and the URA3-selectable marker. The resulting transformant was grown, pulse-labeled for 5 min with [35S]methionine and [35S]cysteine, and then chased for the times indicated as described in MATERIALS AND METHODS. The hac1Δ strain carrying either the 230-type or 238-type mutant HAC1 gene was pulse-labeled for 5 min and shown for comparison. Hac1p and Pdi1p (internal control) were immunoprecipitated and subjected to SDS-PAGE (12% gel). The positions of molecular mass markers, Pdi1p, and Hac1p of 238 aa and 230 aa are indicated. Radioactivities of each band were determined by using a BioImaging Analyzer BAS-2000 and corrected for Pdi1p values. Data from two separate experiments (indicated by circles and triangles) are plotted after normalization to the 0 time value of 230aa-Hac1p.
Figure 10.
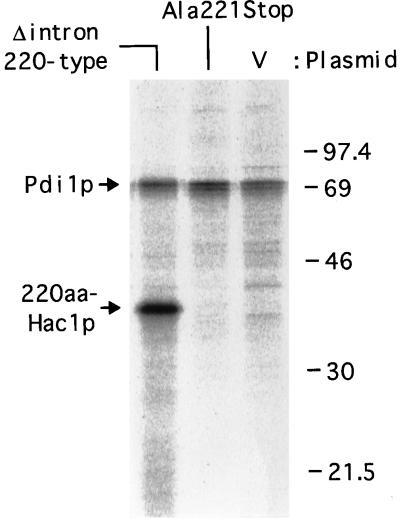
Synthesis rates of 220aa-Hac1p encoded by intron-less and intron-containing mRNAs. The hac1Δ strain (KMY1145) was transformed with a vector alone (V) or a mutant version (Δintron 220-type or Ala221Stop) of the HAC1 gene. Each transformant was grown and pulse-labeled for 5 min with [35S]methionine and [35S]cysteine as described in MATERIALS AND METHODS. Hac1p and Pdi1p were immunoprecipitated and subjected to SDS-PAGE (12% gel). Radioactive bands were visualized by using a BioImaging Analyzer BAS-2000. The positions of molecular mass markers, 220aa-Hac1p, and Pdi1p are indicated in kilodaltons.
Figure 9.
Production of Hac1p only from mature mRNA. At the top, the nucleotide sequence of the wild-type (WT) HAC1 gene around the 5′ splice site and its deduced amino acid sequence are shown. In a mutant HAC1 gene designated WT(XbaI), a XbaI site (underlined) was created at nt 640 between the two MfeI sites in YCp-HAC1WT that changed aa 214 and 215 from Leu-Asp to Ser-Arg. Δintron(XbaI) contains a XbaI site in YCp-HAC1Δintron similarly. The Ala221Stop mutant contains a stop codon TAG (double underlined) instead of the codon for Ala221. The Ala221Stop′ mutant contains a 2-nt (TA) insertion between the codons for Pro220 and Ala221 that creates a stop codon (double underlined) immediately after Pro220 but maintains nucleotide sequences at the 5′ splice site from the position −2 (AGCCG—). The Asn216Stop mutant contains a stop codon TAG (double underlined) instead of the codon for Asn216. The hac1Δ strain (KMY1145) was transformed with a vector alone (V) or each of these constructs as indicated. Transformants were grown at 30°C in SC(−Ura, Leu) medium to midlogarithmic phase, and aliquots were incubated for 3 h in the presence (hatched bars) or absence (solid bars) of tunicamycin (TM). β-Galactosidase activities in cell extracts were determined, and are presented as the mean ± SD, based on duplicate determinations with three independent transformants. Separate samples taken after 1 h were used for extracting total proteins or total RNAs that were analyzed by immunoblotting using anti-Hac1p antibodies or by Northern blot hybridization using DNA probes specific for HAC1 and ACT1. Hac1p of 238 and 220 aa were also translated in vitro. * denotes a nonspecific band.
Preparation of Cell Extracts
Yeast cell extracts for electrophoretic mobility shift assays and immunoblotting were prepared from approximately 100 ml of midlogarithmic phase cultures (OD600 ∼ 0.8) as described (Mori et al., 1992) with some modifications. Breakage buffer [200 mM Tris(hydroxymethyl)aminomethane (Tris)-HCl, pH8.0, 10 mM MgCl2, 10% glycerol, and 0.5 mM dithiothreitol] contained various protease inhibitors (2 μM pepstatinA, 2 μM leupeptin, 2 mg/l chymostatin, 2 mM benzamidine, and 1 mM phenylmethylsulfonyl fluoride). Total proteins extracted as described (Mori et al., 1992) were precipitated by a final concentration of 2.6 M (NH4)2SO4 by using 4 M instead of saturated (NH4)2SO4 to obtain whole cell extracts. When proteins in extracts were fractionated, proteins were first precipitated by 2.0 M (NH4)2SO4 (final concentration) to obtain fraction A. Proteins that remained in the supernatant were then precipitated by 2.6 M (NH4)2SO4 to obtain fraction B. Precipitated proteins were dissolved in ∼200 μl of buffer and dialyzed extensively against dialysis buffer (20 mM HEPES, pH 7.9, 50 mM KCl, 0.25 mM EDTA, 10% glycerol, and 0.5 mM dithiothreitol) at 4°C. The protein concentration in extracts ranged between 2 and 6 mg/ml, as determined by the Bio-Rad protein assay kit (Bio-Rad, Richmond, CA). Bovine serum albumin was used as a standard.
Electrophoretic Mobility Shift Assays
Electrophoretic mobility shift assays were carried out as described (Mori et al., 1996). When competition experiments were performed, unlabeled competitor DNA was included in the binding buffer before addition of cell extracts.
Immunoblotting
Fifty micrograms of proteins in cell extracts per lane were subjected to SDS-PAGE using 12% (for Hac1p) or 8% gel (for Kar2p and Pdi1p) and transferred to a Hybond-ECL nitrocellulose filter (Amersham, Arlington Heights, IL). Rainbow colored protein molecular weight markers (Amersham) were used as size markers. Rabbit polyclonal antisera against Kar2p (kindly provided by Dr. D. Williams, University of Toronto) or Pdi1p (kindly provided by Dr. H. Tachikawa, Tokyo University of Agriculture and Technology) were used at a 1:5000 dilution. Rabbit polyclonal antibodies were raised against the maltose-binding protein-Hac1p (230-type) fusion protein expressed and purified from Escherichia coli cells (Mori et al., 1996). After antibodies against maltose-binding protein were removed from the antiserum by affinity chromatography through maltose-binding protein immobilized on CH-Sepharose 4B (Pharmacia, Piscataway, NJ), antibodies specific to Hac1p were purified with CH-Sepharose 4B to which the fusion protein had been coupled. Anti-Hac1p antibodies were dialyzed against phosphate-buffered saline and the resulting solution of OD280 ∼ 0.8 was used at a 1:2000 dilution. ECL Western blotting detection kit (Amersham) was used to detect each antigen.
Northern Blot Hybridization Analysis
Northern blot hybridization analysis was carried out as described previously (Mori et al., 1993, 1996). Total RNAs were subjected to 1.5% agarose gel electrophoresis containing formaldehyde. The DNA probe for the yeast LHS1/SSI1/CER1 (Baxter et al., 1996; Craven et al., 1996; Hamilton and Flynn, 1996) or UBC4 (Seufert and Jentsch, 1990) was prepared by PCR.
β-Galactosidase Assays
Assays for β-galactosidase activity in yeast cell extracts were carried out as described previously (Mori et al., 1993).
Pulse–Chase Analysis of Hac1p
The method described by Franzusoff et al. (1991) was basically used. Various transformants were grown at 30°C to midlogarithmic phase in minimal medium containing 100 μM (NH4)2SO4, 2% glucose, and other nutrients as required. Cells were harvested at 8 OD600 units by centrifugation, washed once with distilled water, and resuspended in 2.6 ml of minimal medium containing 2% glucose and other nutrients but lacking (NH4)2SO4. After shaking at 30°C for 30 min, cells were pulse-labeled for 5 min at 30°C with 400 μCi (14.8 MBq) of EXPRE35S35S protein labeling mix (DuPont, Wilmington, DE) and chased by adding a 0.01 volume of concentrated chase solution [100 mM (NH4)2SO4, 0.3% cysteine, and 0.4% methionine]. An aliquot (650 μl) was removed and mixed with 7 ml of ice-cold 0.1 M Tris-HCl, pH 8.0, and 10 mM NaN3 to terminate the labeling. Cells were recovered by centrifugation, washed once with 1 ml of ice-cold 0.1 M Tris-HCl, pH 8.0, 10 mM NaN3, and resuspended in 200 μl of TBS/1% SDS containing 1 mM phenylmethylsulfonyl fluoride and 5 μg/ml leupeptin. Cells were disrupted by vigorous agitation with acid-washed glass beads (0.425–0.6 mm in diameter, Sigma, St. Louis, MO) for four 30-s periods at intervals of 90 s on ice. The resulting lysate was freed of glass beads, boiled for 5 min, and after addition of 800 μl of TBS/2% Triton X-100, clarified by centrifugation at 14,000 rpm for 10 min. The supernatant was treated with 10 μl each of purified anti-Hac1p antibodies and anti-Pdi1p antibodies as an internal control. After standing overnight at 4°C, 50 μl of 50% protein A-Sepharose 4 fast flow (Pharmacia) in TBS were added and the mixture was rotated for 2 h at room temperature. The resin was then washed as described by Franzusoff et al. (1991), and the immunoprecipitates were subjected to SDS-PAGE (12% gel). The amounts of 35S-labeled Hac1p and Pdi1p were analyzed by using BioImaging Analyzer BAS-2000 (Fuji Photo Film, Stamford, CT).
Other Techniques
RT-PCR was carried out using SUPERSCRIPT preamplification system (Life Technologies, BRL, Gaithersburg, MD) and Takara Ex Taq (Takara, Berkeley, CA). Hac1p was translated in vitro using TNT coupled wheat germ extract system (Promega, Madison, WI) and appropriate HAC1-derived DNAs as templates.
RESULTS
Induction of Hac1p by Tunicamycin
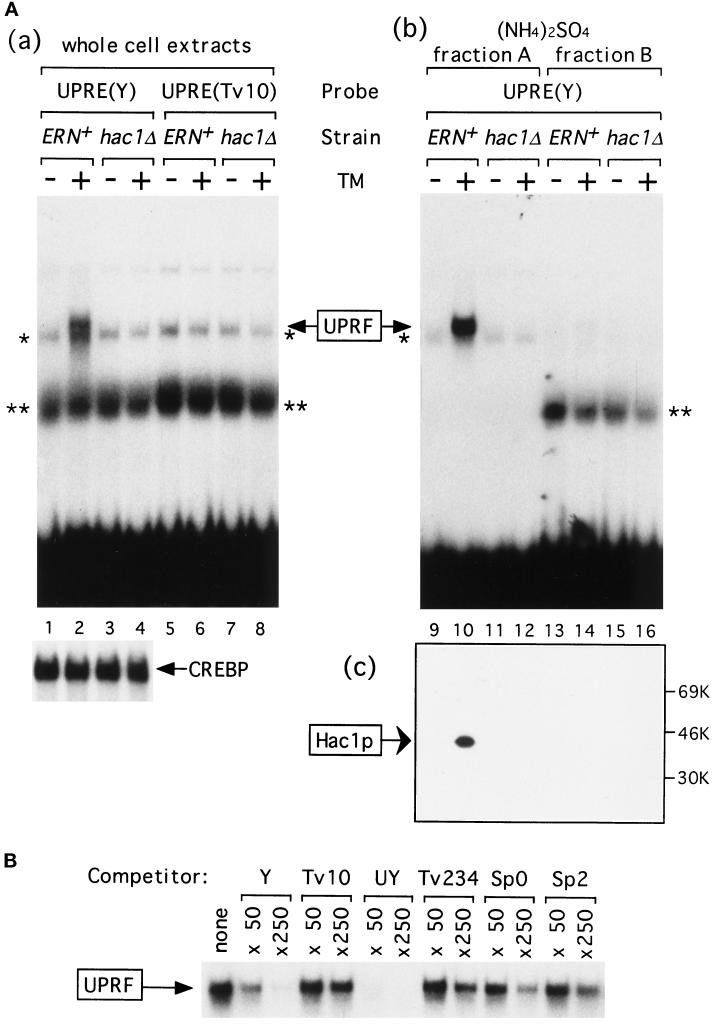
The wild-type (ERN+) and the hac1Δ strains were grown in rich medium (YPD) and incubated for 1 h in the presence or absence of tunicamycin, known to elicit ER stress by inhibiting N-glycosylation of newly synthesized proteins in the ER (Elbein, 1981; Kozutsumi et al., 1988). Whole cell extracts were prepared, and electrophoretic mobility shift assays were carried out to determine whether UPRE-specific DNA-binding activity was affected by unfolded proteins accumulated in the ER. When the wild-type UPRE designated UPRE(Y) was used as a 32P-labeled probe, specific binding activity was detected only with extracts prepared from tunicamycin-treated ERN+ strain (Figure 1Aa, lane 2). This binding was specific because it was not detected when a mutant version of UPRE designated UPRE(Tv10) was used as a probe (lane 6). UPRE(Tv10) contains a single transversion at an essential nucleotide in the palindromic sequence that almost completely abolishes the function of UPRE (Mori et al., 1996). In addition, extracts from the hac1Δ strain did not exhibit any specific binding to UPRE(Y) (lane 4), indicating that the binding obtained in lane 2 represents UPRF of which Hac1p functions as a major component. In contrast, when the cAMP-response element (CRE) was used as a probe, similar amounts of specific binding activity [marked as CREBP in Figure 1Aa] were detected in extracts prepared from the ERN+ or hac1Δ strain that had been treated or untreated with tunicamycin, confirming that Hac1p plays little or no role in cellular CRE-binding activity, contrary to the previous report (Nojima et al., 1994). These results indicated that DNA-binding activity of UPRF was induced by ER stress. Previously, UPRE-specific DNA-binding activity was detected constitutively (Mori et al., 1992), but the assay condition used was not optimal because 10 mM MgCl2 was found to be required for specific binding (Mori et al., 1996). Because significant amounts of nonspecific binding (indicated by * and **) were also detected, proteins in extracts were fractionated by differential precipitation with (NH4)2SO4 as described in MATERIALS AND METHODS. As shown in Figure 1Ab, UPRF was recovered in fraction A (lane 10), whereas nonspecific binding** was recovered in fraction B (lanes 13–16). By using fraction A from tunicamycin-treated ERN+ strain, specific binding of UPRF to 32P-labeled UPRE(Y) was competed by 50- or 250-fold molar excess of various UPRE-like sequences (Figure 1B). The ability of a mutant UPRE to compete the specific binding correlated very well with its ability to mediate the UPR in vivo (Mori et al., 1996).
Figure 1.
Induction of Hac1p by tunicamycin. (A) Whole cell extracts, (NH4)2SO4 fraction A, and (NH4)2SO4 fraction B were prepared from the ERN+ (KMY1105) and hac1Δ (KMY1145) strains that had been grown in YPD medium to a midlogarithmic phase and incubated for 1 h with (+) or without (−) tunicamycin (TM). (a) Forty micrograms of proteins in each of whole cell extracts were mixed with 0.3 ng of 32P-labeled wild-type UPRE [designated UPRE(Y), 9060 cpm], a point mutant of UPRE [designated UPRE(Tv10), 9390 cpm], or CRE (7590 cpm). (b) Forty micrograms of proteins in each of (NH4)2SO4 fraction A or fraction B were mixed with 0.2 ng (9130 cpm) of 32P-labeled UPRE(Y). Protein-bound probes were separated from free probes in a 5% nondenaturing gel. The specific binding to UPRE(Y) is marked as UPRF. * and ** denote nonspecific bindings that were also detected when 32P-labeled CRE was used as a probe. Only specific binding to CRE was shown below and marked as CREBP. (c) Fifty micrograms of proteins in (NH4)2SO4 fraction A or fraction B were subjected to SDS-PAGE (12% gel) and immunoblotted with purified anti-Hac1p antibodies. The positions of molecular mass markers are indicated. (B) The specific binding between 0.3 ng (7870 cpm) of 32P-labeled UPRE(Y) and UPRF in the (NH4)2SO4 fraction A (20 μg of proteins) from tunicamycin-treated ERN+ strain was competed by 50- or 250-fold molar excess of unlabeled UPRE(Y) or various mutant forms of UPRE, whose activity in mediating the UPR was well characterized previously (Mori et al., 1996). Only specific binding is shown. Tv10, a point mutant of Y described in the text, is virtually inactive in mediating the UPR in vivo. UY contains a palindrome of 7 bp that is more active than Y. Tv234 contains three transversions upstream of the palindromic sequence. Sp0 and Sp2 are mutant versions of UPRE in which the half-sites in the palindromic sequence are separated by spacers of 0 and 2 nt, respectively, instead of 1 nt in Y. Tv234, Sp0, and Sp2 exhibit very weak activities in vivo. Neither 5′ nor 3′ termini of competitor oligonucleotides were filled in, and possible formation of concatemers may explain the large molar excess of unlabeled UPRE-Y required for competition under these conditions.
Next we performed immunoblotting analysis to detect Hac1p in cell extracts with purified anti-Hac1p antibodies. As shown in Figure 1Ac, Hac1p was detected only in extracts of tunicamycin-treated ERN+ cells (lane 10), indicating that not only DNA-binding activity but also Hac1p itself was induced by ER stress. Hac1p migrated as a single band of 41 kDa in SDS-PAGE (12% gel), which is appreciably larger than its calculated molecular weight 26,045 but identical with the position of Hac1p translated in vitro (see Figure 6). A high content of charged residues may account for this aberrant behavior.
Induction of Hac1p by Tunicamycin Requires the 3′ Untranslated Region (UTR) of HAC1 mRNA
When the ERN+ strain was grown in synthetic medium selective for plasmid maintenance, a small amount of Hac1p was produced but the amount was markedly enhanced by treatment with tunicamycin for 1 h (Figure 2, lanes 1 and 2; immunoblot). The relatively high basal expression of Hac1p, as compared with cells grown in broth, probably results from increased amounts of unfolded proteins in the ER: it was not detected in the ern1Δ strain lacking functional Ern1p, a key enzyme in the signal transduction across the ER membrane (Figure 2, lane 3). This is consistent with the approximately 2.5-fold higher β-galactosidase expressed from the UPRE-CYC1-lacZ reporter gene in the ERN+ strain as compared with the ern1Δ or hac1Δ strain grown in the absence of tunicamycin (Figure 2, compare bars 1 and 3 or 5). The treatment of the ERN+ strain with tunicamycin for 3 h caused an approximately 10-fold induction of β-galactosidase under these conditions (Figure 2, bar 2). The induction of both Hac1p and β-galactosidase depended entirely on the function of Ern1p (Figure 2, lane 4) and Hac1p (Figure 2, lane 6). Introduction of the wild-type HAC1 gene into the hac1Δ strain fully restored the induction of both proteins (Figure 2, lanes 7 and 8). Interestingly, deletions downstream of the Hac1p-coding region (ΔHN and ΔHA, Figure 2, lanes 9–12) abolished the induction of both proteins, indicating that the induction of Hac1p requires 3′UTR of the mRNA. This also suggested that the induction of Hac1p occurs posttranscriptionally rather than transcriptionally, consistent with the absence of UPRE in the promoter region.
Induction of the 1.2-kb HAC1 mRNA by Tunicamycin
We then used Northern blot hybridization to determine whether ER stress affected the amount or size of mRNA transcribed from the chromosomal HAC1 gene. In the ERN+ strain, 1.4-kb HAC1 mRNA was expressed even in the absence of tunicamycin (Figure 3A). Transcription of the KAR2 gene, a target of the UPR, dramatically increased within 1 h after addition of tunicamycin. At this time, the amount of 1.4-kb HAC1 mRNA decreased and instead, the smaller mRNA (1.2 kb) was observed. The smaller mRNA failed to appear in the ern1Δ strain carrying a vector alone (V) but did appear when the ERN1 gene was provided by a single-copy plasmid (Cen). The overexpression of the ERN1 gene from a multicopy vector (2 μm) is known to activate the UPR (Mori et al., 1993; Shamu and Walter, 1996): the amount of KAR2 mRNA increased by twofold in the absence of tunicamycin. Under these conditions, the smaller HAC1 mRNA was detectable in the absence of tunicamycin. Thus, the induction of 1.2-kb HAC1 mRNA by ER stress correlated well with the cellular UPR.
Because tunicamycin treatment may cause pleiotropic defects in cellular metabolism, we also examined the effect of the temperature-sensitive sec53 mutation on HAC1 mRNA. At nonpermissive temperature, sec53 cells accumulate full-length precursors of secretory proteins that are abnormally glycosylated and malfolded in the ER due to the defect in phosphomannomutase activity (Feldman et al., 1987), leading to activation of the UPR (Normington et al., 1989; Rose et al., 1989). As expected, the 1.4-kb HAC1 mRNA was constitutively expressed in the sec53, ERN+ strain grown at the permissive temperature of 23°C (Figure 3B). Upon a shift to a seminonpermissive temperature of 30°C, the smaller HAC1 mRNA as well as KAR2 mRNA was markedly induced within 1 h. Neither KAR2 mRNA nor the smaller HAC1 mRNA were induced at 30°C in the sec53, ern1Δ strain. These results strongly indicated that 1.2-kb HAC1 mRNA is induced only if unfolded proteins are accumulated in the ER, regardless of the nature of stress conditions employed.
Time-course experiments with the ERN+, SEC+ strain revealed that the amount of 1.4-kb mRNA decreased within 10 min after addition of tunicamycin, concomitant with the appearance of 1.2-kb mRNA (Figure 3C). The sum of the two mRNA species remained almost constant, suggesting that 1.4-kb mRNA was directly converted to 1.2-kb mRNA. On the other hand, the amounts of target mRNAs (KAR2 and PDI1) when normalized with that of ACT1 mRNA, increased only after 10–20 min. Thus, the appearance of the 1.2-kb HAC1 mRNA preceded the induction of target mRNAs, consistent with the notion that the induction of 1.2-kb HAC1 mRNA is required for the UPR.
ER Stress-induced Splicing of HAC1 mRNA
The 1.4-kb and 1.2-kb HAC1 mRNAs were then compared, particularly with respect to the 3′UTR, because a small deletion downstream of the Hac1p-coding region abolished the induction of Hac1p (ΔHA, see Figure 2), and the size of truncated HAC1 mRNA thus produced, in comparable amounts, remained unchanged upon addition of tunicamycin. Total RNAs were extracted from the ERN+ strain grown for 1 h in the presence or absence of tunicamycin, and mRNAs were converted to cDNAs by using an oligo(dT) primer and RT. HAC1 cDNA was amplified by PCR using a 5′ primer just upstream of the start ATG codon and each of the 3′ primers indicated in Figure 4A. Only when the 3′ primer 5 or 6 was used, RNAs from tunicamycin-treated cells gave rise to an additional PCR product shorter, by approximately 250 nt, than those obtained from untreated cells (Figure 4B). Nucleotide sequence of the shorter PCR product obtained with primers 1 and 5 was determined after inserting it into pT7 blue T-vector (Novagen, Madison, WI) and found to lack 252 nt, from nt 662 to 913 (Figure 4A). The absence of this region from the 1.2-kb HAC1 mRNA was confirmed by Northern blot hybridization using the various probes shown in Figure 4A. Both 1.4-kb and 1.2-kb mRNAs were detected in total RNAs from tunicamycin-treated cells with probe A, D, or E, whereas only the 1.4-kb mRNA was detected with probe B or C (Figure 4C). Evidently, HAC1 mRNA becomes spliced in response to ER stress.
The putative 5′ and 3′ splice sites did not match the consensus sequence found in S. cerevisiae and higher eukaryotes (GT-AG or AT-AC; Kreivi and Lamond, 1996). Inclusion of G661 but not G913 in the 1.2-kb mRNA was established by mutating either of these Gs to A and sequencing the resulting RT-PCR product obtained from tunicamycin-treated cells. Since the 5′ splice site was located in the coding region, this unusual splicing was expected to replace the C-terminal portion of Hac1p. Whereas 1.4-kb pre-mRNA should encode a protein of 230 aa, the 1.2-kb mature mRNA should encode a protein of 238 aa, because the C-terminal 10 aa should be removed by the splicing, and the remaining 220 aa should be fused to a stretch of 18 aa encoded by the sequence starting with nt 914 (Figure 4A).
Constitutive Activation of the UPR by the Intron-less HAC1 Gene
We then prepared and expressed cDNA corresponding to the 1.2-kb mRNA in the hac1Δ strain under the control of the HAC1 promoter by using a single-copy expression plasmid (YCp-HAC1Δintron) and compared its effects with those of the wild-type gene (YCp-HAC1WT; Figure 5). Total RNAs and total proteins were extracted from cells grown for 1 h in the presence or absence of tunicamycin. In hac1Δ cells carrying YCp-HAC1WT, HAC1 pre-mRNA was spliced and Hac1p was induced by tunicamycin as in the ERN+ strain. As a result, transcription of all of the six target genes (KAR2, LHS1/SSI1/CER1 [Baxter et al., 1996; Craven et al., 1996; Hamilton and Flynn, 1996], SCJ1 [Schlenstedt et al., 1995], PDI1, EUG1 [Tachibana and Stevens, 1992], FKB2 [Partaledis and Berlin, 1993]) was well induced. In contrast, 1.2-kb HAC1 mRNA was constitutively present in hac1Δ cells carrying YCp-HAC1Δintron and was little affected by tunicamycin as expected. This resulted in constitutive synthesis of Hac1p and constitutively high levels of transcripts of all target genes (Figure 5A). The levels of two target proteins (Kar2p and Pdi1p) were also constitutively elevated in these cells (see Figure 6B, lane 9 in the Inset). Cells carrying YCp-HAC1Δintron grew at significantly slower rates than those carrying YCp-HAC1WT (doubling time 3 h versus 2 h), indicating that excess synthesis of these target proteins is toxic to the cell.
Figure 5.
Expression of cDNA corresponding to 1.2-kb HAC1 mRNA lacking the intron constitutively activates the UPR. (A) The hac1Δ strain (KMY1145) was transformed with a single-copy expression plasmid (YCp-HAC1) carrying the wild-type (WT) or intron-less (Δintron) HAC1 gene. Transformants were grown at 30°C in SC(−Ura, Leu) medium to midlogarithmic phase, and aliquots were incubated for 1 h in the presence (+) or absence (−) of tunicamycin (TM). Total proteins and total RNAs were extracted and analyzed by immunoblotting using anti-Hac1p antibodies and by Northern blot hybridization using DNA probes specific for HAC1, KAR2, LHS1/SSI1/CER1, SCJ1, PDI1, EUG1, FKB2, and ACT1, respectively. (B) The ERN+, ern1Δ, and hac1Δ strains (KMY1105, 1115, and 1145, respectively) were transformed with a vector alone (V), YCp-HAC1WT, or YCp-HAC1Δintron. Transformants were grown at 30°C in SC(−Ura, Leu) medium to midlogarithmic phase, and aliquots were incubated for 3 h in the presence (hatched bars) or absence (solid bars) of tunicamycin (TM). β-Galactosidase activities in cell extracts were determined and are presented as the mean ± SD, based on duplicate determinations with three independent transformants.
Consistent with these results, β-galactosidase expressed from the UPRE-CYC1-lacZ reporter gene was markedly induced in the hac1Δ strain carrying YCp-HAC1WT (Figure 5B, lanes 11 and 12) as in the ERN+ strain (Figure 5B, lanes 1 and 2), and the induction depended on the function of Ern1p (Figure 5B, lanes 5 and 6). In sharp contrast, β-galactosidase was constitutively expressed in the hac1Δ strain carrying YCp-HAC1Δintron (Figure 5B, lanes 13 and 14). The high level expression in the latter case did not require the function of Ern1p (Figure 5B, lanes 7 and 8). Thus, the expression of 1.2-kb HAC1 mRNA lacking the intron constitutively activates the UPR without involvement of the Ern1p kinase. From these results, we concluded that the UPR is controlled by the regulated splicing of HAC1 pre-mRNA.
Constitutive Expression of Hac1p of 238, 220, and 230 aa from Mutant Versions of the HAC1 Gene
The results so far presented are consistent with those reported previously (Cox and Walter, 1996). Hac1p produced from the intron-less HAC1 gene (referred as 238-type in Figure 6A) in the hac1Δ strain migrated at the position identical with that of 238aa-Hac1p translated in vitro but faster than that of 230aa-Hac1p translated in vitro (Figure 6C, lanes 9 and 10). The apparent anomaly in migration may result from the fact that the last 10 aa of 230aa-Hac1p contain two basic and no acidic residues but the last 18 aa of 238aa-Hac1p contain no basic and three acidic residues. This provided additional confirmation of the previous observation cited above that all of Hac1p so far detected in cell extracts was composed of 238 aa and thus translated from mature mRNA. Therefore, we next focused on the question of why 230aa-Hac1p possibly synthesized from pre-mRNA was not detected.
As mentioned earlier, Cox and Walter (1996) proposed that pre-mRNA is also translated but its product of 230 aa is degraded as soon as it is made due to the C-terminal 10 aa that confer extreme instability to the otherwise stable N-terminal portion of 220 aa. However, the data on differential stability among different types of Hac1p were lacking. We thus tried to express the three types of Hac1p and test their stability individually. To express 220aa-Hac1p, the 18-aa segment and the whole intron were deleted from the HAC1 gene, and the resulting plasmid was introduced into the hac1Δ strain (Figure 6A, construct 220-type). This strain produced a large amount of protein regardless of tunicamycin treatment (Figure 6C, lanes 11 and 12), which migrated at the position identical with that of 220aa-Hac1p translated in vitro. The construct 220-type also caused production of β-galactosidase in both ern1Δ (Figure 6B, bars 3 and 4) and hac1Δ (Figure 6B, bars 11 and 12) strains, whose activities were approximately 40% of those for 238aa-Hac1p (Figure 6B, bars 1, 2, 9, and 10) with or without tunicamycin.
To express 230aa-Hac1p, we removed the region of nt 691–966 containing the first stop codon, the majority of the intron, and the 18-aa segment from the HAC1 gene (see Figure 4A) and transformed the hac1Δ strain with the resulting plasmid (Figure 6A, construct 230-type). Doublet protein bands were detected with or without tunicamycin (Figure 6C, lanes 13 and 14) that migrated at positions around that of 230aa-Hac1p translated in vitro. The results were quite unexpected and suggested that the stability of 230aa-Hac1p may not differ appreciably from that of 238aa-Hac1p.
It was also noteworthy that the β-galactosidase activity detected in tunicamycin-untreated ern1Δ or hac1Δ cells expressing 238aa-Hac1p (Figure 6B, bar 1 or 9) was approximately 12-fold higher than that in cells expressing 230aa-Hac1p (Figure 6B, bar 5 or 13) when compared after subtracting the activity with vector alone (Figure 6B, bar 7 or 15), whereas the amount of 230aa-Hac1p detected by immunoblotting was nearly half as much as 238aa-Hac1p (Figure 6C, compare lanes 9 with 13). We therefore examined proteins from tunicamycin-untreated hac1Δ cells by immunoblotting with anti-Kar2p and anti-Pdi1p antibodies: the levels of two target proteins were again found to be well correlated with those of β-galactosidase activity (Figure 6B, inset). The anti-Hac1p antibodies used were raised against 230aa-Hac1p fused to maltose-binding protein as described in MATERIALS AND METHODS and appeared to recognize mainly the bZIP region (our unpublished results). If we assume the same immunoreactivities of the two proteins, the specific transcriptional activator activity of 238aa-Hac1p appeared to be severalfold higher than that of 230aa-Hac1p. Thus, the splicing-mediated replacement of the C terminus may generate a transcription factor with significantly higher activity. We are currently characterizing transcriptional activation domains found in Hac1p of 238, 220, and 230 aa.
Hac1p Is Highly Unstable
We then examined stability of three types of Hac1p after treating cells with cycloheximide, an inhibitor of protein synthesis. Cycloheximide at 10 to 300 μg/ml completely inhibited the induction by tunicamycin of β-galactosidase from the UPRE-CYC1-lacZ reporter gene without affecting cell viability. Total proteins were isolated from hac1Δ cells carrying each of the mutant versions of the HAC1 gene described in Figure 6A at various times after addition of 20 μg/ml cycloheximide and immunoblotted with anti-Hac1p or anti-Kar2p antibodies (Figure 7A). Whereas the amount of Kar2p remained virtually unchanged, both the 238aa- and 230aa-Hac1p totally disappeared within 5 min after addition of cycloheximide; the 220aa-Hac1p seemed to be slightly more stable than the others. A separate experiment with hac1Δ cells expressing both 238aa- and 230aa-Hac1p showed that the amounts of both Hac1p decreased with similar time course (half-life of 1 min or less), indicating that both proteins are highly unstable in vivo in the absence of protein synthesis.
To further examine possible differential stability, we conducted pulse–chase experiments with [35S]methionine and [35S]cysteine and hac1Δ cells that were made to produce both 238aa- and 230aa-Hac1p (Figure 7B). We adopted this approach particularly to avoid possible complications arising from differential growth rates: hac1Δ cells expressing 238aa-Hac1p grew significantly more slowly than those expressing 230aa-Hac1p (doubling time 3 h versus 2 h), perhaps reflecting the differential ability to transactivate the UPR. The amount of pulse-labeled 238aa-Hac1p obtained by immunoprecipitation was 1.6-fold higher than that of 230aa-Hac1p, which might be due, at least in part, to their differential mRNA levels (see Figure 6C). Both types of Hac1p were chased by unlabeled amino acids with similar time course, whereas Pdi1p remained virtually unchanged. The quantification of radioactivities followed by correction with Pdi1p values revealed essentially no difference in stability between 238aa- and 230aa-Hac1p, the half-life being approximately 2 min. Thus, the differential protein stability is unlikely to explain the absence of 230aa-Hac1p in cells carrying the wild-type HAC1 gene.
Constitutive Activation of the UPR in ubc Mutants Results from Increased but Low Levels of Splicing
Constitutive activation of the UPR in mutants defective in some of the ubiquitin-conjugating enzymes provided strong support to the proposal by Cox and Walter (1996). The β-galactosidase activities expressed from the reporter plasmid were approximately 4.5-, 2-, 10-, 5-, and 4-fold higher in the ubc4Δ, ubc5Δ, ubc4Δ5Δ, ubc7Δ, and ubc8Δ strains, respectively, than in the wild-type (UBC+) and the increased basal activity in the ubc4Δ5Δ strain was independent of the mRNA splicing, suggesting stabilization of 230aa-Hac1p translated from pre-mRNA in these mutants. Unfortunately, however, the presence of stabilized 230aa-Hac1p in the mutant extracts was not shown directly. We thus carried out similar experiments using the same set of strains and the isogenic UBC+ strain (see Table 1). These strains (MATα) were transformed with the UPRE-CYC1-lacZ reporter gene carried on either the single-copy or multicopy vector, and β-galactosidase activities in the resulting transformants were determined in the absence of tunicamycin.
Unexpectedly, β-galactosidase activity in the ubc4Δ strain was lower, rather than higher, than that in the UBC+ strain (Figure 8A). We also examined the ubc4Δ strain of opposite mating type (MATa) but the results were almost identical. The absence of UBC4 mRNA in the ubc4Δ strain was confirmed by Northern blot hybridization (Figure 8B), and the levels of two target proteins of the UPR (Kar2p and Pdi1p) in the ubc4Δ strain were not significantly higher than those in the UBC+ strain (see Figure 8B, bottom). Currently, we have no explanation for this discrepancy. Similarly, β-galactosidase activity in the ubc4Δ5Δ double mutant was lower than that in the UBC+ strain (Figure 8A) unlike the previous report, although its extremely long doubling time (10-fold of control; Seufert and Jentsch, 1990) posed a question in interpreting the results. On the other hand, β-galactosidase activities in the ubc5Δ and ubc7Δ strains were 3.1- and 3.4-fold higher than that in the UBC+ strain, respectively, when the multicopy reporter plasmid was used (Figure 8A). However, the increase was marginal when the single-copy reporter plasmid was used (1.4- and 1.6-fold, respectively), and the levels of Kar2p and Pdi1p were enhanced only slightly (Figure 8B, bottom).
Figure 8.
Effects of various ubc mutations on the UPR. (A) Various strains indicated on the abscissa were transformed with the UPRE-CYC1-lacZ reporter gene carried on a single-copy vector (pSCZ-Y, hatched bars) or a multicopy vector (pMCZ-Y, stippled bars) and transformants were grown at 30°C in SC(−Ura) medium. The amounts of β-galactosidase expressed without tunicamycin treatment were determined with midlogarithmic-phase cells and are presented as the mean ± SD, based on duplicate determinations with three independent transformants after normalization to the values for the UBC+ strain of MATα. (B) The UBC+, ubc4Δ, ubc5Δ, and ubc7Δ strains carrying pMCZ-Y were grown at 30°C in SC(−Ura) medium to midlogarithmic phase, and aliquots were incubated in the presence (+) or absence (−) of tunicamycin (TM). Samples taken after 1 h were used for extracting total RNAs that were analyzed by Northern blot hybridization using DNA probes specific for UBC4, HAC1, and ACT1. Total proteins were extracted from the UBC+, ubc4Δ, ubc5Δ, and ubc7Δ strains that had been grown in YPD medium to midlogarithmic phase, then incubated for 1 h in the presence (+) or absence (−) of tunicamycin, and analyzed by immunoblotting using anti-Hac1p, anti-Kar2p, and anti-Pdi1p antibodies. (C) The ERN1 and HAC1 loci in the ubc5Δ and ubc7Δ strains were disrupted as described previously (Mori et al., 1996). Various strains indicated on the abscissa were transformed with pMCZ-Y. Transformants were grown at 30°C in SC(−Ura) medium to midlogarithmic phase, and aliquots were incubated in the presence (hatched bars) or absence (solid bars) of tunicamycin. Samples taken after 3 h were used for β-galactosidase assays, and the activities are presented as the mean ± SD, based on duplicate determinations with three independent transformants.
We then asked which type of Hac1p was responsible for the weak but constitutive activation of the UPR in the ubc5Δ and ubc7Δ strains. As in the ERN+ strain, HAC1 pre-mRNA was spliced within 1 h after addition of tunicamycin in the three ubc mutant as well as UBC+ strains (Figure 8B). Total proteins were isolated from cells grown in YPD medium to avoid small but significant amounts of 238aa-Hac1p that would be produced when cells were grown in synthetic medium without tunicamycin (see Figures 2 and 5). The normal levels of 238aa-Hac1p were found after but not before addition of tunicamycin in all the strains tested (Figure 8B). Upon longer exposure of the film, low but significantly enhanced levels of 238aa- but not 230aa-Hac1p were detected in the ubc5Δ and ubc7Δ strains in the absence of tunicamycin (Figure 8B, bottom). This indicated that the splicing of HAC1 pre-mRNA occurred constitutively at very low levels in these two strains, though no appreciable amounts of mature mRNA were detected by Northern blot hybridization. We then disrupted the ERN1 and HAC1 loci in the ubc5Δ and ubc7Δ strains and examined their effects: not only the induction by tunicamycin but also the increased basal expression of β-galactosidase were abolished (Figure 8C). These results demonstrated that the constitutively activated UPR in some of the ubc mutants results from enhanced splicing of HAC1 pre-mRNA rather than stabilization of the pre-mRNA product.
Hac1p Is Translated Only from Mature mRNA
The results shown in Figures 7 and 8 clearly demonstrated that differential protein degradation cannot be the main mechanism regulating the expression of Hac1p, contrary to the previous proposal (Cox and Walter, 1996). We then tested the alternative possibility that the absence of 230aa-Hac1p in cell extracts is due to the lack of translation of HAC1 pre-mRNA. We showed above that Hac1p of 220aa was expressed constitutively in the hac1Δ strain carrying the mutant HAC1 gene lacking the entire intron and the 18-aa segment (Figure 6, 220-type). It was then asked whether we could similarly express 220aa-Hac1p from a mutant version of the HAC1 gene containing the intron. For convenience in plasmid construction, a XbaI site was introduced at nt 640 between the two MfeI sites in YCp-HAC1WT and YCp-HAC1Δintron [Figure 9, WT(XbaI) and Δintron(XbaI), respectively] that changed Leu214-Asp215 to Ser-Arg (see MATERIALS AND METHODS). The two amino acid changes did not affect the level of either HAC1 mRNA or Hac1p but slightly reduced the transcriptional activator activity of 238aa-Hac1p; only slightly lower levels of β-galactosidase were produced from the UPRE-CYC1-lacZ reporter gene in the hac1Δ strain (compare Figure 9, lanes 1–4, with Figure 5B, lanes 11–14). Three mutants were constructed by using YCp-HAC1WT(XbaI). The Ala221Stop mutant contained a stop codon at aa 221 instead of the codon for Ala221 without affecting other nucleotide sequences in the intron. When this mutant was introduced into the hac1Δ strain, similar level of HAC1 pre-mRNA was expressed as the wild-type without tunicamycin (Figure 9, compare lane 1 with 5 and also lane 13 with 17). Strikingly, however, this mutant mRNA was not translated to produce 220aa-Hac1p (Figure 9, lane 17). Addition of tunicamycin also failed to induce Hac1p (Figure 9, lane 18), probably because cleavage of the mRNA at the 5′ splice site was inhibited due to the alteration of the 3 nt, causing degradation of the mRNA cleaved only at the 3′ splice site. These results should be contrasted with those shown in Figure 6C, lanes 11 and 12, where a large amount of 220aa-Hac1p was constitutively expressed from the mutant HAC1 gene (Δintron 220-type). To examine synthesis rates of Hac1p, and Pdi1p as a control, cells were pulse-labeled with [35S]methionine and [35S]cysteine for 5 min and immunoprecipitates were analyzed by SDS-PAGE. As shown in Figure 10, the labeled 220aa-Hac1p was detected from hac1Δ cells carrying the Δintron 220-type mutant but not from those carrying the Ala221Stop mutant; synthesis of Hac1p from the latter mRNA was estimated to be less than 4% of that from the intron-less mRNA.
To further examine the effects of splicing on Hac1p synthesis, we constructed another mutant Ala221Stop′ by inserting 2 nt, TA, between the codons for Pro220 and Ala221, so that a stop codon was created immediately after Pro220 but the nucleotide sequences at the 5′ splice site remained identical from the position −2. However, the results obtained with Ala221Stop′ (Figure 9, lanes 7 and 8) were similar to those with Ala221Stop, indicating that the sequence upstream of the position −2 is also important for recognition of HAC1 pre-mRNA by putative endonuclease(s). We thus made another construct in which a stop codon was created at aa 216 next to the XbaI site. In this case (Asn216Stop), the mutation site was placed far enough from the 5′ splice site to allow tunicamycin-inducible pre-mRNA splicing albeit at reduced efficiencies (Figure 9, lanes 15 and 16). From this mutant mRNA, Hac1p of probably 215 aa migrating slightly faster than the 220aa-Hac1p translated in vitro was produced in a tunicamycin-inducible manner. The pattern of protein expression was indistinguishable from the case of 238aa-Hac1p produced from the wild-type mRNA (Figure 9, lanes 13 and 14). These results clearly demonstrated that ER stress-induced splicing of HAC1 pre-mRNA can effectively lead to the synthesis of Hac1p.
DISCUSSION
Hac1p is a trans-acting factor responsible for the UPR, transcriptional induction of genes encoding ER-localized molecular chaperones and folding enzymes in response to ER stress, in S. cerevisiae (Cox and Walter, 1996; Mori et al., 1996; Nikawa et al., 1996). Hac1p itself is also induced by ER stress and the induction is mediated by ER stress-induced unconventional mRNA splicing as reported previously (Cox and Walter, 1996) and further substantiated by the present study. In addition, we demonstrated inclusion of G661 but not G913 in mature mRNA (see Figure 4) and involvement of the splicing in induction of all known target genes of the UPR (Figure 5). The results from two laboratories agreed that only Hac1p translated from mature mRNA was detected in extracts prepared from cells carrying the wild-type HAC1 gene under a variety of conditions tested. However, the model proposed by Cox and Walter (1996) to explain significance and consequence of the splicing differed from ours in some important respects. They proposed effective translation of pre-mRNA and mature mRNA and ascribed the absence of the pre-mRNA product to its extreme instability, essentially on the basis of three lines of evidence.
Their first result on pre-mRNA being associated with polysomes was not pursued in this report; however, its significance in the regulated expression of Hac1p is discussed below in connection with our model. Their second result related to the ubc mutants could not be reproduced by our hands, although we analyzed the same strains under similar conditions (Figure 8). The levels of β-galactosidase activity expressed from the UPRE-CYC1-lacZ reporter gene and two target proteins (Kar2p and Pdi1p) were not elevated in the ubc4Δ strain. In addition, the weak but constitutive activation of the UPR observed in the ubc5Δ and ubc7Δ strains resulted from enhanced splicing of HAC1 pre-mRNA. Since the specific transcriptional activator activity of 230aa-Hac1p seemed to be lower than that of 238aa-Hac1p (Figure 6), our immunoblotting system should have been sensitive enough to detect 230aa-Hac1p in the ubc mutant extracts, if the UPR were indeed activated by stabilized 230aa-Hac1p. However, we actually detected increased amounts of only 238aa-Hac1p in extracts from tunicamycin-untreated ubc5Δ and ubc7Δ cells. Furthermore, the introduction of the ern1Δ as well as hac1Δ mutation completely abolished the increased basal expression of β-galactosidase from the reporter gene. We can offer no explanation for these discrepancies at this time. In this connection, it was recently reported that the ubiquitin–proteasome pathway is involved in the “ER degradation,” degradation of malfolded proteins in the ER (reviewed by Brodsky and McCracken, 1997; Kopito, 1997). Specifically, UBC7 was found to be identical with DER2, which was isolated as a gene that complemented a mutant defective in degrading mutant soluble vacuolar proteins, carboxypeptidase yscY (CPY*) and proteinase yscA (PrA*), malfolded in the ER (Hiller et al., 1996). Perhaps, unfolded proteins can be formed at low levels even under normal growth conditions, and some remain bound to molecular chaperones or folding enzymes longer in the ER, whereas others are degraded by the ubiquitin–proteasome pathway in the cytoplasm. Thus, the defects in some Ubc proteins may enhance levels of unfolded proteins in the ER and activate the Ern1p-mediated signaling from the ER, resulting in increased splicing of HAC1 pre-mRNA.
Their third result, namely, the constitutive activation of the UPR by 220aa-Hac1p, could be an argument based on the result using an inappropriate plasmid construct. In the present study, the UPR was constitutively activated by 220aa-Hac1p as reported by Cox and Walter (1996) when it was expressed in large amounts from the mutant (220-type) HAC1 gene lacking the whole intron, although the level of activation was approximately 40% of that of 238aa-Hac1p (Figure 6). However, 220aa-Hac1p was not produced from the HAC1 mutant (Ala221Stop) containing the intron, indicating that HAC1 pre-mRNA is not translated (Figure 9). The 220aa-Hac1p appeared most stable among the three types of Hac1p examined (Figure 7A). Therefore, if both pre-mRNA and mature mRNA were translated effectively as they proposed, the presence or absence of intron should not make any difference. However, synthesis of 220aa-Hac1p from the intron-containing mRNA was not detected, where 4% of synthesis from the intron-less mRNA would have been detected (Figure 10). Moreover, the intron-containing HAC1 mutant (Asn216Stop) produced a slightly smaller protein (215aa-Hac1p) in a tunicamycin-inducible manner (Figure 9). These results clearly demonstrated the correlation between pre-mRNA splicing and synthesis of Hac1p.
Finally, we directly compared the stability of 230aa- and 238aa-Hac1p in cells that were made to produce both. We found that both these proteins were highly unstable in the absence of protein synthesis (Figure 7A) and that there was essentially no difference in their stability under the conditions employed (Figure 7B). These results made the proposal by Cox and Walter (1996) quite unlikely. On the other hand, the C-terminal half of Hac1p is rich in Pro, Asp, Glu, Ser, and Thr residues, and this so-called PEST domain may be responsible for the instability of Hac1p observed (Rechsteiner and Rogers, 1996).
It now seems clear that Hac1p is synthesized only after the mRNA splicing takes place, leading to a new model for the regulated expression of Hac1p in response to ER stress. When yeast cells do not require Hac1p such as those grown in rich medium, its synthesis is blocked at the level of pre-mRNA. Thus, yeast cells would not need to waste energy to synthesize and degrade Hac1p when not required. When cells are placed under ER stress, an unconventional type of mRNA splicing is induced and the resulting mature mRNA is translated. The synthesized Hac1p activates transcription of target genes encoding ER-localized molecular chaperones such as Kar2p, Lhs1p/Ssi1p/Cer1p (E. coli DnaK homologues), and Scj1p (a DnaJ homologue), as well as folding enzymes such as Pdi1p (protein disulfide isomerase), Eug1p (a multicopy suppressor of the pdi1Δ strain), and Fkb2p (peptidyl-prolyl cis–trans isomerase). Yeast cells must be equipped with this signaling system from the ER to the nucleus to cope with deleterious effects of excessive unfolded proteins accumulated in the ER.
Then, what is the mechanism by which HAC1 pre-mRNA is prevented from being translated? Although Cox and Walter (1996) showed substantial loading of pre-mRNA onto polysomes, it is difficult to see how ribosomes can distinguish mature mRNA from pre-mRNA and translate only mature mRNA if both types of mRNA are associated with ribosomes. Interestingly, the same group proposed that the splicing of HAC1 pre-mRNA occurs in the nucleus. This proposal is consistent with the nuclear localization of tRNA ligase encoded by the RLG1 gene (Clark and Abelson, 1987), which was demonstrated to be involved in joining the two exons after ER stress induced cleavage of HAC1 pre-mRNA (Sidrauski et al., 1996). When we consider the fact that as much as 70% of pre-mRNA were converted to mature mRNA within 20 min after addition of tunicamycin (Figure 3C), we speculate that a majority of pre-mRNA is retained in the nucleus that provides a pool “waiting” for cleavage by putative endonuclease(s) activated by Ern1p-mediated signaling from the ER followed by tRNA ligase-dependent ligation: only mature mRNA thus produced is allowed to leave the nucleus to be translated in the cytoplasm. Unfortunately, our attempt to determine the localization of HAC1 pre-mRNA by in situ hybridization was unsuccessful, presumably because of its low abundance even when expressed from a multicopy plasmid. Therefore, we do not rule out the possibility that HAC1 pre-mRNA is exported out from the nucleus but prevented from being translated by yet unknown mechanisms. However, in this case, HAC1 mRNA must be brought back into the nucleus before or after cleavage by putative endonuclease(s) to be ligated by the action of tRNA ligase.
This response appears to be quite unique among biological signal transduction systems in the sense that mRNA but not protein “waits” for a signal to come. Then, what is the advantage for the cell to do so? We imagine that this mechanism would allow the cell to avoid “cross-talk” among various signal transduction systems. S. cerevisiae is known to possess multiple signal transduction and stress response systems and in most cases, signals arise in the cytoplasm and activate a protein kinase(s) responsible for respective pathway (reviewed by Thevelein, 1994; Mager and De Kruijff, 1995; Ruis and Schuller, 1995). Therefore, if Hac1p is constitutively expressed under normal growth conditions and activated by phosphorylation directly or indirectly by Ern1p protein kinase, there would be a fair chance of being activated accidentally by other protein kinases in the absence of excess unfolded proteins in the ER. This situation should be unfavorable for the cell: constitutive activation of the UPR by the intron-less HAC1 cDNA without ER stress (Figures 5 and 6) caused significantly slower growth, presumably because excess levels of target proteins are also toxic to the cell. Evidently, this response is rapid and sensitive: the splicing takes place within 10 min after the addition of tunicamycin (Figure 3C), and even a shift from rich (YPD) to poor (synthetic) medium (compare Figure 1Ac, lane 9, with Figure 2, lane 1) or defects in some of Ubc proteins (Figure 8) induces the splicing, leading to synthesis of small amounts of 238aa-Hac1p. In addition, Hac1p is highly unstable in vivo (Figure 7), allowing the response to be shut off immediately after recovery from stress conditions. This mechanism may therefore provide a means for specific and tight adjustment of levels of molecular chaperones and folding enzymes, solely to meet the requirement in the ER. The present results also provide a new insight into biological significance and consequences of an mRNA splicing event that has not been addressed in this or other eukaryotic systems.
ACKNOWLEDGMENTS
We are grateful to Dr. Peter Walter for communicating their results prior to publication, Dr. Mark Hochstrasser for providing yeast strains, and Dr. Akihiko Nakano for helpful discussions. We thank Masako Nakayama, Mayumi Ueda, and Hideaki Kanazawa for technical assistance. This work was supported in part by grants from the Japan Health Sciences Foundation, Tokyo.
Footnotes
Abbreviations used: bZIP, basic leucine zipper; ER, endoplasmic reticulum; CRE, cAMP response element; nt, nucleotide; PCR, polymerase chain reaction; RT, reverse transcriptase; UPR, unfolded protein response; UPRE, unfolded protein-response element; UPRF, unfolded protein-response factor.
The HAC1 gene was originally isolated as a multicopy suppressor of a cdc10 mutant of Schizosaccharomyces pombe (Nojima et al., 1994), but its nucleotide sequence was found to contain multiple deletions and insertions in the coding region. The corresponding YFL031w gene identified by a genome project (Murakami et al., 1995) has also a deletion of 1 nt in the coding region. The nucleotide sequence of the HAC1 gene reported by Cox and Walter (1996) seems to be identical with that reported previously (Mori et. al., 1996) and deposited in the GenBank/EMBL/DDBJ DNA database as the ERN4 gene (accession number D86413).
REFERENCES
- Baxter BK, James P, Evans T, Craig EA. SSI1 encodes a novel Hsp70 of the Saccharomyces cerevisiae endoplasmic reticulum. Mol Cell Biol. 1996;16:6444–6456. doi: 10.1128/mcb.16.11.6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky JL, McCracken AA. ER-associated and proteasome-mediated protein degradation: how two topologically restricted events came together. Trends Cell Biol. 1997;7:151–156. doi: 10.1016/S0962-8924(97)01020-9. [DOI] [PubMed] [Google Scholar]
- Chen P, Johnson P, Sommer T, Jentsch S, Hochstrasser M. Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MATα2 repressor. Cell. 1993;74:357–69. doi: 10.1016/0092-8674(93)90426-q. [DOI] [PubMed] [Google Scholar]
- Clark MW, Abelson J. The subnuclear localization of tRNA ligase in yeast. J Cell Biol. 1987;105:1515–1526. doi: 10.1083/jcb.105.4.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox JS, Shamu CE, Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- Cox JS, Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87:391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- Craven RA, Egerton M, Stirling CJ. A novel Hsp70 of the yeast ER lumen is required for the efficient translocation of a number of protein precursors. EMBO J. 1996;15:2640–2650. [PMC free article] [PubMed] [Google Scholar]
- Elbein AD. The tunicamycins—useful tools for studies on glycoproteins. Trends Biochem Sci. 1981;6:219–221. [Google Scholar]
- Feldman RI, Bernstein M, Schekman R. Product of SEC53 is required for folding and glycosylation of secretory proteins in the lumen of the yeast endoplasmic reticulum. J Biol Chem. 1987;262:9332–9339. [PubMed] [Google Scholar]
- Franzusoff A, Rothblatt J, Schekman R. Analysis of polypeptide transit through yeast secretory pathway. Methods Enzymol. 1991;194:662–674. doi: 10.1016/0076-6879(91)94048-h. [DOI] [PubMed] [Google Scholar]
- Gething MJ, Sambrook J. Protein folding in the cell. Nature. 1992;355:33–45. doi: 10.1038/355033a0. [DOI] [PubMed] [Google Scholar]
- Hamilton TG, Flynn GC. Cer1p, a novel Hsp70-related protein required for posttranslational endoplasmic reticulum translocation in yeast. J Biol Chem. 1996;271:30610–30613. doi: 10.1074/jbc.271.48.30610. [DOI] [PubMed] [Google Scholar]
- Helenius A, Tatu U, Marquardt T, Braakman I. Protein folding in the endoplasmic reticulum. In: Rupp RG, Oka MS, editors. Cell Biology and Biotechnology. Berlin/Heidelberg: Springer Verlag; 1992. [Google Scholar]
- Hiller MM, Finger A, Schweiger M, Wolf DH. ER degradation of a misfolded luminal protein by the cytosolic ubiquitin-proteasome pathway. Science. 1996;273:1725–1728. doi: 10.1126/science.273.5282.1725. [DOI] [PubMed] [Google Scholar]
- Hurst H. Protein Profile Series-Transcription Factors 1: bZIP proteins. Vol. 2. London: Academic Press; 1995. [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno K, Normington K, Sambrook J, Gething MJ, Mori K. The promoter region of the yeast KAR2 (BiP) gene contains a regulatory domain that responds to the presence of unfolded proteins in the endoplasmic reticulum. Mol Cell Biol. 1993;13:877–890. doi: 10.1128/mcb.13.2.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopito RR. ER quality control: the cytoplasmic connection. Cell. 1997;88:427–430. doi: 10.1016/s0092-8674(00)81881-4. [DOI] [PubMed] [Google Scholar]
- Kozutsumi Y, Segal M, Normington K, Gething MJ, Sambrook J. The presence of malfolded proteins in the endoplasmic reticulum signals the induction of glucose-regulated proteins. Nature. 1988;332:462–464. doi: 10.1038/332462a0. [DOI] [PubMed] [Google Scholar]
- Kreivi J-P, Lamond AI. RNA splicing: unexpected spliceosome diversity. Curr Biol. 1996;6:802–805. doi: 10.1016/s0960-9822(02)00599-7. [DOI] [PubMed] [Google Scholar]
- Kunkel TA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMantia M, Miura T, Tachikawa H, Kaplan HA, Lennarz WJ, Mizunaga T. Glycosylation site binding protein and protein disulfide isomerase are identical and essential for cell viability in yeast. Proc Natl Acad Sci USA. 1991;88:4453–4457. doi: 10.1073/pnas.88.10.4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AS. Coordinated regulation of a set of genes by glucose and calcium ionophores in mammalian cells. Trends Biochem Sci. 1987;12:20–23. [Google Scholar]
- Littlewood T, Evan G. Protein Profile Series—Transcription Factors 2: Helix–Loop–Helix. Vol. 2. London: Academic Press; 1995. [PubMed] [Google Scholar]
- Mager WH, De Kruijff AJJ. Stress-induced transcriptional activation. Microbiol Rev. 1995;59:506–531. doi: 10.1128/mr.59.3.506-531.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan DR, Gething MJ, Sambrook J. The cellular response to unfolded proteins: intercompartmental signaling. Curr Opin Biotechnol. 1994;5:540–545. doi: 10.1016/0958-1669(94)90071-x. [DOI] [PubMed] [Google Scholar]
- Mori K, Kawahara T, Yoshida H, Yanagi H, Yura T. Signalling from endoplasmic reticulum to nucleus: transcription factor with a basic-leucine zipper motif is required for the unfolded protein-response pathway. Genes Cells. 1996;1:803–817. doi: 10.1046/j.1365-2443.1996.d01-274.x. [DOI] [PubMed] [Google Scholar]
- Mori K, Ma W, Gething MJ, Sambrook J. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell. 1993;74:743–756. doi: 10.1016/0092-8674(93)90521-q. [DOI] [PubMed] [Google Scholar]
- Mori K, Sant A, Kohno K, Normington K, Gething MJ, Sambrook JF. A 22 bp cis-acting element is necessary and sufficient for the induction of the yeast KAR2 (BiP) gene by unfolded proteins. EMBO J. 1992;11:2583–2593. doi: 10.1002/j.1460-2075.1992.tb05323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y, Naitou M, Hagiwara H, Shibata T, Ozawa M, Sasanuma S, Sasanuma M, Tsuchiya Y, Soeda E, Yokoyama K, Yamazaki M, Tashiro H, Eki T. Analysis of the nucleotide sequence of chromosome VI from Saccharomyces cerevisiae. Nat Genet. 1995;10:261–268. doi: 10.1038/ng0795-261. [DOI] [PubMed] [Google Scholar]
- Nikawa J, Akiyoshi M, Hirata S, Fukuda T. Saccharomyces cerevisiae IRE2/HAC1 is involved in IRE1-mediated KAR2 expression. Nucleic Acids Res. 1996;24:4222–4226. doi: 10.1093/nar/24.21.4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojima H, Leem SH, Araki H, Sakai A, Nakashima N, Kanaoka Y, Ono Y. Hac1: a novel yeast bZIP protein binding to the CRE motif is a multicopy suppressor for cdc10 mutant of Schizosaccharomyces pombe. Nucleic Acids Res. 1994;22:5279–5288. doi: 10.1093/nar/22.24.5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normington K, Kohno K, Kozutsumi Y, Gething MJ, Sambrook J. S. cerevisiae encodes an essential protein homologous in sequence and function to mammalian BiP. Cell. 1989;57:1223–1236. doi: 10.1016/0092-8674(89)90059-7. [DOI] [PubMed] [Google Scholar]
- Pahl HL, Baeuerle PA. Endoplasmic reticulum-induced signal transduction and gene expression. Trends Cell Biol. 1997;7:50–55. doi: 10.1016/S0962-8924(96)10050-7. [DOI] [PubMed] [Google Scholar]
- Partaledis JA, Berlin V. The FKB2 gene of Saccharomyces cerevisiae, encoding the immunosuppressant-binding protein FKBP-13, is regulated in response to accumulation of unfolded proteins in the endoplasmic reticulum. Proc Natl Acad Sci USA. 1993;90:5450–5454. doi: 10.1073/pnas.90.12.5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechsteiner M, Rogers SW. PEST sequences and regulation by proteolysis. Trends Biochem Sci. 1996;21:267–271. [PubMed] [Google Scholar]
- Rose MD, Broach JR. Cloning genes by complementation in yeast. Methods Enzymol. 1991;194:195–230. doi: 10.1016/0076-6879(91)94017-7. [DOI] [PubMed] [Google Scholar]
- Rose MD, Misra LM, Vogel JP. KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell. 1989;57:1211–1221. doi: 10.1016/0092-8674(89)90058-5. [DOI] [PubMed] [Google Scholar]
- Ruis H, Schuller C. Stress signaling in yeast. Bioessays, 1995;17:959–965. doi: 10.1002/bies.950171109. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schlenstedt G, Harris S, Risse B, Lill R, Silver PA. A yeast DnaJ homologue, Scj1p, can function in the endoplasmic reticulum with BiP/Kar2p via a conserved domain that specifies interactions with Hsp70s. J Cell Biol. 1995;129:979–988. doi: 10.1083/jcb.129.4.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seufert W, Jentsch S. Ubiquitin-conjugating enzymes UBC4 and UBC5 mediate selective degradation of short-lived and abnormal proteins. EMBO J. 1990;9:543–550. doi: 10.1002/j.1460-2075.1990.tb08141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamu CE, Cox JS, Walter P. The unfolded-protein-response pathway in yeast. Trends Cell Biol. 1994;4:56–60. doi: 10.1016/0962-8924(94)90011-6. [DOI] [PubMed] [Google Scholar]
- Shamu CE, Walter P. Oligomerization and phosphorylation of the Ire1p kinase during intracellular signaling from the endoplasmic reticulum to the nucleus. EMBO J. 1996;15:3028–3039. [PMC free article] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Hicks JB. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- Sidrauski C, Cox JS, Walter P. tRNA ligase is required for regulated mRNA splicing in the unfolded protein response. Cell. 1996;87:405–413. doi: 10.1016/s0092-8674(00)81361-6. [DOI] [PubMed] [Google Scholar]
- Tachibana C, Stevens TH. The yeast EUG1 gene encodes an endoplasmic reticulum protein that is functionally related to protein disulfide isomerase. Mol Cell Biol. 1992;12:4601–4611. doi: 10.1128/mcb.12.10.4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachikawa H, Miura T, Katakura Y, Mizunaga T. Molecular structure of a yeast gene, PDI1, encoding protein disulfide isomerase that is essential for cell growth. J Biochem. 1991;110:306–313. doi: 10.1093/oxfordjournals.jbchem.a123576. [DOI] [PubMed] [Google Scholar]
- Thevelein JM. Signal transduction in yeast. Yeast, 1994;10:1753–1790. doi: 10.1002/yea.320101308. [DOI] [PubMed] [Google Scholar]
- Welihinda AA, Kaufman RJ. The unfolded protein response pathway in Saccharomyces cerevisiae. Oligomerization and trans-phosphorylation of Ire1p(Ern1p) are required for kinase activation. J Biol Chem. 1996;271:18181–18187. doi: 10.1074/jbc.271.30.18181. [DOI] [PubMed] [Google Scholar]