Abstract
Cell cycle progression during oocyte maturation requires the strict temporal regulation of maternal mRNA translation. The intrinsic basis of this temporal control has not been fully elucidated but appears to involve distinct mRNA 3′ UTR regulatory elements. In this study, we identify a novel translational control sequence (TCS) that exerts repression of target mRNAs in immature oocytes of the frog, Xenopus laevis, and can direct early cytoplasmic polyadenylation and translational activation during oocyte maturation. The TCS is functionally distinct from the previously characterized Musashi/polyadenylation response element (PRE) and the cytoplasmic polyadenylation element (CPE). We report that TCS elements exert translational repression in both the Wee1 mRNA 3′ UTR and the pericentriolar material-1 (Pcm-1) mRNA 3′ UTR in immature oocytes. During oocyte maturation, TCS function directs the early translational activation of the Pcm-1 mRNA. By contrast, we demonstrate that CPE sequences flanking the TCS elements in the Wee1 3′ UTR suppress the ability of the TCS to direct early translational activation. Our results indicate that a functional hierarchy exists between these distinct 3′ UTR regulatory elements to control the timing of maternal mRNA translational activation during oocyte maturation.
Introduction
Fully grown (immature) vertebrate oocytes are typically arrested in prophase I of the first meiotic division. In response to the appropriate external stimulus, immature oocytes re-enter the cell cycle and resume meiotic progression before arresting again at metaphase of Meiosis II. This progression through meiosis is termed oocyte maturation and results in oocytes that can be fertilized by sperm. During oocyte maturation and the developmental period immediately following fertilization, gene transcription is actively repressed and altered patterns of protein expression are determined by translation of pre-existing mRNAs (maternal mRNAs) present in the oocyte (Heikinheimo and Gibbons, 1998; Newport and Kirschner, 1982). The immature oocyte contains a pool of nontranslated, maternal derived mRNAs that are stored as ribonucleoprotein complexes. At various stages in development, specific mRNAs are recruited for translation (Davidson, 1986). This regulated translation of maternal mRNAs is critical for early developmental processes (Colegrove-Otero et al., 2005; de Moor et al., 2005; Kuersten and Goodwin, 2003).
Regulation of maternal mRNA translation has been best characterized in oocytes of the frog, Xenopus laevis. Maternally derived mRNAs may be classed as “early” or “late” based on the order of their translational activation during oocyte maturation. Late class mRNAs are activated coincident with or after breakdown of the oocyte nucleus (termed the germinal vesicle) and completion of Meiosis I. Early class mRNAs are translationally activated prior to germinal vesicle breakdown (GVBD). Activation of the late class mRNAs requires early class mRNA translation (Ballantyne et al., 1997; Charlesworth et al., 2006; de Moor and Richter, 1997). The strict temporal order of early and late class mRNA translation is essential to ensure hormone-dependent progression through oocyte meiotic maturation (Ferby et al., 1999; Freeman et al., 1991; Howard et al., 1999; Murakami and Vande Woude, 1998; Nakajo et al., 2000; Roy et al., 1991; Sheets et al., 1995). The mechanisms that specifically recognize early and late class mRNA species and induce translation differentially have not been fully elucidated.
Recent work has underscored the role of regulatory elements in the 3′ untranslated region (3′ UTR) of targeted mRNAs for orchestrated developmental changes in proteomic profiles (reviewed in (de Moor et al., 2005; Kuersten and Goodwin, 2003)). Two distinct 3′ UTR regulatory elements have been described which have unique temporal induction properties during Xenopus oocyte maturation. Cytoplasmic polyadenylation elements (CPEs) can enforce late mRNA translational activation (Charlesworth et al., 2000; Tung et al., 2007) while Musashi/polyadenylation response elements (PREs) direct early mRNA translational activation (Charlesworth et al., 2002). However, some early class mRNAs do not contain consensus Musashi/PRE or CPE sequences suggesting that other, as yet undefined, 3′ UTR regulatory elements may contribute to early mRNA translational activation.
In this study, we have characterized a novel regulatory element that confers early mRNA polyadenylation and translational activation. This element, designated a translational control sequence (TCS), mediates both mRNA translational repression in immature Xenopus oocytes and directs early mRNA translational activation in response to progesterone stimulation. We show that TCS elements contribute to the repression in immature oocytes of both the late class Wee1 mRNA (which encodes a protein involved in regulation of CDK1 activity; McGowan and Russell, 1993; Mueller et al., 1995; Parker and Piwnica-Worms, 1992) and the Pcm-1 mRNA (which encodes a protein involved in the assembly of centrosomes and micro-tubule networks; Balczon et al., 1994; Dammermann and Merdes, 2002). We demonstrate that TCS function directs early class mRNA translational activation exerted by the 3′ UTR of the Pcm-1 mRNA. Interestingly, CPE sequences flanking the TCS elements in the Wee1 3′ UTR suppress the ability of the TCS to exert early translational activation. These findings indicate that multiple mechanisms exist to direct both repression and early translational activation of mRNAs and a functional hierarchy between distinct 3′ UTR regulatory elements may refine the temporal patterns of maternal mRNA translational activation during oocyte maturation.
Materials and methods
Plasmid constructions and RNA synthesis
Standard PCR mutagenesis was used to generate the 3′ UTR reporter constructs utilized in this study. A detailed methodology of plasmid construction is provided as Supplementary data.
The sequence integrity of all 3′ UTR constructs was confirmed by DNA sequencing. For in vitro transcription, all plasmids were linearized with Pst1 and 5′ capped RNA synthesized in vitro with SP6 RNA polymerase as previously described (Melton et al., 1984). The resulting RNAs lacked poly[A] tails but became oligoadenylated following injection into immature oocytes (typically receiving 5′ 10 adenylate residues), consistent with prior studies (Charlesworth et al., 2000; de Moor and Richter, 1997; Gillian-Daniel et al., 1998; Sheets et al., 1994).
Oocyte culture and injection
Oocytes were defolliculated by collagenase digestion as previously described (Charlesworth et al., 2000). Dumont stage VI immature oocytes (≥ 1200 μm in diameter) were isolated and injected with 1 ng of reporter RNA. Where indicated, real-time PCR was employed to verify the levels of reporter mRNAs present in the injected oocyte samples. The levels of GST reporter mRNA were normalized to the levels of the endogenous cyclin B1 mRNA (see Supplementary data for real-time PCR primer sequence and reaction parameters). To reveal progesterone-inducible polyadenylation, oocytes were stimulated with 2 μg/ml progesterone and the rate of germinal vesicle breakdown (GVBD) was monitored morphologically by the appearance of a white spot on the animal hemisphere. Pools of 10 oocytes were harvested at each time point, along with time-matched immature oocyte controls. Because oocytes from different frogs mature at different rates in response to progesterone, the culture times were standardized between experiments to the time taken for 50% of oocytes to undergo germinal vesicle breakdown (GVBD50).
Western blot analyses
Oocytes were lysed in 10 μl of ice cold Nonidet P-40 (NP-40) lysis buffer per oocyte (MacNicol et al., 1993), and insoluble material and lipid were separated by centrifugation at 13,000×g for 10 min at 4 °C. RNA and protein lysate were prepared from the same oocyte samples, as previously described (Charlesworth et al., 2000). The lysates were normalized for the amount of total protein, separated on sodium dodecyl sulfate (SDS)-12% polyacrylamide gels, and transferred to a 0.2-μm-pore-size nitrocellulose filter. The filter was blocked with 5% nonfat dried milk in TBS. Filters were incubated with antibody and visualized with an appropriate horseradish peroxidase-linked secondary antibody by enhanced chemiluminescence (ECL). Antiserum to GST was obtained from Santa Cruz Biotechnology, antiserum to tubulin was obtained from Sigma, and antiserum to Pcm-1 was a gift from Dr. Andreas Merdes (Dammermann and Merdes, 2002). GST accumulation was visualized and quantitated by ECL western blotting as previously described (Charlesworth et al., 2000, 2002, 2004) using ChemiGlow West, a ChemiImager 5500 and AlphaEaseFC software (AlphaInnotech Corp.). One-way analysis of variance and post hoc Newman–Keuls multiple comparison test were performed to analyze differences between the means and differences in P-value less than 0.05 were considered statistically significant.
Polyadenylation assays
Reporter mRNA polyadenylation was assessed by northern blotting or RNA ligation-coupled PCR as previously described (Charlesworth et al., 2000, 2002; Rassa et al., 2000). For RNA ligation PCR, a forward primer to the β-globin 3′ UTR was used (GCG GAA TTC ACA CTT ACA AAA TGT TGT) to analyze polyadenylation of reporter constructs, except in the case of the 5′ boundary mutants where a primer to the GST coding region was used instead (Prasad et al., 2008). For analysis of endogenous mRNA polyadenylation, the following gene-specific forward primers were employed: Pcm-1, AAG CCT GTC TTT TTC CTC TC; Cyclin A1, CAT TGA ACT GCT TCA TTT TCC CAG; and Cyclin B1, GTG GCATTC CAATTG TGTATT GTT. All PCR products were resolved on a 3% agarose gel and visualized using ethidium bromide staining. An increase in PCR product size is indicative of poly[A] tail extension (Charlesworth et al., 2002) and where noted polyadenylation was verified by direct sequencing of the PCR products. Where indicated, the mode of the PCR product population poly[A] tail length extension was determined using AlphaEaseFC Software (Alpha Innotech) snap-to-peak analyses.
Results
We have previously reported that CPE sequences contribute to the repression of the Wee1 mRNA in immature oocytes and enforce late translational activation in response to progesterone stimulation (Charlesworth et al., 2000). In the absence of functional CPE sequences, the CPE-binding protein (CPEB1) does not interact with the mutant Wee1 3′ UTR and yet the mutant Wee1 3′ UTR can nonetheless direct repression in immature oocytes (Charlesworth et al., 2000). Moreover, progesterone-initiated polyadenylation and translational activation of the mutant Wee1 3′ UTR occur prematurely, with early class mRNA behavior (Charlesworth et al., 2000). The observation that the CPE-disrupted Wee1 3′ UTR does not behave like an unregulated mRNA (which would direct equivalent translation in both immature and progesterone-stimulated oocytes) indicates the presence of an early acting regulatory element that is functionally distinct from, and yet suppressed by, the Wee1 CPE sequences (Charlesworth et al., 2000). The purpose of this study was to identify and characterize the novel regulatory element in the Wee1 3′ UTR.
An examination of the Wee1 3′ UTR revealed that no consensus Musashi/PRE sequences were present to account for this early translational control. We thus utilized a series of deletions to localize the region conferring the early progester-one-dependent polyadenylation within the context of a GST reporter RNA fused to the CPE-disrupted Wee1 3′ UTR (Fig. 1A). A 28 nucleotide region was found capable of directing progesterone-dependent polyadenylation (Fig. 1B, 256′ 283). In these experiments, progesterone-dependent polyadenylation results in the addition of heterogeneous poly[A] tail length extensions and a consequently broader distribution of PCR products than that seen in immature oocytes. After discounting the polyadenylation hexanucleotide and the disrupted CPE sequences, a 17 nucleotide Wee1 region, residing between the two CPEs upstream of the polyadenylation hexanucleotide, was considered the likely element responsible for directing progesterone-dependent polyadenylation (Fig. 1C).
Fig. 1.
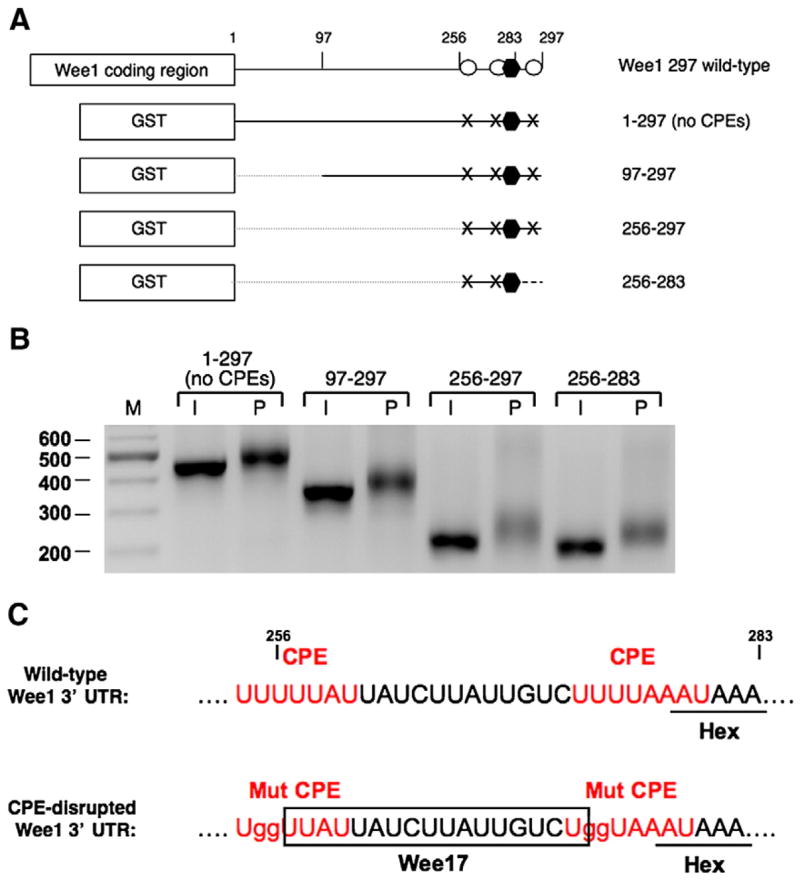
Localization of a novel regulatory element in the Wee1 3′ UTR. (A) Schematic diagram showing the constructs used in the experiment. The GST coding region was fused to various Wee1 deletion 3′ UTRs, as indicated. The polyadenylation hexanucleotide is represented by a black hexagon; CPEs are represented by open circles; mutated CPEs are represented by “X”s; and the broken line indicates deletion of the 3′ UTR. In the 256–283 reporter construct, sequences 3′ of the Wee1 polyadenylation hexanucleotide were replaced by β-globin 3′ UTR sequence (dashed line). (B) Polyadenylation of the injected RNAs was assessed in time-matched immature (I) or progesterone-stimulated (P) oocytes by RNA ligation-coupled PCR using a forward primer specific to the GST coding region of the injected reporter mRNAs. Retarded mobility is indicative of polyadenylation. (C) Sequence and regulatory element composition of the wild-type and CPE-disrupted 256–283 region of the Wee1 3′ UTR. A candidate 17 nucleotide region (Wee17) responsible for progesterone-regulated polyadenylation is indicated.
To verify the functionality of the 17 nucleotide region (Wee17), we tested the ability of Wee17 to confer translational regulation to a heterologous 3′ UTR. The Wee17 region was cloned into the unregulated β-globin 3′ UTR fused downstream of a GST reporter mRNA. The position of the 17 nucleotide Wee1 region in the heterologous 3′ UTR matched the position within the wild type Wee1 3′ UTR relative to the polyadenylation hexanucleotide. The Wee17 region directed early, progesterone-dependent polyadenylation of the heterologous β-globin 3′ UTR (Fig. 2A, designated β-globin/Wee17). Elongation of the poly[A] tail was initiated within 3 h of progesterone stimulation and prior to oocytes completing GVBD and Meiosis I. After GVBD, some deadenylation of the β-globin/Wee17 3′ UTR reporter mRNA occurred. As expected, the β-globin 3′ UTR was not subject to significant cytoplasmic polyadenylation (Fig. 2A).
Fig. 2.
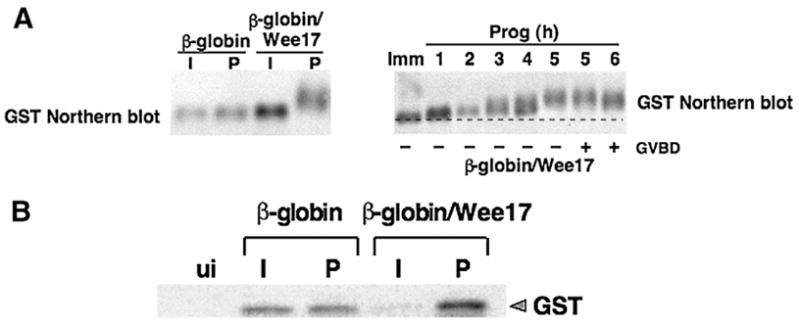
The 17 nucleotide Wee1 region exerts translational regulation on a heterologous reporter mRNA. (A) The indicated reporter mRNAs were injected into immature oocytes and cultured in the presence (P) or absence (I) of progesterone for 16 h, and polyadenylation of the reporter mRNAs was assessed by northern blot (left panel). The time course of polyadenylation of β-globin/Wee17 reporter mRNAwas assessed in immature (Imm) or progesterone-stimulated (Prog) at the indicated times (hours) by northern blot (right panel). In this experiment, GVBD50 occurred at 5 h. (B) Immature oocytes were injected with the indicated GST reporter mRNAs and time matched immature (I) and progesterone-stimulated (P) samples analyzed for GST protein accumulation by western blot after 15 h of culture.
In addition to controlling mRNA polyadenylation, the Wee17 region was also able to exert translational control of the GST reporter mRNA. The Wee17 region directed repression of translation of the GST reporter mRNA (β-globin/Wee17) in immature oocytes (relative to the levels of GST controlled by the unregulated β-globin 3′ UTR) and directed accumulation of GST protein (above the levels controlled by the β-globin 3′ UTR) in progesterone-stimulated oocytes (Fig. 2B). Although the Wee17 region acts as an early regulatory element, we prepared protein samples from oocytes which had completed GVBD because differences in GST protein accumulation between the β-globin/Wee17 and β-globin 3′ UTR reporter mRNAs were more pronounced at later time points. Equivalent GST protein accumulation was observed in progesterone-stimulated and time-matched immature oocytes expressing the GST reporter RNA under the control of the unregulated β-globin 3′ UTR, as described previously (Charlesworth et al., 2000). These results indicate that the Wee17 region is sufficient to mimic the behavior of the CPE-disrupted Wee1 3′ UTR, by exerting mRNA translational repression in immature oocytes and early progesterone-dependent polyadenylation and translational activation in maturing oocytes.
Examination of the Wee17 sequence revealed two imperfect repeats in proximity to each other (Fig. 3A, arrows). To determine the minimal functional sequence within the Wee17 region, a series of deletion mutants were employed (Fig. 3A). Deletion mutants that disrupted either repeat sequence alone in the Wee17 region did not prevent progesterone-dependent polyadenylation, suggesting that each imperfect repeat sequence is sufficient to direct progesterone-dependent polyadenylation (Fig. 3B, 3′ Δ6 and 5′ Δ6). Indeed, analysis of polyadenylation of additional deletion mutants demonstrated that a 7 nucleotide sequence within one repeat was sufficient to direct progesterone-dependent polyadenylation (Fig. 3B, 5′ Δ10). DNA sequencing of the RNA ligation-coupled PCR products verified that the progesterone-induced increase in PCR product size was specifically due to polyadenylation of the 3′ UTR.
Fig. 3.
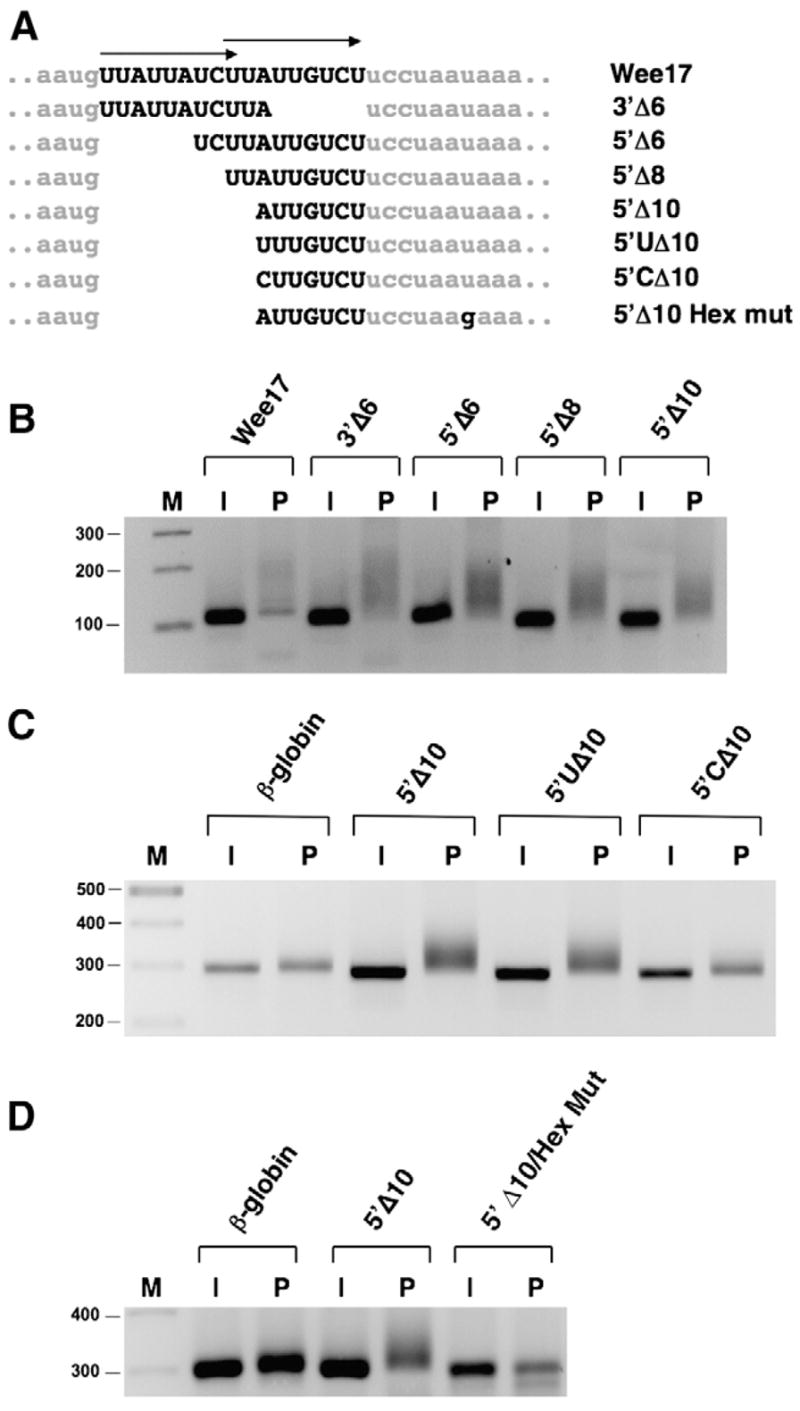
The Wee17 region contains two functional translational control sequences (TCS). (A) Schematic representation of β-globin/Wee17 deletion and mutant constructs. Arrows above the Wee17 region indicate the position of two imperfect repeat sequences. Lower case nucleotides represent β-globin 3′ UTR sequence and upper case nucleotides indicate Wee1 3′ UTR sequence. (B) Analysis of progesterone-dependent polyadenylation of the β-globin/Wee17 deletion mutant reporter RNAs. Reporter mRNAs were injected into immature (I) oocytes and left untreated or stimulated with progesterone (P) for 4 h 40 min. Polyadenylation was assessed by RNA ligation PCR using a forward primer specific to the β-globin 3′ UTR of the injected reporter mRNAs. (C) Analysis of progesterone-dependent polyadenylation of 5′ boundary substitution mutations was determined as described for (B), except a forward primer specific to the GST coding region of the injected reporter mRNAs was employed. Progesterone was added for 4 h 20 min. (D) The requirement for a functional polyadenylation hexanucleotide for progesterone-dependent polyadenylation was assessed using the indicated reporter mRNAs as described for (panel C) above, except oocytes were cultured in progesterone for 3 h 40 min.
Fig. 5.
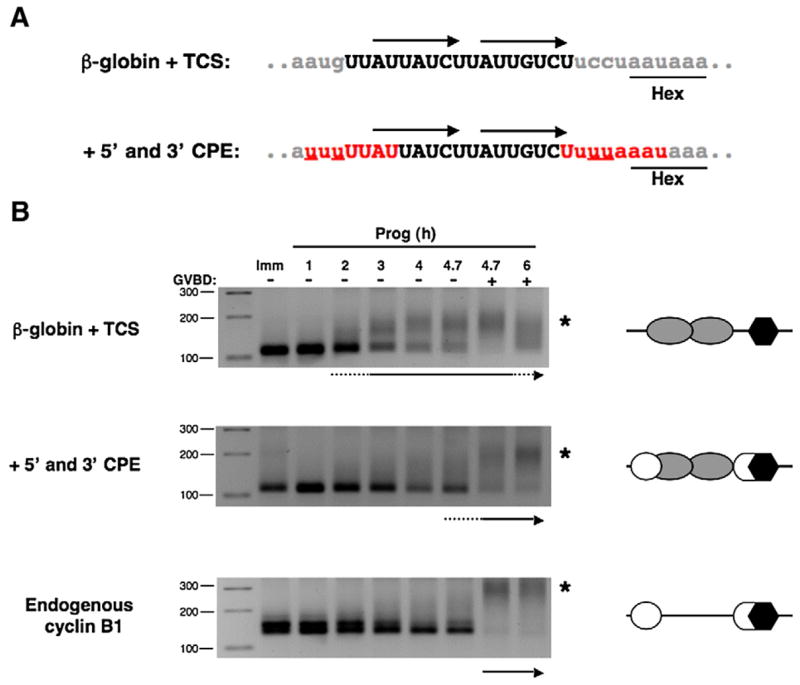
The presence of flanking CPE sequences suppresses TCS function. (A) Sequence and regulatory element composition of β-globin 3′ UTR reporter constructs employed. The β-globin 3′ UTR containing the two TCS elements (upper panel, arrows) was mutated (underlined red nucleotides) to generate 5′ and 3′ CPE sequences found in the wild-type Wee1 3′ UTR (lower panel). Note that the 3′ CPE overlaps the polyadenylation hexanucleotide. Lower case gray letters represent β-globin sequence, upper case black nucleotides represent Wee1 sequence, and red nucleotides represent reconstituted CPE sequences. (B) Immature oocytes were separately injected with GST reporter mRNAs fused to the β-globin UTR containing a Wee1 TCS (β-globin+TCS) or the Wee1 TCS flanked with both a 5′ CPE and 3′ hexanucleotide overlapping CPE (+5′ and 3′ CPE) that replicate the sequence and position of the CPE sequences within the endogenous Wee1 3′ UTR. RNA samples were taken from progesterone-stimulated oocytes at the indicated times and the polyadenylation of the reporter mRNAs and the endogenous cyclin B1 mRNA was assessed by RNA ligation PCR. Reporter RNA polyadenylation was assessed using a forward primer specific to the β-globin 3′ UTR of the injected reporter mRNAs. A schematic of the regulatory element arrangement within the 3′ UTRs is depicted beside each polyadenylation assay, where open circles represent CPEs, gray ovals are TCS elements, and polyadenylation hexanucleotides are shown as black hexagons. The initiation of polyadenylation for each mRNA examined is indicated by an arrow beneath each panel and the extent of GVBD indicated above the top panel. GVBD50 occurred after 4.7 h of stimulation. An asterisk indicates the position of polyadenylated mRNA species.
To test if the 7 nucleotide Wee1 sequence remaining in the 5′ Δ10 mutant was the minimal functional regulatory unit, 5′ boundary substitution mutants were assessed (Fig. 3C). A substitution of the 5′ A with U did not prevent progesterone-dependent polyadenylation indicating that either A or U can be tolerated at this position. However, substitution of the 5′ A with a C significantly inhibited progesterone-dependent extension of the poly[A] tail. These observations indicate that the choice of nucleotide at the 5′ position of the 7 nucleotide region is important for directing polyadenylation in response to progesterone stimulation.
To determine if the ability of the 7 nucleotide region (5′ Δ10) to exert progesterone-dependent poly[A] tail extension required the canonical polyadenylation hexanucleotide, a mutant 3′ UTR was employed (5′ Δ10/Hex Mut) where the AAUAAA sequence was changed to AAgAAA to disrupt function (Fox et al., 1989). The 5′ Δ10 region was not able to direct polyadenylation of the hexanucleotide disrupted 3′ UTR (Fig. 3D), suggesting that the 7 nucleotide region acts in conjunction with the AAUAAA sequence to mediate translational activation.
Since both the 5′ and 3′ imperfect repeats in the Wee17 region can individually direct progesterone-dependent polyadenylation (Fig. 3B, 3′ Δ6 and 5′ Δ6), we conclude that the Wee17 region contains two functionally equivalent elements. The derived regulatory unit in this region appears to be a 7 nucleotide sequence (A/U)UU(G/A)UCU, which we term a translational control sequence (TCS). When compared to the previously characterized consensus or atypical CPE (Barkoff et al., 2000; Charlesworth et al., 2004; Richter, 1999) and Musashi/PRE (Charlesworth et al., 2006; Imai et al., 2001) consensus sequences, the TCS element has a distinct linear nucleotide composition that may reflect distinct regulatory properties.
To verify that the TCS element regulated both repression and activation of the reporter mRNA, a double guanosine substitution was introduced into a single 7 nucleotide TCS (AUUGUCU to AggGUCU, designated TCS/gg; see Fig. 4A) and inserted in the β-globin 3′ UTR. The double guanosine substitution ablated progesterone-stimulated polyadenylation directed by the TCS (Fig. 4B, TCS/gg) and prevented TCS-directed repression in immature oocytes and translational activation in response to progesterone stimulation (Fig. 4C). Disruption of the TCS resulted in equal GST protein expression in immature oocytes and progesterone-stimulated oocytes, similar to that seen with the parental GST β-globin 3′ UTR reporter RNA (Fig. 4C). The wild-type TCS directed the addition of approximately 55 adenylate residues in progesterone-stimulated oocytes (Fig. 4B). A small increase in the length of the β-globin 3′ UTR (approximately 5 adenylate residues) was observed in maturing oocytes, but this addition was not sufficient to confer translational induction (Fig. 4C). These results indicate that TCS function regulates mRNA translation as well as polyadenylation in the heterologous β-globin 3′ UTR.
Fig. 4.
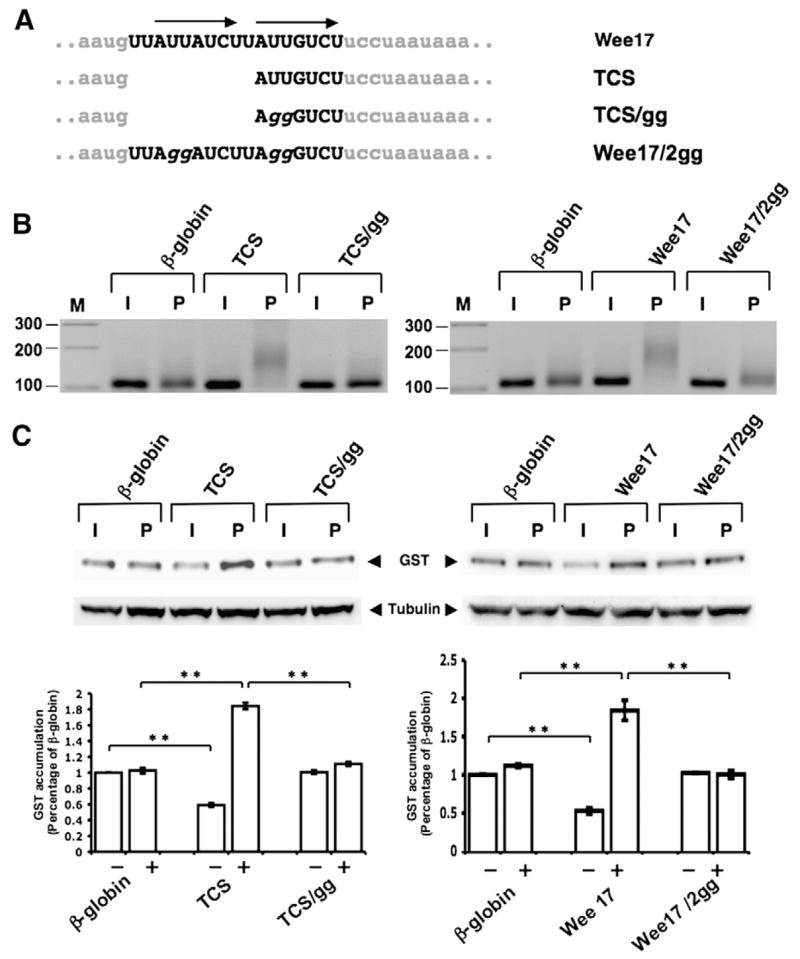
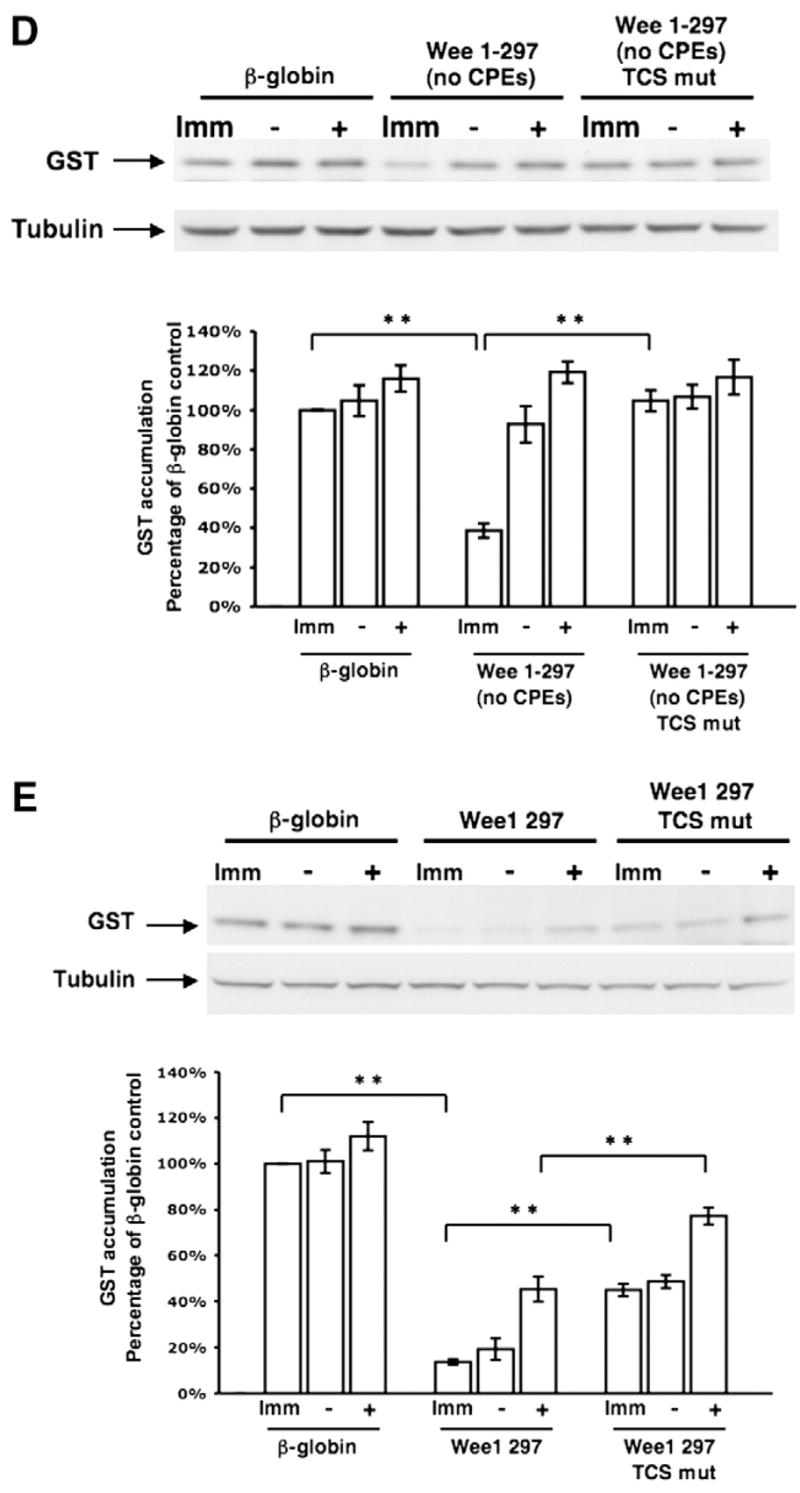
Disruption of translational control sequence (TCS) function within the Wee17 region. (A) Schematic representation of the nucleotide disruptions used to inactivate the two TCS elements (arrows) within the Wee17 region. (B) Progesterone-dependent polyadenylation of wild-type or mutant TCS reporter mRNAs. Experiments were performed essentially as described in the legend to Fig. 3, using a forward primer specific to the β-globin 3′ UTR of the injected reporter mRNAs. Oocytes were cultured in the presence of progesterone for 5 h. (C) Translational regulation of the wild-type or mutant TCS-containing β-globin 3′ UTR GST reporter mRNAs was assessed by western blotting (the β-globin 3′ UTRs analyzed had either a single TCS element (left panels) or two TCS elements (right panels)). The same lysates were analyzed for tubulin protein levels as an internal control for protein loading. The levels of GST accumulation were quantitated over three independent experiments (graph). Error bars represent SEM. One-way analysis of variance and post hoc Newman–Keuls multiple comparison test were performed. Differences between the means were scored as statistically significant, P<0.001 (**). Oocytes were cultured in the presence of progesterone for 5 h 30 min (left panel) or 5 h (right panel). (D) Translational regulation of GST reporters fused to either the entire Wee1 3′ UTR that lacked CPE sequences (Wee 1–297 (no CPEs); see Fig. 1) or the Wee 1–297 (no CPE) 3′ UTR that also had mutations in both TCS elements (Wee 1–297 (no CPEs) TCS mut) was assessed by western blotting. The same lysates were analyzed for tubulin protein levels as an internal control for protein loading. The levels of GST accumulation were quantitated over three independent experiments (graph) as described for panel C. Oocytes were cultured in progesterone for 5 h (when they reached GVBD50) and segregated into those oocytes that had (+) or had not (−) completed GVBD. (E) Translational regulation of GST reporters fused to either the entire wild type Wee1 3′ UTR (Wee1 297, see Fig. 1) or the Wee1 297 3′ UTR that also had mutations in the two TCS elements (Wee1 297 TCS mut) was assessed by western blotting. The same lysates were analyzed for tubulin protein levels as an internal control for protein loading. The levels of GST accumulation were quantitated over three independent experiments (graph) as described for panel C. Oocytes were cultured in progesterone for 5 h (when they reached GVBD50) and segregated into those oocytes that had (+) or had not (−) completed GVBD.
While these experiments define the function of a single TCS element, the identified Wee17 region contains two TCS elements and we aimed to determine if they were functionally redundant in the control of mRNA translation. The extent of repression in immature oocytes and activation in progesterone-stimulated oocytes (relative to the β-globin 3′ UTR) was indistinguishable between the Wee17 and single TCS reporter 3′ UTRs (Fig. 4C). These findings suggest that the two TCS elements in the Wee17 region function redundantly to exert translational control. As expected, double guanosine substitutions in both TCS elements attenuated Wee17-mediated polyadenylation and abolished translational regulation of the reporter mRNA (Figs. 4B and C, respectively).
To verify that the identified Wee17 region was indeed responsible for the early class mRNA translational regulation of the CPE-disrupted Wee1 3′ UTR (Charlesworth et al., 2000), we compared the translational regulation exerted by the full length Wee1 UTR lacking CPE sequences (Wee 1–297 (no CPEs)) and the same 3′ UTR but with additional mutational disruption of both TCS elements (Wee 1–297 (no CPEs) TCS mut). The Wee 1–297 (no CPE) 3′ UTR exerted repression in immature oocytes and de-repression occurred prior to GVBD as expected (Fig. 4D). However, the ability of the CPE-disrupted Wee1 3′ UTR to repress mRNA translation in immature oocytes was abolished when the TCS elements were mutated. Indeed, the TCS and CPE-disrupted Wee1 3′ UTR (Wee 1–297 (no CPEs) TCS mut) behaved indistinguishably from the unregulated β-globin 3′ UTR in the same experiment. These findings indicate that the translational regulation exerted by the CPE-disrupted Wee1 3′ UTR was attributable to the identified TCS elements.
Since the TCS elements were identified in a CPE-disrupted Wee1 3′ UTR, the physiological contribution of the TCS elements to translational regulation exerted by the wild-type Wee1 3′ UTR had not been assessed. Consequently, we compared the translational regulation exerted by the full length wild-type Wee1 3′ UTR and the TCS-disrupted version of the same 3′ UTR. Disruption of the TCS elements in the otherwise wild-type Wee1 3′ UTR attenuated the translational repression of the reporter mRNA in both immature and progesterone-stimulated oocytes prior to GVBD (Fig. 4E). Mutational disruption of the two TCS elements in the context of the wild-type Wee1 3′ UTR reduced the ability of the Wee1 3′ UTR to mediate repression by approximately 33%. These findings indicate that TCS elements contribute physiologically to Wee1 3′ UTR repression in immature oocytes over and above that exerted by the CPE sequences. De-repression of the wild-type and TCS-disrupted Wee1 3′ UTRs occurred after GVBD (Fig. 4E) due to the action of the CPE sequences which enforce late translational activation (Charlesworth et al., 2000). In these experiments, no significant induction of GST reporter mRNA (over that exerted by the control β-globin 3′ UTR) was observed after 5 h of progesterone stimulation. This is consistent with earlier findings that the wild-type and CPE-disrupted Wee1 3′ UTRs require more extended incubations for enhanced GST accumulation (Charlesworth et al., 2000).
Although the Wee 3′ UTR possesses two early-acting TCS regulatory elements, the wild-type Wee1 3′ UTR does not direct early translation in response to progesterone. Indeed, our prior data demonstrated that CPE sequences within the Wee1 3′ UTR enforce late mRNA translational activation (Charlesworth et al., 2000). To test if CPE sequences act dominantly in cis to override TCS activation function, CPEs were introduced to flank the two TCS sequences in the Wee17 region within the context of the heterologous β-globin 3′ UTR (designated +5′ and 3′ CPE), such that the contextual arrangement of the elements mimicked that found in the wild-type Wee1 3′ UTR (Fig. 5A). Initiation of progesterone-dependent polyadenylation of the injected β-globin+TCS 3′ UTR reporter mRNA was observed after 3 h of stimulation as evidenced by a distinct slower migrating polyadenylated mRNA population (Fig. 5B, asterisk). By contrast, when the Wee17 region was flanked by CPE sequences, the appearance of a discrete polyadenylated mRNA population was not observed until after completion of GVBD (Fig. 5B, +5′ and 3′ CPE). The polyadenylation profile of the CPE-containing reporter mRNA (+5′ and 3′ CPE) was similar to that of the endogenous late class, cyclin B1 mRNA (Fig. 5B). We conclude that CPE sequences exert temporal dominance over TCS function in cis and enforce late-class mRNA polyadenylation behavior.
Since the Wee1 CPE sequences prevent TCS-directed early translational activation, are there other mRNAs in which the TCS can direct early translation? For this analysis, we presumed that TCS function may be best revealed in mRNA 3′ UTRs that lack a CPE sequence. To identify candidate TCS-regulated mRNAs, the derived TCS consensus sequence (A/U)UU(A/G) UCU was used to search the 3′ UTR database (Mignone et al., 2005). TCS consensus elements were found in 12.3% (1159 mRNAs) of the nonredundant Xenopus mRNA 3′ UTR entries. However, since CPE sequences appear to only direct progesterone-dependent polyadenylation if they are positioned within 100 nucleotides of the polyadenylation hexanucleotide (de Moor et al., 2005), the number of candidate mRNAs was reduced to 147 (1.6% of the nonredundant 3′ UTR entries) by assuming the same requirement for TCS function. We then selected the mRNA encoding pericentriolar material-1 (Pcm-1) for further characterization because this mRNA 3′ UTR is reasonably short (313 nucleotides), contains two TCS elements, lacks consensus or atypical CPEs (Charlesworth et al., 2004; de Moor et al., 2005; Mendez and Richter, 2001) or Musashi/PRE binding sites (Charlesworth et al., 2006), and an antibody to the endogenous Xenopus Pcm-1 protein has been reported (Dammermann and Merdes, 2002). Pcm-1 is a cell cycle control protein involved in the assembly of centrosomes and micro-tubule networks (Balczon et al., 1994; Dammermann and Merdes, 2002).
The Pcm-1 3′ UTR contains a TCS positioned 45 nucleotides 5′ of the polyadenylation hexanucleotide and a second TCS 7 nucleotides 3′ of the polyadenylation hexanucleotide (Fig. 6A). To assess the possible role of translational regulation of the Pcm-1 mRNA, we first determined if the levels of Pcm-1 protein changed during progesterone-stimulated oocyte maturation (Fig. 6B). Endogenous Pcm-1 protein levels are low in immature oocytes but increase in response to progesterone stimulation. Indeed, significant accumulation of Pcm-1 occurs prior to GVBD. These observations suggest that the Pcm-1 mRNA is translationally regulated in response to progesterone. Consistent with a possible role for TCS-directed control, the endogenous Pcm-1 mRNA displays early class mRNA polyadenylation behavior in response to progesterone stimulation (Fig. 6C). The Pcm-1 mRNA has a short poly[A] tail in immature oocytes and undergoes progesterone-dependent polyadenylation prior GVBD. By contrast, polyadenylation of the endogenous CPE-dependent, late class cyclin A1 mRNA occurs after GVBD in the same samples (Fig. 6C).
Fig. 6.
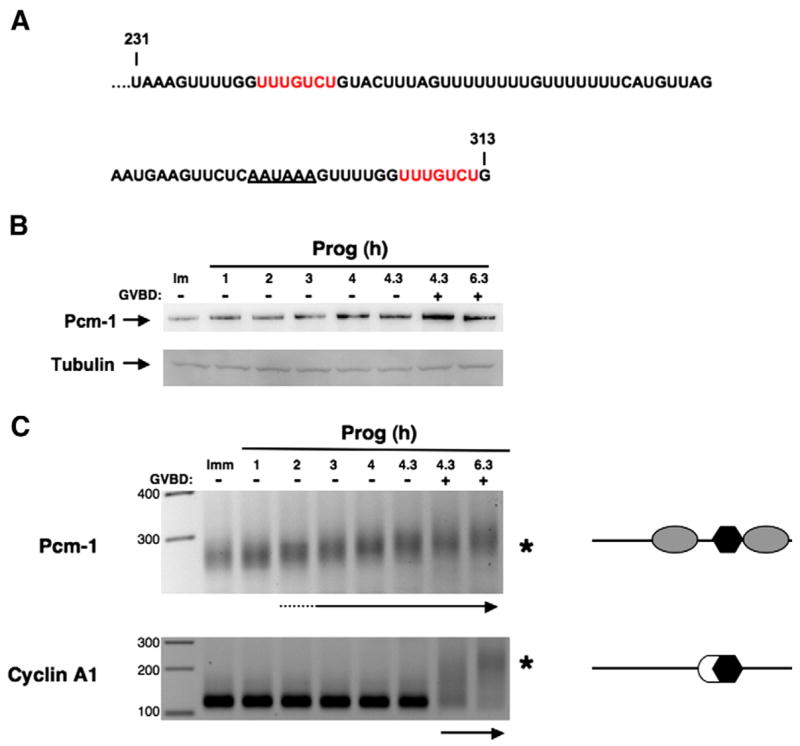
The Pcm-1 3′ UTR contains two TCS elements and displays early class mRNA polyadenylation in response to progesterone stimulation. (A) Sequence of the terminal 82 nucleotides of the Xenopus Pcm-1 3′ UTR. Two candidate TCS elements are indicated by red nucleotides and the polyadenylation hexanucleotide is underlined. (B) Immature oocytes (Imm) or oocytes treated with progesterone for the indicated times were analyzed for endogenous Pcm-1 protein levels (230 kDa) by western blot analysis. Equivalent protein was loaded for each time point as verified by a tubulin western blot of the same samples (lower panel). (C) Immature oocytes (Imm) or oocytes treated with progesterone were analyzed for endogenous Pcm-1 and cyclin A1 polyadenylation from the same samples used for the western blot analysis (B). An increase in PCR product size is indicative of polyadenylation (maximal polyadenylation is indicated by an asterisk for each mRNA). The arrows below each panel indicate when polyadenylation was first detected. The schematics to the right of each panel reflect the regulatory element composition of each 3′ UTR (open circles represent CPEs, gray ovals are the TCS elements, and polyadenylation hexanucleotides are shown as black hexagons). GVBD50 occurred after 4.3 h of stimulation.
To examine the role of the Pcm-1 TCS elements to the regulation of mRNA translation in immature oocytes, we designed GST reporter mRNAs containing the full length Pcm-1 3′ UTR (Pcm-1) or a TCS mutant Pcm-1 3′ UTR (5′ +3′ TCS Mut), where both TCS elements were disrupted by diguanosine nucleotide substitution (UUUGUCU to UggGUCU; Fig. 7A). Following injection and 18 h of culture, the Pcm-1 3′ UTR exerted significant repression of GST reporter mRNA translation when compared to the β-globin 3′ UTR (Fig. 7B, upper panels). Disruption of the TCS elements reduced, but did not eliminate, repression exerted by the Pcm-1 3′ UTR. To verify this subtle effect of TCS disruption, quantitative PCR was used to determine relative reporter mRNA levels in the injected oocytes and normalize the levels of GST accumulation quantitated after western blotting. These results confirmed a specific contribution of the TCS elements to the translational repression exerted by the Pcm-1 3′ UTR (Fig. 7B, graph), and indicated that approximately 25% of the overall repression is attributable to the TCS elements. Since significant repression still occurs in the presence of the disrupted TCS elements, however, one or more additional regulatory elements must also be present in the Pcm-1 3′ UTR.
Fig. 7.
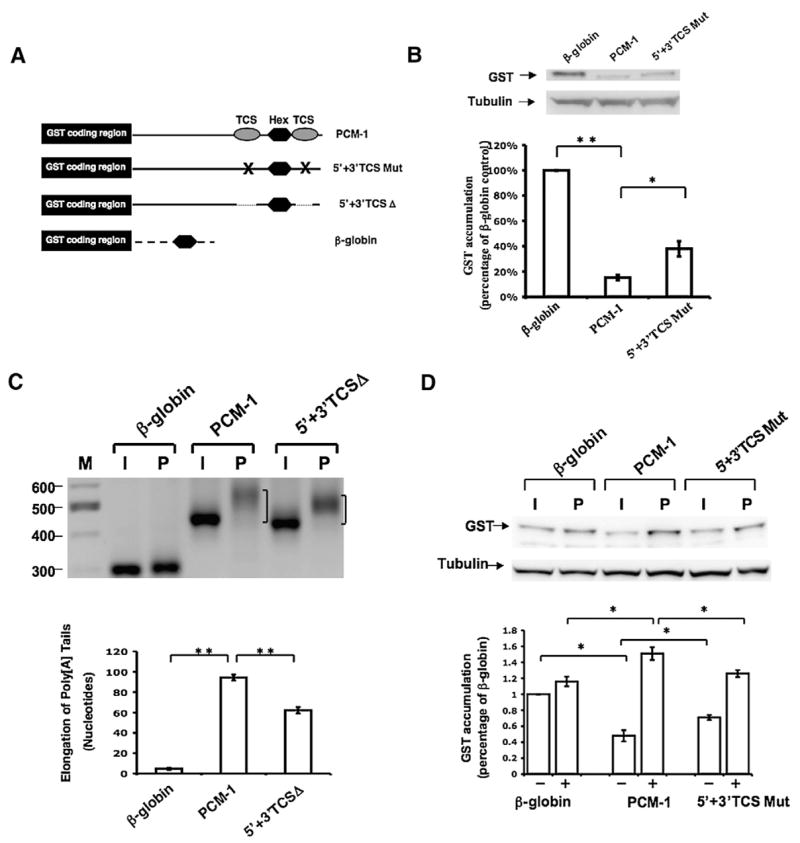
TCS-directed translational regulation contributes to both repression and translational activation exerted by the Pcm-1 3′ UTR. (A) Schematic representation of the Pcm-1 3′ UTR reporter mRNAs employed. Symbols are the same as used in Figs. 1, 5, and 6. (B) Immature oocytes were injected with the indicated reporter mRNAs and the levels of GST reporter mRNA translation assessed by western blot after 18 h of culture. The levels of GST accumulation were normalized for the levels of each reporter mRNA by real-time PCR (see Materials and methods) and quantitated over three independent experiments (graph). One-way analysis of variance and post hoc Newman–Keuls multiple comparison test were performed to analyze differences between the means. Means were scored as statistically significant, P<0.05 (*), P<0.001 (**). Error bars represent SEM. (C) Progesterone-dependent polyadenylation of wild-type or mutant TCS reporter mRNAs. Experiments were performed essentially as described in the legend to Fig. 3, using a forward primer specific to the GST coding region of the injected reporter mRNAs. The mode of the PCR product population was determined using AlphaEaseFC Software (Alpha Innotech) snap-to-peak analyses. Oocytes were cultured in the presence of progesterone for 4 h 20 min. Statistical differences between the mode length of poly[A] tail extension were assessed over three trials as described in panel B. (D) Immature oocytes were injected with the indicated reporter mRNAs and the levels of GST reporter mRNA translation in progesterone-stimulated oocytes and time-matched immature oocytes were assessed by GST western blot (upper panel) after 5 h of culture. The levels of GST accumulation were quantitated over four independent experiments (graph). Error bars represent SEM. One-way analysis of variance and post hoc Newman–Keuls multiple comparison test were performed. Differences between the means were scored as statistically significant, P<0.05 (*).
To determine the contribution of the TCS elements to translational activation, we first assessed the progesterone-dependent polyadenylation controlled by the Pcm-1 3′ UTR. The wild-type Pcm-1 3′ UTR conferred progesterone-dependent cytoplasmic polyadenylation to the GST reporter mRNA, where the mode of poly[A] tail length extension of the reporter mRNA population was approximately 90 adenylate residues (Fig. 7C). When averaged over three independent experiments, deletion of both TCS elements consistently reduced the mode of the progesterone-dependent poly[A] tail extension to approximately 60 adenylate residues (Fig. 7C). Although the resolution of PCR products was less pronounced, similar results were obtained with the Pcm-1 3′ UTR encoding disruptive diguanosine nucleotide substitutions (5′ +3′ TCS mut, data not shown). DNA sequencing of the RNA ligation-coupled PCR products verified that the progesterone-induced increase in PCR product size was specifically due to poly[A] tail extension. We conclude that the TCS elements within the Pcm-1 3′ UTR contribute to the early polyadenylation in maturing oocytes (directing approximately 33% of the total poly[A] tail extension), although an additional element must also be present to account for the remaining TCS-independent polyadenylation observed.
Since the TCS elements only account for part of the polyadenylation directed by the Pcm-1 3′ UTR, we aimed to determine the contribution of the TCS elements to early (pre-GVBD) translational induction. Progesterone-stimulated oocytes were thus analyzed prior to GVBD (and so were still in the early phase of maturation). When compared to the β-globin 3′ UTR, the wild-type Pcm-1 3′ UTR directed a significant translational induction of the GST reporter mRNA following 5 h of progesterone stimulation (Fig. 7D, 1.51±0.08-fold, P<0.05). By contrast, mutation of the TCS elements attenuated the ability of the Pcm-1 3′ UTR to direct translational induction in progesterone-stimulated oocytes prior to GVBD (Fig. 7D). Indeed, the levels of progesterone-stimulated GST reporter mRNA translation were statistically indistinguishable from the levels controlled by the β-globin 3′ UTR. We conclude that the TCS elements direct the early translational activation of the Pcm-1 3′ UTR. In these experiments, the Pcm-1 3′ UTR exerted translational repression in time-matched immature oocytes, although it should be noted that the magnitude of translational repression (52% of the levels of GST translation controlled by the β-globin 3′ UTR) was less than that seen in Fig. 7B (where the Pcm-1 3′ UTR repressed GST translation to 85% of the levels controlled by the β-globin 3′ UTR). A likely explanation for the differences in magnitude of repression is the different incubation times employed in the two experiments (18 h in Fig. 7B vs. 7 h in Fig. 7D). Consistent with the results of Fig. 7B, mutational disruption of the TCS elements reduced the ability of the Pcm-1 3′ UTR to exert repression in immature oocytes (Fig. 7D). Taken together, our results indicate that TCS elements contribute to the translational control exerted by the Pcm-1 3′ UTR in both immature and progesterone-stimulated oocytes.
Discussion
In this study, we have identified a novel mRNA translational regulatory element in the Wee1 and Pcm-1 3′ UTRs, herein designated a translational control sequence (TCS). We demonstrate that the TCS sequence confers both mRNA translational repression in immature oocytes and cytoplasmic polyadenylation and mRNA translational induction in progesterone-stimulated oocytes. The TCS is similar to the Musashi/PRE regulatory sequence in that it directs early polyadenylation and translational induction. Unlike the Musashi/PRE, however, the TCS can also exert translational repression in immature oocytes. While the duality of TCS element function is similar to the previously characterized CPE regulatory sequence, the TCS does not appear to enforce late polyadenylation and translational induction in response to progesterone stimulation as CPE sequences can in certain mRNA 3′ UTRs. Our findings indicate that TCS-directed control exerts a unique combination of temporal regulation of maternal mRNA translation compared to that enforced by CPE or Musashi-directed translational control pathways. This diversity of regulatory element function would facilitate a greater range of proteomic control than that achieved by any one element alone. Indeed, the pattern of protein expression exerted by the TCS would not be recapitulated by CPE and Musashi/PRE sequences acting in a combinatorial manner if present in the same 3′ UTR. In such an arrangement, the CPE would mediate repression in the immature oocyte and the Musashi/PRE would direct early mRNA translational activation in response to progesterone stimulation. However, the presence of a CPE would maintain extension of the poly[A] tail after GVBD, whereas TCS-directed polyadenylation would be attenuated after GVBD (see Figs. 2A and 5 and Charlesworth et al., 2000). We speculate that the pattern of protein expression directed by a TCS element may be necessary to control the synthesis of certain proteins required for early maturation but which are either unnecessary for, or may inhibit, later stages of maturation or embryogenesis.
The timing of TCS- and Musashi/PRE-initiated polyadenylation and translation appears similar, occurring 2–3 h after progesterone stimulation and 1–2 h prior to GVBD. Indeed, when assessed in 1 h intervals after progesterone stimulation, initiation of polyadenylation of endogenous Musashi/PRE-containing Mos mRNA and TCS-containing Pcm-1 mRNA appeared coincident (data not shown). It remains to be determined if the co-incident timing of Musashi/PRE- and TCS-mediated translational control pathways is mediated independently or whether they share a common regulatory step that coordinates initiation of all early class maternal mRNA translation. The role of CPE sequences in the control of early class mRNA translation has not been fully resolved. Conversion of the CPE-binding protein (CPEB1) from a repressor to an activator of translation is downstream of the early translation of the Ringo mRNA (Padmanabhan and Richter, 2006) and MAP kinase signaling (Keady et al., 2007). CPEB1 activation is associated with the phosphorylation of serine 174 and occurs prior to progesterone-stimulated GVBD, consistent with a role for CPEB1 in facilitating early class mRNA translation (Mendez et al., 2000). Indeed, CPEB1 has been implicated in the polyadenylation and translational activation of the early class Mos mRNA (Mendez et al., 2000). However, Mos mRNA translation occurs with similar kinetics in the presence or absence of MAP kinase signaling (Gross et al., 2000) and subsequent work has demonstrated that neither CPEB1 activation nor the CPE in the Mos 3′ UTR is essential for the initial polyadenylation and translational activation of the Mos mRNA (Charlesworth et al., 2002). By contrast, disruption of CPE sequences in the early class mRNAs D7, G10, and histone-like B4 reduces the length of progesterone-stimulated poly[A] tail extension (Charlesworth et al., 2004) and while not addressed directly, these findings suggest that CPE-dependent regulation contributes to the early translation of the D7, G10, and histone-like B4 mRNAs. An additional issue in considering the role of CPEs in the temporal control of maternal mRNA translation is the position of the CPE sequences within a 3′ UTR. The position can dramatically affect the timing of mRNA translation, since a CPE overlapping the polyadenylation hexanucleotide is a defining feature of late class mRNAs (Charlesworth et al., 2004; Tung et al., 2007). Further studies will be required to investigate the temporal interrelationships between the multiple cis-elements and their trans-acting factors in determining the timing of early class mRNA translation.
The observation that mutational disruption of the TCS elements reduces but does not eliminate Pcm-1-directed translational repression in immature oocytes (Figs. 7B and D) suggests that multiple distinct regulatory elements within the Pcm-1 3′ UTR contribute to translational control. The translational repression of the Xenopus cyclin B1 3′ UTR has also been reported to be subject to combinatorial control, involving both CPE and Pumilio regulatory elements (Nakahata et al., 2003). As we report in this study, Wee1 mRNA repression involves the combinatorial function of both TCS and CPE sequences. In addition, we have reported previously that the Xenopus Mos mRNA is subject to a combinatorial control of polyadenylation and translational activation where a Musashi/PRE directs early polyadenylation and translational activation, while a CPE maintains the poly[A] tail extension and contributes to full translational induction later in maturation (Charlesworth et al., 2002, 2006). Taken together, these observations suggest that the regulation of maternal mRNA translation may not be simply attributable to a single type of element in any given 3′ UTR, but in many instances may involve complex combinatorial control by multiple elements which are regulated independently. The mechanistic basis of this differential control is unknown but may include upstream activation by different signaling pathways, downstream recruitment of distinct poly[A] polymerases and/or differential access of deadenylases to the Musashi/PRE- and TCS-containing 3′ UTRs compared to 3′ UTRs containing CPE sequences.
Our findings demonstrating that TCS elements in the Pcm-1 3′ UTR contribute to translational activation are consistent with a physiological role for the TCS in the progesterone-dependent early accumulation of Pcm-1 protein (Fig. 6). Pcm-1 function has been shown to be necessary for assembly of centrosomes and microtubule networks during mitotic cell cycle progression (Balczon et al., 1994; Dammermann and Merdes, 2002). The early translational activation of the Pcm-1 mRNA may contribute to Xenopus meiotic cell progression or be necessary to accumulate sufficient levels of the protein to facilitate mitotic cell cycle progression after fertilization, as has been reported for murine embryos (Balczon et al., 2002). Further studies will be necessary to determine the role of early class Pcm-1 mRNA translational activation in Xenopus meiotic and/or mitotic cell cycle progression.
Our data indicate that the presence of CPEs in the wild-type Wee1 3′ UTR suppresses TCS-directed early translational activation but does not interfere with TCS-directed repression in immature oocytes. The mechanistic basis of the dominant CPE effect on TCS-directed translational activation is unknown but may be a consequence of interaction of the Wee1 3′ UTR with CPEB1 and CPEB1-associated proteins which preclude disruption of the TCS repressor complex. While the molecular nature of the presumed TCS repressor complex remains to be determined, we have recently identified a TCS-specific RNA binding protein by a yeast three hybrid screen (data not shown). This protein contains a plant homeodomain that appears necessary for TCS-specific RNA binding and is related to zygotic arrest 1, a murine protein implicated in activation of the zygotic genome after fertilization (Wu et al., 2003). The identification of a candidate TCS-binding protein will now facilitate future studies on the interactions between TCS and CPE sequences in the same 3′ UTR and will allow a determination of the role of TCS-mediated mRNA translational regulation during vertebrate oocyte maturation.
Supplementary Material
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.ydbio.2008.02.033.
Acknowledgments
This work was supported by ACS RPG 101279 and NIH RO1 HD35688 to A.M.M.; NIH grant RR020146, a UAMS Medical Research Endowment and a UAMS Alzheimer Disease Center Pilot study grant to M.C.M.; an NCRR pilot study award to A.C.; and NIH grant P2RR-16460 from the IDeA Networks of Biomedical Research Excellence (INBRE) Program of the National Center for Research Resources to R.G. We thank Dr. Andreas Merdes (CNRS-Pierre Fabre) for the generous gift of Xenopus Pcm-1 antiserum.
Footnotes
Publisher's Disclaimer: This article appeared in a journal published by Elsevier. The attached copy is furnished to the author for internal non-commercial research and education use, including for instruction at the authors institution and sharing with colleagues.
Other uses, including reproduction and distribution, or selling or licensing copies, or posting to personal, institutional or third party websites are prohibited.
In most cases authors are permitted to post their version of the article (e.g. in Word or Tex form) to their personal website or institutional repository. Authors requiring further information regarding Elsevier’s archiving and manuscript policies are encouraged to visit: http://www.elsevier.com/copyright
References
- Balczon R, et al. PCM-1, a 228-kD centrosome autoantigen with a distinct cell cycle distribution. J Cell Biol. 1994;124:783–793. doi: 10.1083/jcb.124.5.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balczon R, et al. Arrest of cell cycle progression during first interphase in murine zygotes microinjected with anti-PCM-1 antibodies. Cell Motil Cytoskeleton. 2002;52:183–192. doi: 10.1002/cm.10043. [DOI] [PubMed] [Google Scholar]
- Ballantyne S, et al. A dependent pathway of cytoplasmic polyadenylation reactions linked to cell cycle control by c-mos and CDK1 activation. Mol Biol Cell. 1997;8:1633–1648. doi: 10.1091/mbc.8.8.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkoff AF, et al. Translational control of cyclin B1 mRNA during meiotic maturation: coordinated repression and cytoplasmic polyadenylation. Dev Biol. 2000;220:97–109. doi: 10.1006/dbio.2000.9613. [DOI] [PubMed] [Google Scholar]
- Charlesworth A, et al. The temporal control of Wee1 mRNA translation during Xenopus oocyte maturation is regulated by cytoplasmic polyadenylation elements within the 3′ untranslated region. Dev Biol. 2000;227:706–719. doi: 10.1006/dbio.2000.9922. [DOI] [PubMed] [Google Scholar]
- Charlesworth A, et al. A novel regulatory element determines the timing of Mos mRNA translation during Xenopus oocyte maturation. EMBO J. 2002;21:2798–2806. doi: 10.1093/emboj/21.11.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth A, et al. Cytoplasmic polyadenylation element (CPE)- and CPE-binding protein (CPEB)-independent mechanisms regulate early class maternal mRNA translational activation in Xenopus oocytes. J Biol Chem. 2004;279:17650–17659. doi: 10.1074/jbc.M313837200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth A, et al. Musashi regulates the temporal order of mRNA translation during Xenopus oocyte maturation. EMBO J. 2006;25:2792–2801. doi: 10.1038/sj.emboj.7601159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colegrove-Otero LJ, et al. RNA-binding proteins in early development. Crit Rev Biochem Mol Biol. 2005;40:21–73. doi: 10.1080/10409230590918612. [DOI] [PubMed] [Google Scholar]
- Dammermann A, Merdes A. Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J Cell Biol. 2002;159:255–266. doi: 10.1083/jcb.200204023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson E. Gene Activity in Early Development. Academic Press; 1986. [Google Scholar]
- de Moor CH, Richter JD. The Mos pathway regulates cytoplasmic polyadenylation in Xenopus oocytes. Mol Cell Biol. 1997;17:6419–6426. doi: 10.1128/mcb.17.11.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Moor CH, et al. Mechanisms of translational control by the 3′ UTR in development and differentiation. Semin Cell Dev Biol. 2005;16:49–58. doi: 10.1016/j.semcdb.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Ferby I, et al. A novel p34(cdc2)-binding and activating protein that is necessary and sufficient to trigger G(2)/M progression in Xenopus oocytes. Genes Dev. 1999;13:2177–2189. doi: 10.1101/gad.13.16.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox CA, et al. Poly(A) addition during maturation of frog oocytes: distinct nuclear and cytoplasmic activities and regulation by the sequence UUUUUAU. Genes Dev. 1989;3:2151–2162. doi: 10.1101/gad.3.12b.2151. [DOI] [PubMed] [Google Scholar]
- Freeman RS. Meiotic induction by Xenopus cyclin B is accelerated by coexpression with mosXe. Mol Cell Biol. 1991;11:1713–1717. doi: 10.1128/mcb.11.3.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillian-Daniel DL, et al. Modifications of the 5′ cap of mRNAs during Xenopus oocyte maturation: independence from changes in poly(A) length and impact on translation. Mol Cell Biol. 1998;18:6152–6163. doi: 10.1128/mcb.18.10.6152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross SD, et al. The critical role of the MAP kinase pathway in meiosis II in Xenopus oocytes is mediated by p90(Rsk) Curr Biol. 2000;10:430–438. doi: 10.1016/s0960-9822(00)00425-5. [DOI] [PubMed] [Google Scholar]
- Heikinheimo O, Gibbons WE. The molecular mechanisms of oocyte maturation and early embryonic development are unveiling new insights into reproductive medicine. Mol Hum Reprod. 1998;4:745–756. doi: 10.1093/molehr/4.8.745. [DOI] [PubMed] [Google Scholar]
- Howard EL, et al. The mitogen-activated protein kinase signaling pathway stimulates mos mRNA cytoplasmic polyadenylation during Xenopus oocyte maturation. Mol Cell Biol. 1999;19:1990–1999. doi: 10.1128/mcb.19.3.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai T, et al. The neural RNA-binding protein Musashi1 translationally regulates mammalian numb gene expression by interacting with its mRNA. Mol Cell Biol. 2001;21:3888–3900. doi: 10.1128/MCB.21.12.3888-3900.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keady BT, et al. MAPK interacts with XGef and is required for CPEB activation during meiosis in Xenopus oocytes. J Cell Sci. 2007;120:1093–1103. doi: 10.1242/jcs.03416. [DOI] [PubMed] [Google Scholar]
- Kuersten S, Goodwin EB. The power of the 3′ UTR: translational control and development. Nat Rev, Genet. 2003;4:626–637. doi: 10.1038/nrg1125. [DOI] [PubMed] [Google Scholar]
- MacNicol AM, et al. Raf-1 kinase is essential for early Xenopus development and mediates the induction of mesoderm by FGF. Cell. 1993;73:571–583. doi: 10.1016/0092-8674(93)90143-e. [DOI] [PubMed] [Google Scholar]
- McGowan CH, Russell P. Human Wee1 kinase inhibits cell division by phosphorylating p34cdc2 exclusively on Tyr15. EMBO J. 1993;12:75–85. doi: 10.1002/j.1460-2075.1993.tb05633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton DA, et al. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984;12:7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez R, Richter JD. Translational control by CPEB: a means to the end. Nat Rev, Mol Cell Biol. 2001;2:521–529. doi: 10.1038/35080081. [DOI] [PubMed] [Google Scholar]
- Mendez R, et al. Phosphorylation of CPE binding factor by Eg2 regulates translation of c-mos mRNA. Nature. 2000;404:302–307. doi: 10.1038/35005126. [DOI] [PubMed] [Google Scholar]
- Mignone F, et al. UTRdb and UTRsite: a collection of sequences and regulatory motifs of the untranslated regions of eukaryotic mRNAs. Nucleic Acids Res. 2005;33:D141–D146. doi: 10.1093/nar/gki021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller PR, et al. Cell cycle regulation of a Xenopus Wee1-like kinase. Mol Biol Cell. 1995;6:119–134. doi: 10.1091/mbc.6.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami MS, Vande Woude GF. Analysis of the early embryonic cell cycles of Xenopus; regulation of cell cycle length by Xe-wee1 and Mos. Development. 1998;125:237–248. doi: 10.1242/dev.125.2.237. [DOI] [PubMed] [Google Scholar]
- Nakahata S, et al. Involvement of Xenopus Pumilio in the translational regulation that is specific to cyclin B1 mRNA during oocyte maturation. Mech Dev. 2003;120:865–880. doi: 10.1016/s0925-4773(03)00160-6. [DOI] [PubMed] [Google Scholar]
- Nakajo N, et al. Absence of wee1 ensures the meiotic cell cycle in Xenopus oocytes. Genes Dev. 2000;14:328–338. [PMC free article] [PubMed] [Google Scholar]
- Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: I. Characterization and timing of cellular changes at the midblastula stage. Cell. 1982;30:675–686. doi: 10.1016/0092-8674(82)90272-0. [DOI] [PubMed] [Google Scholar]
- Padmanabhan K, Richter JD. Regulated Pumilio-2 binding controls RINGO/Spy mRNA translation and CPEB activation. Genes Dev. 2006;20:199–209. doi: 10.1101/gad.1383106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker LL, Piwnica-Worms H. Inactivation of the p34cdc2-cyclin B complex by the human WEE1 tyrosine kinase. Science. 1992;257:1955–1957. doi: 10.1126/science.1384126. [DOI] [PubMed] [Google Scholar]
- Prasad CK, et al. Mos 3′ UTR regulatory differences underlie species-specific temporal patterns of Mos mRNA cytoplasmic polyadenylation and translational recruitment during oocyte maturation. Molecular Reproduction and Development. 2008 doi: 10.1002/mrd.20877. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rassa JC, et al. Spacing constraints on reinitiation of paramyxovirus transcription: the gene end U tract acts as a spacer to separate gene end from gene start sites. Virology. 2000;274:438–449. doi: 10.1006/viro.2000.0494. [DOI] [PubMed] [Google Scholar]
- Richter JD. Cytoplasmic polyadenylation in development and beyond. Microbiol Mol Biol Rev. 1999;63:446–456. doi: 10.1128/mmbr.63.2.446-456.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy LM. Activation of p34cdc2 kinase by cyclin A. J Cell Biol. 1991;113:507–514. doi: 10.1083/jcb.113.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheets MD, et al. The 3′-untranslated regions of c-mos and cyclin mRNAs stimulate translation by regulating cytoplasmic polyadenylation. Genes Dev. 1994;8:926–938. doi: 10.1101/gad.8.8.926. [DOI] [PubMed] [Google Scholar]
- Sheets MD, et al. Polyadenylation of c-mos mRNA as a control point in Xenopus meiotic maturation. Nature. 1995;374:511–516. doi: 10.1038/374511a0. [DOI] [PubMed] [Google Scholar]
- Tung JJ. Translational unmasking of Emi2 directs cytostatic factor arrest in meiosis II. Cell Cycle. 2007;6:725–731. doi: 10.4161/cc.6.6.3936. [DOI] [PubMed] [Google Scholar]
- Wu X, et al. Zygote arrest 1 (Zar1) is a novel maternal-effect gene critical for the oocyte-to-embryo transition. Nat Genet. 2003;33:187–191. doi: 10.1038/ng1079. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.ydbio.2008.02.033.


