Abstract
The evolutionary-conserved interactions between KASH and SUN domain-containing proteins within the perinuclear space establish physical connections, called LINC complexes, between the nucleus and the cytoskeleton. Here, we show that the KASH domains of Nesprins 1, 2 and 3 interact promiscuously with luminal domains of Sun1 and Sun2. These constructs disrupt endogenous LINC complexes as indicated by the displacement of endogenous Nesprins from the nuclear envelope. We also provide evidence that KASH domains most probably fit a pocket provided by SUN domains and that post-translational modifications are dispensable for that interaction. We demonstrate that the disruption of endogenous LINC complexes affect cellular mechanical stiffness to an extent that compares to the loss of mechanical stiffness previously reported in embryonic fibroblasts derived from mouse lacking A-type lamins, a mouse model of muscular dystrophies and cardiomyopathies. These findings support a model whereby physical connections between the nucleus and the cytoskeleton are mediated by interactions between diverse combinations of Sun proteins and Nesprins through their respective evolutionary-conserved domains. Furthermore, they emphasize, for the first time, the relevance of LINC complexes in cellular mechanical stiffness suggesting a possible involvement of their disruption in various laminopathies, a group of human diseases linked to mutations of A-type lamins.
Keywords: Sun1, Sun2, Nesprin, LINC complexes, Laminopathies, Ballistic intracellular nanorheology
Introduction
The nuclear envelope (NE) comprises two lipid bilayers, the inner and the outer nuclear membrane (INM and ONM), which connect at nuclear pores and delineate the perinuclear space. While the ONM is an extension of the rough endoplasmic reticulum (ER), the INM adheres to the nuclear lamina, a meshwork of intermediate filaments composed of A- and B-type lamins [1–3].
Evolutionary-conserved physical connections between the nuclear lamina and the peripheral cytoskeleton, called LINC complexes, were recently uncovered. They form through interactions between luminal domains of two families of transmembrane proteins of the nuclear envelope: Sun proteins and Nesprins [4–7].
Human Sun1 and Sun2 are integral type II transmembrane proteins (824 and 717 amino acids, respectively) with a transmembrane domain separating the N-terminal nucleoplasmic region from the C-terminal luminal region [8–11]. Their luminal regions protrude into the perinuclear space and contain two coiled-coil domains as well as a conserved C-terminal region of approximately 150 amino acids (Fig. 1A). This stretch of amino acids, initially characterized within the C-terminus of the Caenorhabditis elegans protein UNC-84 [12] and the Schizosaccharomyces pombe Sad1 protein [13], defines the so-called SUN (Sad1 and UNC-84 homology) domain. In addition to Sun1 and 2, two other mammalian proteins, Sun3 (NP_001025190) and Spag4 (AAF75268) also display the overall structure of Sun proteins. However, Sun3 does not localize at the NE and, based on examination of EST databases, expression levels and tissue distributions of Sun3 and Spag4 are much more restricted than Sun1 and Sun2. We recently demonstrated that the nucleoplasmic regions of Sun1 and Sun2 interact directly with both A- and B-type lamins [9]; this interaction also occurs in C. elegans where the retention of UNC-84 at the NE requires Ce-lamin [14]. The localization of mammalian Sun proteins at the NE, however, seems to involve additional lamin-independent retention mechanisms [15].
Fig. 1.
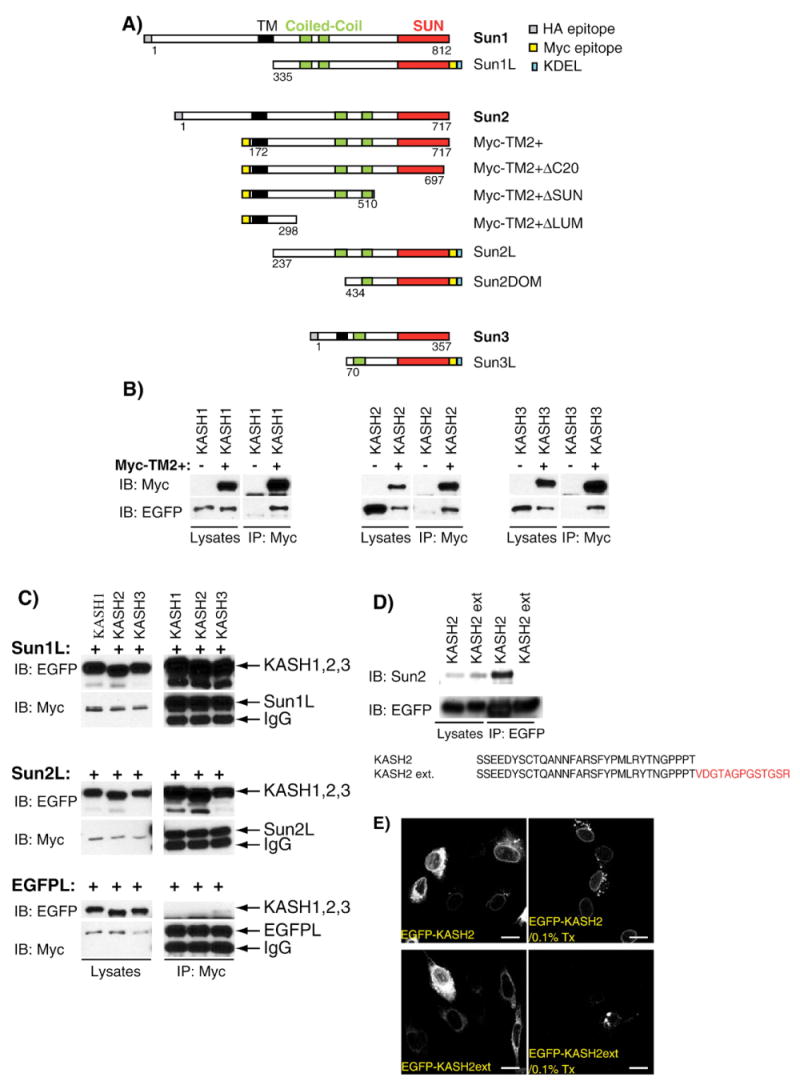
A) Depiction of the structural organization of human Sun1, Sun2 and Sun3 and the corresponding recombinant constructs used in this study. The relative position of the transmembrane domain (TM, black), coiled-coils (green) and SUN domain (red) as well as the N- or C-terminal localization of HA (gray) and Myc epitopes (yellow) and KDEL domains (blue) is also indicated. B) The luminal domain of Sun2 interacts with the KASH domains of Nesprins 1, 2 and 3. EGFP-KASH1 (left), EGFP-KASH2 (middle) or EGFP-KASH3 (right) were either singly transfected or co-transfected with Myc-TM2+ in HeLa cells. Lysates were immunoprecipitated with an anti-Myc antibody and immunoblotted with either anti-EGFP or anti-Myc antibodies. For each experiment, the amount of initial recombinant proteins in whole lysates (left panels) and immunoprecipitation products (right panels) are shown. C) Sun1L and Sun2L indiscriminately interact with the KASH domain of Nesprins 1, 2 and 3. Sun1L or Sun2L were co-transfected with EGFP-KASH1, 2 or 3. The cotransfection of EGFPL with KASH constructs was used as a negative control. Lysates were immunoprecipitated with an anti-Myc antibody and analyzed by immunoblot with either anti-EGFP (upper panels) or anti-Myc (lower panels) antibodies. For each experiment, the amount of initial recombinant proteins in whole lysates (left panels) and immunoprecipitation products (right panels) are shown. D) Recombinant EGFP-KASH2 but not EGFP-KASH2ext co-immunoprecipitate endogenous Sun2. HeLa cells were transfected with the indicated constructs and lysates were immunoprecipitated with an anti-EGFP antibody. Endogenous Sun2 was detected with a specific anti-Sun2 serum. For each experiment, the amount of recombinant protein and endogenous Sun2 in whole lysates (left panel) and immunoprecipitates (right panel) are shown. E) EGFP-KASH2, but not EGFP-KASH2ext, is anchored at the NE. C2C12 cells were transfected with both constructs and analyzed in direct immunofluorescence with or without a 0.1% Triton X-100 (Tx) pretreatment before the fixation step. Scale bars: 100 μm.
Nesprins (Nuclear Envelope SPectRIN repeats) are also type II transmembrane proteins [16,17]. Several isoforms of Nesprin 1 (also called Syne1 [18], Myne1 [19], and Enaptin [20]) and Nesprin 2 (also called Syne2 [18] and NUANCE [21]) are encoded by the alternative transcription and splicing of two distinct genes [22]. Nesprins are characterized by a variable number of spectrin repeats and most isoforms share a conserved C-terminal KASH domain (Klarsicht-Ancl-Syne1 Homology) comprising ∼50 amino acids. The KASH domain consists of a transmembrane domain followed by an evolutionary-conserved stretch of ∼30 amino acids that protrude into the perinuclear space [5,6]. While Nesprin isoforms display a complex subcellular distribution, giant Nesprin 1 (> 700 kDa) and Nesprin 2 (> 1 MDa) both localize to the ONM and extend as rod-like structures of up to 300–400 nm into the cytoplasm. Through their N-terminal actin-binding domains (ABD), giant Nesprins 1 and 2 bind to actin microfilaments [20,21,23]. Nesprin 3α and β proteins, both encoded by the alternative splicing of a separate gene, each contain a KASH domain and localize to the ONM but are more modest in size (∼100 kDa) than giant Nesprins 1 and 2. Interestingly, the N-terminal region of Nesprin 3α contains a plectin-binding domain [24] suggesting that KASH domain-containing proteins can connect the NE to different cytoskeletal networks. Because of the increasing number of characterized SUN and KASH domain-containing proteins in mammalian cells, we addressed whether Nesprins and Sun proteins form specific interacting pairs or assemble in a promiscuous manner and identified new structural and post-translational determinants required for the Nesprins/Sun proteins interaction.
In lower organisms, different studies reviewed in [5,6] clearly point to the involvement of SUN and KASH domain-containing proteins in nuclear migration, nuclear anchorage and coupling to the centrosome. In mammalian cells, the involvement of Nesprins in nuclear anchorage has recently been reported [25,26]. Overall, the current model strongly suggests that interaction between Sun proteins and Nesprins provide a tight connection between the nuclear lamina to the perinuclear cytoskeleton [5–7]. To test that model and motivated by the hypothesis that disruption of LINC complexes observed in fibroblasts lacking A-type lamins are responsible for the loss of cellular stiffness measured in these cells [27], we demonstrate the direct involvement of LINC complexes in mechanical stiffness suggesting that disruption of LINC complexes could be involved in the etiology of some laminopathies.
Materials and methods
Antibodies
Rabbit Sun2 and Nesprin 3 (provided by Dr. A. Sonnenberg, The Netherlands Cancer Institute, Amsterdam, The Netherlands) antisera have been described [10,24]. Rabbit anti-Nesprin 2 giant, kindly provided by Drs. E. Gomes and G. Gundersen (Columbia University, New York), was raised against the actin-binding domain of Nesprin 2 giant (E.R. Gomes et al., manuscript in preparation). Monoclonal rat anti-HA (3F10) antibody was purchased from Roche. Monoclonal mouse anti-myc (9E10) and anti-GFP (B-2) antibodies were purchased from Santa Cruz Biotechnology, Inc.
Recombinant constructs
The region covered by each construct described below is depicted in Fig. 1. Myc-TM2+, Myc-TM2+N636Q, Myc-TM2+ΔC20, Myc-TM2+ΔSUN and Myc-TM2+ΔLUM fragments were amplified by PCR from full-length Sun2 in pBlueScript (KIAA0668, Kasuza DNA Research Institute) and cloned into EcoRI/NotI sites of pCMV-myc (Clontech Laboratories, Inc.). KASH1 (74 C-term amino acids of Nesprin 1), KASH2 (65 C-term amino acids of Nesprin 2) and KASH3 (67 C-term amino acids of Nesprin 3) fragments were amplified by RT-PCR from HeLa cells total RNA and cloned into pEGFP-Cl (Clontech Laboratories, Inc.) at BglI/BamHI sites. pEGFP::KASH2ext was a gift from Catherine M. Shanahan (University of Cambridge, Cambridge, UK). KASH2Y40F/Y58F (tyrosine residues are numbered according to the KASH2 fragment length) and KASH2dupl. were obtained by the mutation pEGFP-Cl::KASH2 using QuikChange Site-Directed Mutagenesis (Stratagene). Sun2L and SUN2DOM fragments were PCR-amplified from the KIAA0668 cDNA and cloned into pShooter/myc/ER (Invitrogen) at Sall/NotI sites. Sun1L and Sun3L fragments were amplified by PCR from Sun1 and Sun3 cDNAs and cloned into pShooter/myc/ER at the Sail site. In addition to providing a C-terminal myc tag, pShooter/myc/ER contains in-frame signal and retention (KDEL) sequences. EGFP cloned in pShooter/myc/ER (Invitrogen) was used as a negative control. HA-Sun1 and HA-Sun3 in pcDNA3.1 were gifts from Brian Burke (University of Florida, Gainesville, FL, USA). HA-Sun2 was amplified by PCR and cloned into pcDNA3.1 (Invitrogen) by TOPO/TA ligation.
Cell culture and transfection
HeLa and C2C12 cells were grown in 10% bovine growth serum with DMEM (GIBCO Life Technologies). Transient transfection was carried out using Mirus TransIT-LT1 Transfection Reagent (Mirus Bio Corp.) according to the manufacturer's instructions.
Deglycosylation
Cells (20–24 h post-transfection with the indicated constructs) were lysed with either radio-immunoprecipitation assay (RIPA) lysis buffer (1% Triton X-100, 0.5% deoxycholate, 0.1% SDS, 2.5 mM Na4P2O7, 1 mM β-glycerophosphate, 1 mM Na3VO4, 1 mM EGTA, 1 mM EDTA, 1× PBS, protease and phosphatase inhibitor cocktails (Roche)) or 2× Laemmli buffer. Lysates were processed with either EndoH or PNGaseF (New England Biolabs) according to the manufacturer's instructions. Negative controls were run in parallel without addition of endoglycanases.
Immunofluorescence microscopy
Where indicated, cells were treated with 1× phosphate-buffered saline (PBS) containing 0.1% Triton X-100 and protease inhibitors for 5 min on ice prior to the fixation step. Cells (20–24 h post-transfection) plated on coverslips were routinely fixed with 2.5% paraformaldehyde in PBS for 15 min at room temperature (RT), permeabilized with PBS containing 0.1% Triton X-100 and 10% goat serum for 5 min and stained with primary antibodies diluted in PBS containing 10% goat serum for 1 h at RT. After three washes in PBS, cells were stained with fluorescently labeled (Alexa488 or Alexa594) secondary antibodies (Molecular Probes) for 1 h at RT. Coverslips were then rinsed three times for 5 min and mounted. Images were taken with a 63× oil immersion objective (1.4 N.A., Plan-Achromat, Zeiss) on an Axioplan microscope (Zeiss) outfitted with a confocal laser scanning head (MRC 1024, Bio-Rad). The Lasersharp 2000 software package (Bio-Rad) is used to pilot the microscope and format the raw images into TIFF files. The latter were further digitally merged using Photoshop (Adobe).
Immunoprecipitation and pull-down experiments
Cells (20–24 h post-transfection with the indicated construct) were lysed with RIPA buffer and insoluble proteins were removed by centrifugation at 13,000 rpm in a microcentrifuge at 4 °C for 10 min. Myc and EGFP constructs were immunoprecipitated with 2.5 μg monoclonal mouse anti-myc (9E10) antibody (per 35-mm dish) or 10 μg monoclonal mouse anti-GFP (B-2) antibody (per 100-mm dish), respectively. After overnight incubation at 4 °C with end-over-end rotation, immune complexes were pulled down by protein A covalently bound to agarose resin (Sigma-Aldrich, Inc). Immunoprecipitates were washed by end-over-end rotation three times for 10 min in RIPA buffer. Agarose beads were then resuspended in reducing conditions with 2× Laemmli buffer, boiled, and the supernatant loaded on appropriate acrylamide gels. GST-KASH2 fusion protein was obtained by cloning the luminal domain of human KASH2 downstream from the GST moiety of the pGEX-4Tl vector and immobilizing the resulting fusion protein purified from IPTG-induced bacterial lysates on glutathione beads. The C-23 peptide corresponding to the 23 C-terminal amino acids of Nesprin 2 (ALSNNFARSFHPMLRYTNGPPPL) was obtained from Sigma-Genosys and covalently coupled in anhydrous conditions on Affigel-15 (Bio-Rad). Affigel and Glutathione beads (with or without their respective proteins) were incubated with HeLa cell RIPA lysates, washed four times and eluted by boiling 5 min in Laemmli buffer. Immunoprecipitation of Sun2 from these lysates was described previously [10].
Western blots
Proteins were separated by SDS-PAGE and transferred to nitrocellulose membrane. Membranes were blocked in 1× Tris-buffered saline with Tween-20 (TBST), 5% milk for 1 h at RT. Primary antibodies were blotted in 1× TBST, 5% milk either for 1 h at RT or overnight at 4 °C. Secondary antibody-HRP conjugates were blotted in 1× TBST, 5% milk for 1 h at RT and the signals detected with SuperSignal West Pico Chemiluminescent Substrate (Pierce Biotechnologies).
Ballistic intracellular nanorheology (BIN)
The micromechanical properties of the cytoplasm of control and transfected cells were measured using ballistic intracellular nanorheology [28–30]. Briefly, 100-nm diameter polystyrene fluorescent nanoparticles were ballistically injected in the cytoplasm of control and transfected cells using a biolistic gun (Bio-Rad, Hercules, CA). After overnight incubation, the nanoparticles (between 10 and 30 per cell) dispersed uniformly in the cytoplasm and were tracked with high spatial (∼5 nm) and temporal (1/30 s) resolutions using a Cascade 1k CCD camera (Roper Scientific, Tucson AZ) mounted on a Nikon Eclipse TE2000-E epifluorescence microscope and controlled by the software Metaview (Universal Imaging). The mean-squared displacement (MSDs) of individual nanoparticles were computed from the time-dependent (x,y) coordinates of the nanoparticles' centroid displacements. Simple mathematical manipulation detailed in [28] transformed MSDs into elastic modulus (which we report here) and viscous modulus of the cytoplasm. The number of individual cells and total number of nanoparticles tracked for each type of cell is indicated in the caption of Fig. 5. Mean values, standard error of measurement (SEM), and statistical analysis of bead displacements were calculated and plotted using Graphpad Prism (Graphpad Software, San Diego, CA). Two-tailed unpaired t tests were conducted to determine significance, which was indicated using the standard Michelin Guide scale.
Fig. 5.
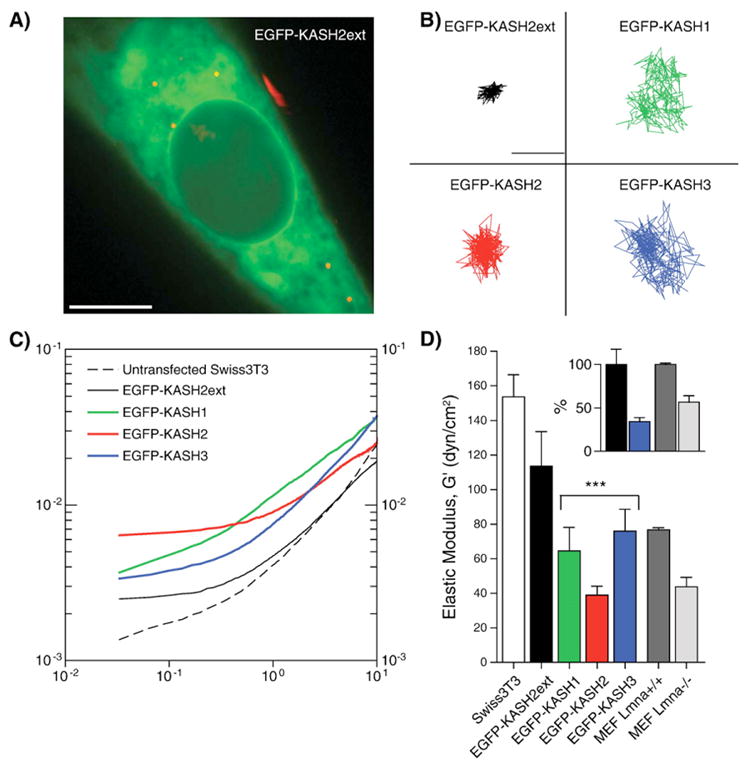
LINC complexes control intracellular mechanics. A) Fluorescent 100-nm diameter polystyrene nanoparticles (red) were ballistically injected into Swiss 3T3 fibroblasts, which were then transfected with EGFP-KASH2ext (displayed), EGFP-KASH1, EGFP-KASH2, or EGFP-KASH3 constructs. The nanoparticles were subsequently tracked with high spatial (<10 nm) and temporal (<1/30 s) resolutions using multi-particle tracking software. Scale bar: 10μm. B) X–Y trajectories of nanoparticles embedded in the cytoplasm of their respective construct-transfected fibroblasts. Scale bar: 0.25μm. C) Ensembled-averaged mean-squared displacements of nanoparticles in untransfected and transfected fibroblasts. D) Mean elasticity of the cytoplasm of untransfected and transfected fibroblasts, as well as Lmna+/+ and Lmna−/− MEFs. Asterisks indicate p values <0.0001 between EGFP-KASH2ext fibroblasts and EGFP-KASH1, EGFP-KASH2, and EGFP-KASH3 transfected fibroblasts. Inset: Normalized mean elasticity between EGFP-KASH2ext and EGFP-KASH2 transfected fibroblasts (black, blue, respectively), and Lmna+/+ and Lmna−/− MEFs (dark gray, light gray, respectively). In all experiments, cells were plated on collagen. Data represents 10 untransfected cells (44 particles), 20 EGFP-KASH2ext transfected cells (63 particles), 9 EGFP-KASH1 transfected cells (37 particles), 20 EGFP-KASH2 transfected cells (110 particles), 9 EGFP-KASH3 transfected cells (85 particles), >35 MEF Lmna+/+ cells (>400 particles), and >35 MEF Lmna−/− cells (>400 particles).
Results
Sun proteins interact promiscuously with Nesprins
The characterization of an increasing number of SUN and KASH domain-containing proteins raises the challenging question of whether LINC complexes exist as specific interacting pairs of both family members or arise through the promiscuous interaction between all members of these protein families. We recently observed co-immunoprecipitation between endogenous Sun2 with endogenous Nesprin 2 giant [9] or endogenous Nesprin 3 (data not shown), However, it is difficult to extend these co-immunoprecipitation experiments to other endogenous Sun proteins and Nesprins due to the lack of suitable reagents as well as the limited solubility of these proteins in mild extraction buffers. To resolve these issues, we analyzed the interaction of recombinant proteins corresponding to the luminal domains of Sun1 and Sun2 with the KASH domains of Nesprins 1, 2 and 3 in mammalian cells. To examine the interaction of the luminal domain of Sun2, we chose to use Myc-TM2+ (Fig. 1A) for two main reasons. First, Myc-TM2+ is largely soluble in mild buffers because it does not include the nucleoplasmic domain that renders Sun2 partially insoluble [10]. Second, as indicated by its glycosylation status (see below), Myc-TM2+ is efficiently translocated within the ER lumen. Constructs encoding the KASH domains of human Nesprins 1, 2 and 3 (termed hereafter KASH1, 2 and 3), which only include the transmembrane and luminal domains, were N-terminally tagged with EGFP.
The interaction between the luminal domain of Sun2 and KASH domains were first tested using a co-transfection/immunoprecipitation/immunoblot analysis approach. HeLa cells were co-transfected with Myc-TM2+ and EGFP-KASH1, 2 or 3 constructs. Anti-Myc immunoprecipitates were immunoblotted with an anti-EGFP antibody to detect EGFP-KASH proteins. Fig. 1B shows that Myc-TM2+ co-immunoprecipitated with EGFP-KASH1, 2 and 3. The amount of input protein was comparable in each case and the extent of co-immunoprecipitation was consistently similar for KASH1, 2 and 3 in different experiments. We conclude that the luminal domain of Sun2 interacts equally well with the KASH domains of Nesprins 1, 2 and 3. To further expand these analyses to the luminal domain of Sun1, the latter was cloned in pShooter/ER (Sun1L, Fig. 1A). This vector enables the translocation of recombinant proteins within the ER lumen by the addition of an N-terminal signal sequence as well as a C-terminal Myc epitope followed by the ER retention sequence (KDEL). The coding sequence of the luminal domain of Sun2 (Sun2L, Fig. 1A) and of EGFP (EGFPL) cloned in pShooter/ER was used as positive and negative controls, respectively. As shown in Fig. 1C, Sun1L and Sun2L co-immunoprecipitated with all EGFP-KASH constructs. Interestingly, the use of the Myc epitope, which is located downstream from the SUN domain of Sun1L and Sun2L (Fig. 1A), to immunoprecipitate Sun1L and Sun2L did not impair the SUN/KASH interaction.
The relevance of this interaction was further indicated by the highly efficient immunoprecipitation of the endogenous Sun2 protein by EGFP-KASH2 (Fig. 1D). These results suggest that Sun proteins and Nesprins interact in a promiscuous manner rather than forming exclusive interacting pairs and implied that LINC complexes can form by pairing between different members of the respective families of proteins. Interestingly, EGFP-KASH2ext, which corresponds to the EGFP-KASH2 protein sequence containing additional C-terminal amino acids originating from the pEGFP expression vector (Fig. 1D), was unable to co-immunoprecipitate with the endogenous Sun2 protein (Fig. 1D). Accordingly, even though both EGFP-KASH2 and EGFP-KASH2ext localized in the ER and the NE (Fig. 1E), a 0.1% Triton X-100 extraction carried out prior to the fixation step clearly indicated that EGFP-KASH2 (as well as EGFP-KASH1 and EGFP-KASH3, data not shown) is anchored at the NE while EGFP-KASH2ext is not (Fig. 1E). These results indicate that anchorage of KASH domains at the NE can be saturated and, as further confirmed below, requires the interaction with Sun proteins.
Disruption of the endogenous LINC complexes
Next, we investigated the physiological relevance of the above findings. As we have just noted, the quantity of KASH domain binding sites at the NE is saturable, and co-immunoprecipitation results suggest promiscuous interactions between mammalian SUN and KASH domain-containing proteins. Therefore, we reasoned that the overexpression of either any EGFP-KASH construct or the luminal domain of any Sun protein would prevent the formation of all possible LINC complex combinations and displace endogenous Nesprins from the NE to the ER. That possibility was examined in C2C12 mouse myoblasts which express both Sun1 and Sun2 (data not shown). Cells were transfected with each of the above constructs and the integrity of endogenous LINC complexes was monitored by immunolocalization of endogenous Nesprin 2 giant and Nesprin 3α and β. The transfection of C2C12 with EGFP-KASH1, 2 and 3 resulted in the complete displacement of Nesprin 2 giant (Fig. 2A) and Nesprin 3 (Fig. 2B) from the NE to the ER while cells transfected with EGFP-KASH2ext displayed a typical NE localization of endogenous Nesprins (Figs. 2A and B). Scoring of these displacements (Fig. 2C) revealed that Nesprin 2 giant and Nesprin 3 were displaced from the NE to the ER in more than 90% of cells transfected with EGFP-KASH1, EGFP-KASH2 or EGFP-KASH3, while less than 5% of cells transfected with EGFP-KASH2ext displayed that phenotype (Fig. 2C).
Fig. 2.
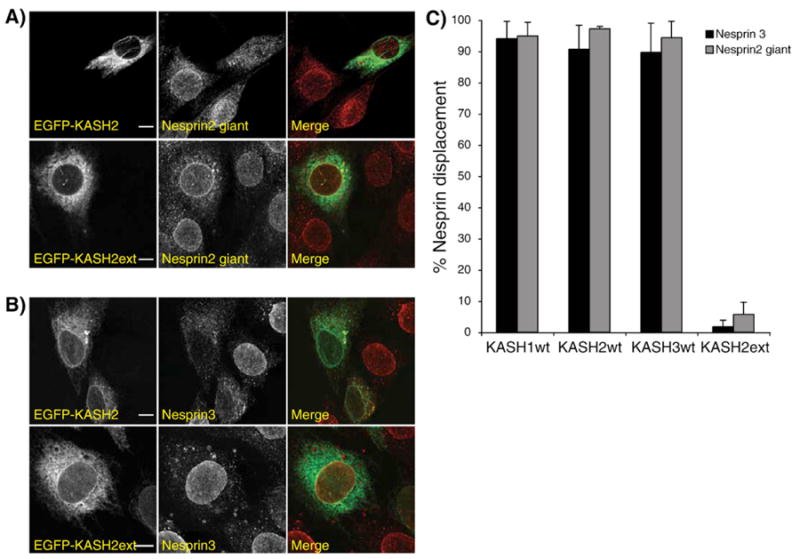
The KASH domain of Nesprins 1, 2 and 3 disrupts endogenous LINC complexes. Immunolocalization of endogenous Nesprin 2 giant (A) and Nesprin 3 (B) in mouse C2C12 cells transfected with either EGFP-KASH2 (upper panels) or EGFP-KASH2ext (lower panels). Specific antisera against Nesprin 2 giant and Nesprin 3 were used in conjunction with a goat anti-rabbit secondary antibody coupled to Alexa594. Scale bars: 50 μm. The same results were obtained with EGFP-KASH1 and EGFP-KASH3. C) Percentage of cells transfected with the indicated construct showing a displacement of endogenous Nesprin 2 giant and Nesprin 3 from the NE to the ER. Only the complete disappearance, as opposed to just a weakened signal, of Nesprin-positive rim structures at the NE was scored as a displacement from the NE to the ER by three independent experimenters. At least 100 transfected cells were examined in each experiment.
These results demonstrate that, as suggested by the promiscuous interactions detected in co-immunoprecipitation experiments, the KASH domains of Nesprins 1, 2 and 3 are each equally potent to compete with pre-formed endogenous LINC complexes. Furthermore, the inability of KASH2ext to displace endogenous Nesprins and to interact with endogenous Sun proteins further demonstrates that the displacement of Nesprins is strictly due to saturation of available SUN proteins at the NE.
To confirm these results, cells were transfected with the luminal domain of Sun1, Sun2 and Sun3 cloned in pShooter/ER (Sun1L, Sun2L and Sun3L are depicted in Fig. 1A). As expected, the LINC complex was disrupted in C2C12 cells transfected with Sun1L and Sun2L (Fig. 3A). The overexpression of Sun3L in the ER lumen, however, did not provoke any significant displacement of Nesprins from the ONM to the ER (Fig. 3A). This result suggested that the SUN domain by itself is not sufficient to compete with endogenous LINC complexes and that upstream regions might also play a role. Since Sun3 only contains a single coiled-coil domain, we cloned SUN2DOM (Fig. 1A), a shortened version of Sun2L that only includes a single predicted coiled-coil domain followed by the SUN domain, into pShooter/ER. As shown in Fig. 3A, SUN2DOM was unable to displace endogenous Nesprin 2 giant and Nesprin 3 (Fig. 3A). Displacements scorings (Fig. 3B) imply that, similarly to what we observed for EGFP-KASH2ext, Sun3L and SUN2DOM are unable to interact with KASH domains. The apparent instability of Sun3L upon cell lysis prevented us from testing its interaction with KASH domains in co-immunoprecipitation experiments. However, in agreement with its failure to displace endogenous Nesprins, SUN2DOM was unable to co-immunoprecipitate with EGFP-KASH2 (Fig. 3C). The inability of Sun3L or SUN2DOM to displace endogenous Nesprins was not due to the failure of these constructs to translocate within the ER lumen since they are glycosylated (see below and Fig. 4A).
Fig. 3.
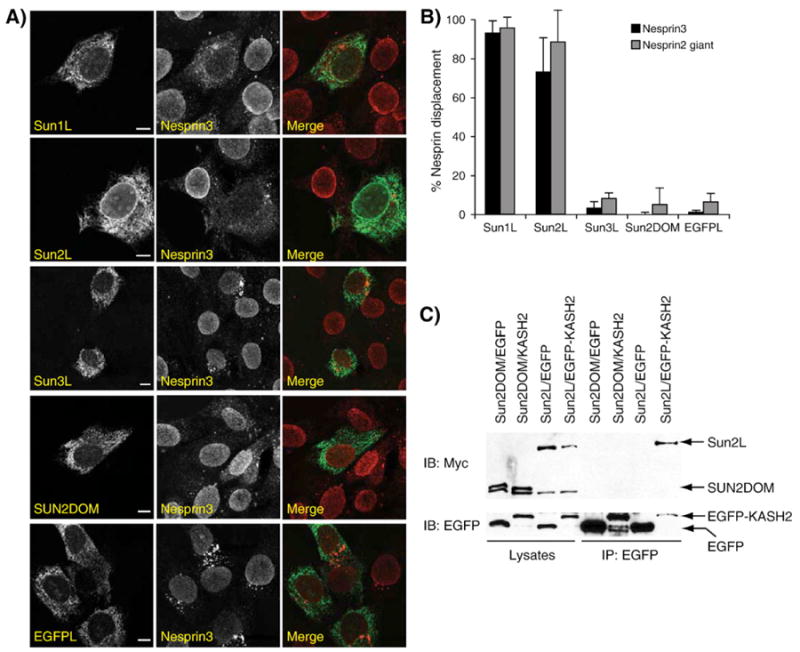
ER-targeted expression of the luminal domain of Sun1 or Sun2 disrupts endogenous LINC complexes. A) Immunolocalization of endogenous Nesprin 3 in C2C12 cells transfected with Sun1L, Sun2L, Sun3L and SUN2DOM. ER-targeted expression of EGFP was used as a negative control. A specific antiserum against Nesprin 3 was used in conjunction with a monoclonal anti-Myc antibody to detect ER-targeted recombinant constructs expressed from the pShooter/ER vector. Primary antibodies were detected with goat-anti-rabbit (Alexa 594) and goat-anti mouse (Alexa 488) antibodies. Scale bars: 50 μm. Nesprin 2 giant was displaced to the same extent than Nesprin 3. B) Percentage of cells transfected with the indicated constructs showing a displacement of endogenous Nesprin 2 giant and Nesprin 3 from the NE to the ER. Only the complete disappearance, as opposed to just a weakened signal, of Nesprin rim structures at the NE was scored as a displacement from the NE to the ER by three independent experimenters (n>200 cells for each experiment). C) SUN2DOM does not interact with EGFP-KASH2. HeLa cells were transfected with SUN2DOM and either EGFP or EGFP-KASH2. The same experiment carried out with Sun2L was used as a positive control. Cell lysates were immunoprecipitated with a polyclonal antiserum against EGFP and analyzed with either anti-Myc (upper panel) or anti-EGFP (lower panel) antibodies. For each experiment, the amounts of initial recombinant proteins in whole lysates (left) and immunoprecipitation products (right) are shown.
Fig. 4.
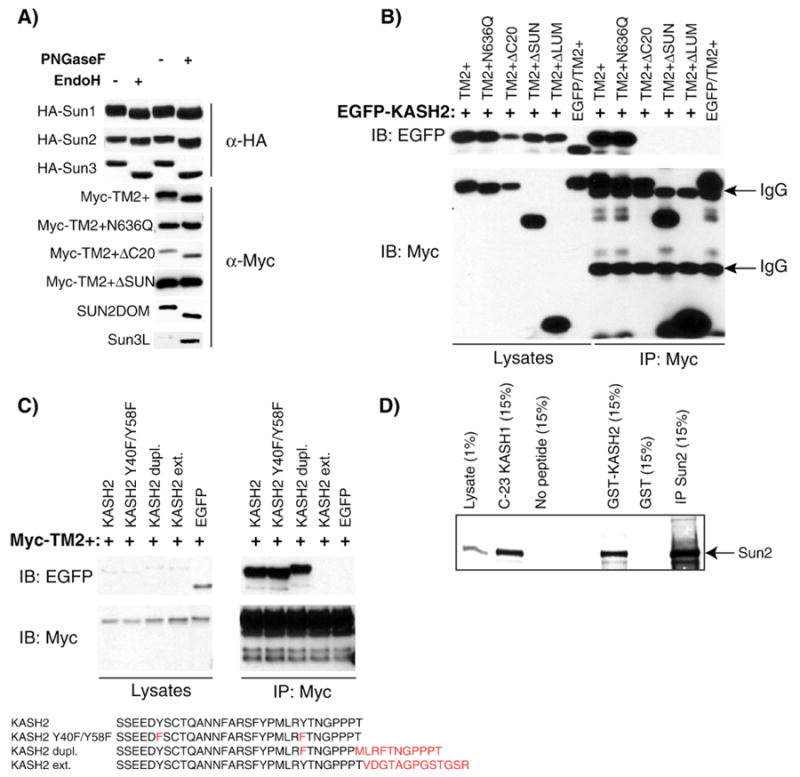
Key determinant mediating the SUN/KASH interaction. A) Sun proteins are glycosylated. HeLa cell lysates transfected with the indicated constructs were processed for endoglycanase digestion with (+) or without (−) PNGaseF or EndoH. The glycosylation status was determined by examining mobility shifts of recombinant constructs in immunoblot with either anti-HA or anti-Myc antibodies. B) HeLa cells were co-transfected with EGFP-KASH2 and the indicated recombinant construct corresponding to different mutations or deletions of the luminal domain of Sun2. A control experiment consisted in the cotransfection of EGFP with Myc-TM2+. Cell lysates were immunoprecipitated with an anti-Myc antibody and analyzed with either anti-EGFP (upper panel) or an anti-Myc antibodies (lower panel). For each experiment, the amounts of initial recombinant proteins in whole lysates (left panels) and immunoprecipitates (right panels) are shown. C) HeLa cells were co-transfected with Myc-TM2+ and either EGFP fused to different sequence modifications of the luminal domain of Nesprin 2 or EGFP alone used as a negative control. Cell lysates were immunoprecipitated with an anti-Myc antibody and analyzed with either anti-EGFP (upper panels) or an anti-Myc antibody (lower panels). For each experiment, the amounts of initial recombinant proteins in whole lysates (left panels) and immunoprecipitation products (right panels) are shown. D) KASH domains do not require post-translational modifications to interact with endogenous Sun2. HeLa cell lysates were incubated with either with a GST fusion protein of the luminal domain of KASH2 immobilized on glutathione beads or with a synthetic peptide corresponding to the 23 C-terminal residues of KASH1 covalently coupled to an Affigel resin. Glutathione or Affigel beads alone were used as negative controls. Endogenous Sun2 was detected with a specific rabbit antiserum. Immunoprecipitated endogenous Sun2 is also shown for comparison. The percentage of either the original cell lysate used in each pull-down experiment or of the respective elutions loaded on the gel is indicated.
Overall and in agreement with the co-immunoprecipitation results, this set of experiments indicate that the whole luminal domain of Sun1 and Sun2 competes with endogenous LINC complexes while, remarkably, the whole luminal domain of Sun3 or SUN2DOM, a truncation of Sun2L, show no such effect. These observations stress the importance of sequences located upstream from the SUN domain, most likely the coiled-coil domains, on either the conformation of the SUN domain or the functionality of Sun proteins as a whole.
Determinants of SUN/KASH interactions
In an effort to identify essential determinants that mediate the interaction between Sun proteins and Nesprins, we tested the effect of post-translational modifications as well as various truncations of the luminal domain of Sun proteins and KASH domains on the SUN/KASH interaction. We previously showed that the C-terminal region of Sun2, which contains the coiled-coil domain and the SUN domain (Fig. 1A), localizes within the perinuclear space [10]. Because the latter is continuous with the ER lumen, we examined the glycosylation status of Sun1, Sun2 and Sun3.
NetNGlyc was used to scan for the presence of putative glycosylation sites within the coding sequence of human Sun1, Sun2 and Sun3. For Sun2 (NP_056189, 717 amino acids), a single asparagine, N636, was predicted to be glycosylated while two potential glycosylation sites were predicted in the luminal domain of Sun1 (O94901, N588 and N732) and Sun3 (NP_001025190, N276 and N306). Interestingly, residues N732 of Sun1, N636 of Sun2 and N276 of Sun3 are all located 80 or 81 amino acids upstream from the C-terminus, i.e., right in the middle of the SUN domain. A predicted glycosylation site (N1031) is similarly positioned within the SUN domain of UNC-84. To examine the glycosylation status of recombinant human Sun proteins, PNGaseF and EndoH digestions were performed on whole cell lysates of HeLa cells transfected with recombinant HA-tagged constructs of Sun1, 2 and 3 (Fig. 1A). These constructs displayed a lower apparent molecular weight upon enzymatic treatment (Fig. 4A), which indicates that Sun1, 2 and 3 are indeed glycosylated. In agreement with the prediction of a single luminal glycosylation site within the luminal domain of Sun2 at N636, endoglycanase treatment of Myc-TM2+ΔSUN, which lacks the SUN domain (Fig. 1A), did not alter the apparent molecular weight of the recombinant protein (Fig. 4A). Furthermore, the introduction of a N636Q mutation within Myc-TM2+ confirmed that N636 is the only N-glycosylation site within Sun2, since the gel mobility of Myc-TM2+N636Q was unaffected by endoglycanase treatment (Fig. 4A). Overall, these results demonstrate that human Sun proteins 1, 2 and 3 are glycosylated and that SUN domains are exclusively located within the ER lumen/perinuclear space.
To examine if the glycosylation of the SUN domain of Sun2 plays any role in the SUN/KASH interaction, we tested whether Myc-TM2+N636Q was able to interact with EGFP/KASH2. Myc-TM2+N636Q interacted as efficiently as Myc-TM2+ with EGFP/KASH2 (Fig. 4B). These findings therefore preclude any role for glycosylation in the SUN/KASH interaction.
Using the same assay, we tested the effect of deleting most of the luminal domain (Myc-TM2+ΔLUM, Fig. 1A), the SUN domain (Myc-TM2+ΔSUN, Fig. 1A), or the last 20 C-terminal amino acids of the SUN domain (Myc-TM2+ΔC20, Fig. 1A). The ΔC20 deletion experiment was motivated by the observation, using ENSEMBL [31], that this stretch contains three amino acids whose identities are strictly conserved among SUN domains from lower eukaryotes to mammals. The absence of any interaction between Myc-TM2+ΔSUN and Myc-TM2+ΔLUM with EGFP-KASH2 indicated that the SUN domain is required for the SUN/KASH interaction. More specifically, there is a strict requirement for the last 20 C-terminal amino acids of the SUN domain for the SUN/KASH interaction to take place. Examination of the glycosylation status of Myc-TM2+ΔC20 (Fig. 4A) indicated that it is translocated within the ER lumen as efficiently as Myc-TM2+, a prerequisite for its interaction with KASH constructs in the perinuclear space.
To probe the determinants within the KASH domain necessary for the SUN/KASH interaction, three modified constructs were engineered. The luminal domain of KASH2 was modified by the mutation of two conserved tyrosine residues (KASH2 Y40F/Y58F). In Nesprins 1 and 2, both residues were embedded within an amino acid context that strongly predicts their phosphorylation. In Nesprin 3, however, Y40 is replaced by an arginine residue and residue Y58 is not in an amino acid context that predicts its phosphorylation by NetPhos [32]. Additional KASH domain modifications (Fig. 4C) included either the extension of the C-terminal end of the luminal domain with 13 amino acids derived from the expression vector by omission of the endogenous stop codon (KASH2ext) or the duplication of the last 11 C-terminal amino acids of KASH2 (KASH2dupl). We found that mutation of luminal tyrosine residues 40 and 58 has no adverse effect on the binding of Myc-TM2+. This observation indicates that the interaction between KASH domains and Sun proteins is independent of the two potentially phosphorylated tyrosine residues. As expected from its failure to interact with endogenous Sun2 (Fig. 1D), KASH2ext did not interact with Myc-TM2+. However, duplication of the last 11 C-terminal amino acids of the KASH2 domain preserved the interaction with Myc-TM2+ (Fig. 4C). These results therefore demonstrate that a free KASH2 C-terminal end is required for the SUN/KASH interaction to take place.
Finally, to test whether KASH domains require any post-translational modification in order to mediate the SUN/KASH interaction, a GST fusion protein containing the luminal domain of Nesprin 2 was incubated with HeLa cell lysates. This construct effectively pulled down endogenous Sun2 (Fig. 4D). Endogenous Sun2 was also pulled down by an immobilized synthetic peptide corresponding to the C-terminal 23 amino acids of human KASH1 (Fig. 4D). The latter observation indicates that post-translational modifications of KASH domains are not strictly required for the SUN/KASH interaction to occur and narrows down the interacting site to within the last 23 C-terminal amino acids of KASH domains.
Overall, these results indicate that the last 20 C-terminal amino acids of the SUN domain and a free C-terminal end of the KASH domain are required for the formation of LINC complexes. Furthermore, neither post-translational modifications of KASH domains nor the glycosylation of SUN domains are required for SUN/KASH interactions to occur.
The integrity of LINC complexes is essential to cytoskeletal mechanical properties
Various biophysical approaches were recently used to demonstrate that mechanical properties of embryonic fibroblasts derived from mice lacking A-type lamins (Lmna−/− MEFs), a model for muscular dystrophies, are greatly affected [33,34]. We recently used Ballistic Intracellular Nanorheology (BIN), a high-throughput approach allowing the rigorous quantification of mechanical properties of living cells [29], to confirm the loss of mechanical stiffness of Lmna−/− MEFs [27]. Lmna−/− MEFs also display abnormal localization of several LINC complexes components [9,35]. Hence, we wanted to address whether disruption of LINC complexes directly alters cellular mechanical properties. To that effect, LINC complexes of Swiss 3T3 fibroblasts were disrupted by transfection with EGFP-KASH 1, 2 and 3. EGFP-KASH2ext, which does not alter LINC complexes integrity (Fig. 2), was used as a negative control. The mechanical stiffness of these cells was then probed using BIN: fluorescent nanobeads were ballistically introduced in cultured cells (Fig. 5A) and their displacement within the cytoskeleton (Fig. 5B) was analyzed with the appropriate software (see Materials and methods section). The mean square displacement (MSD) of fluorescent nanobeads was significantly increased in cells transfected with EGFP-KASH1, 2 and 3 in comparison to EGFP-KASH2ext (Fig. 5C), indicating a significant softening of their cytoplasm. Elastic moduli (G′, expressed in dyn/cm2), which quantify the local resistance of the cytoplasm against small random forces acting on the surface of the nanoparticles, can be derived from MSD curves to quantify cellular mechanical properties. The elastic modulus of the cytoplasm of cells transfected with EGFP-KASH constructs were significantly lower (p < 0.0001) than that of cells transfected with EGFP-KASH2ext (Fig. 5D). The latter value compared well with the elastic modulus of untransfected Swiss 3T3 cells. Interestingly, the extent of cell softening induced by the disruption of LINC complexes with EGFP-KASH's constructs was comparable to cytoskeletal softening measured in Lrnna−/− MEF vs. Lmna+/+ MEF (Fig. 5D, inset). Along with the current model of LINC complexes that physically connect the nuclear lamina to cytoskeletal networks, these results support a major role for nucleo-cytoskeletal connections in cellular mechanical stiffness. Furthermore, they suggest a possible involvement of LINC complexes disruption in pathological pathways associated to A-type lamin mutations and involving defects in cellular mechanical stiffness.
Discussion
Major progress has been made in understanding the overall mechanisms governing nuclear dynamics in metazoans [36,37]. However, clues about the actual tethering mechanisms of mammalian nuclei to their surrounding cytoskeleton during nuclear migration and anchoring have only started to emerge. A role for SUN and KASH domain-containing proteins in nuclear dynamics was originally demonstrated in C. elegans and Drosophila melanogaster [5,6]. In mammalian cells, specific orthologs form macromolecular assemblies, called LINC complexes, spanning the nuclear envelope to connect the nuclear lamina to the peripheral cytoskeleton [8,9,11,38].
Sun proteins interact promiscuously with Nesprins
Co-immunoprecipitation experiments indicate that the luminal domain of both Sun1 and Sun2 interacts with the KASH domains of Nesprins 1, 2 or 3. Accordingly, these recombinant proteins invariably displace endogenous Nesprin 2 giant and Nesprin 3 from the ONM. Even though an antiserum against Nesprin 1 giant was not available, the efficient co-immunoprecipitation of EGFP-KASH1 with the luminal region of Sun1 and Sun2 strongly suggests that endogenous Nesprin 1 giant is displaced as well. These results provide strong supporting evidence that interactions between Sun proteins and Nesprins at the NE are promiscuous. These observations also provide direct evidence that Sun1 and Sun2 are redundant in their anchoring functions of Nesprins at the NE, a conclusion that was anticipated from our previous work [9]. Hence, we expect that LINC complexes of various composition form at the NE. It is important to note, however, that while Sun proteins and Nesprins are ubiquitously expressed, their relative expression levels greatly differ among tissues [9,20,24]. Therefore, the compositional nature of LINC complexes may significantly vary among tissues. In C. elegans, KASH domain-containing proteins Zyg-12, UNC83 and ANC-1 exert distinct functions in nuclear dynamics [5]. Distinct functions of KASH domain-containing proteins in mammals is further suggested by the observation that despite the structural similarity between murine Nesprin 1 (Syne-1) and Nesprin 2 (syne-2), the former is essential for the anchorage of synaptic and extrasynaptic nuclei in skeletal muscle while the latter is not [25]. We therefore hypothesize that, even though the SUN/KASH interaction is promiscuous, the variation (or the modulation) in composition of LINC complexes among different tissues (or within the same tissue at different developmental stage) could provide different functionalities to these macromolecular assemblies.
Structural and post-translational requirements of the SUN/KASH interaction
In addition to confirming the promiscuity of the interaction between Sun proteins and Nesprins family members, displacement studies of endogenous Nesprin 2 giant and Nesprin 3 in cultured cells were also instrumental in probing determinants of the interaction between these proteins. While Sun1L and Sun2L efficiently disrupted the formation of endogenous LINC complexes, Sun3L and SUN2DOM did not. The lack of effect of SUN2DOM was correlated to its failure to interact with EGFP-KASH fusion proteins. These results suggest that while the SUN domain is strictly required for the formation of LINC complexes, additional regions located upstream from the SUN domain of Sun1 and Sun2 could play a regulatory role in the SUN domain by regulating its ability to interact with KASH domains. We anticipate that the coiled-coil regions of the luminal region fulfill such a role. Indeed, we observed that Myc/TM2+ forms stable dimers and tetramers while we were unable to detect any oligomerization of SUN2DOM (data not shown) and the lack of oligomerization of a similar Sun2 recombinant construct lacking the first coiled-coil domain was also reported [39]. Collectively, these observations suggest a role of Sun proteins oligomerization in their interaction with Nesprins. Alternatively, another undefined region conserved within the luminal region of Sun1 and Sun2 but absent from the luminal region of Sun3 could also play a role.
In the same assay, EGFP-KASH1, 2 and 3 efficiently displaced endogenous Nesprins from the NE to the ER (Fig. 6), while KASH2ext did not and further failed to interact with recombinant and endogenous Sun2. These findings indicate that the SUN/KASH interaction requires a free C-terminal end. It has been demonstrated that the four C-terminal amino acids (PPPX) are required for the SUN/KASH interaction to occur [11]. Taken together, these data suggest that the C-terminal region of KASH most likely fits a pocket provided by the luminal domain of Sun proteins. The PPPX motif of Nesprins would interact with that pocket but the addition of unrelated C-terminal amino acids could sterically hinder the interacting region from reaching that pocket. This concept is further reinforced by the observation that a duplication of the C-terminal region of KASH2 still interacts with the luminal domain of Sun protein. We also showed that a synthetic peptide corresponding to the 23 C-terminal amino acid residues of KASH1 is sufficient to fit that putative pocket because endogenous Sun2 from HeLa cell lysates were efficiently pulled down using the C-23 peptide. In addition to narrowing down the interacting domain of KASH domains to that region, this result also indicates that post-translational modifications of the KASH domain are not required for mediation of the SUN/KASH interactions.
Fig. 6.
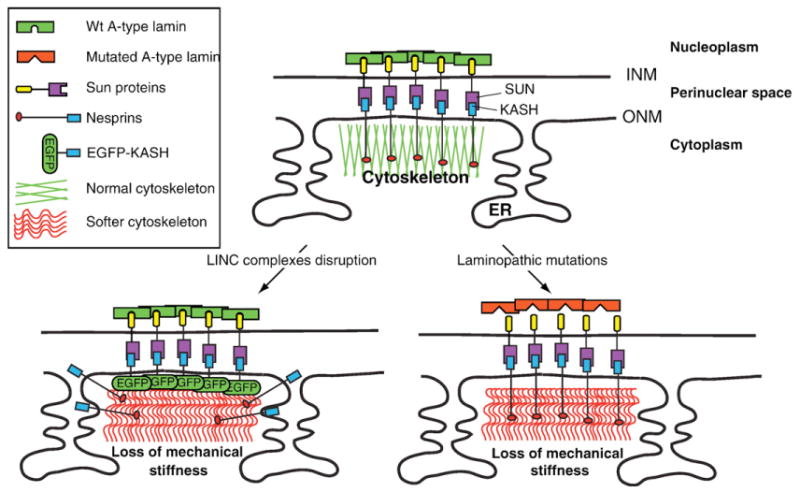
Putative involvement of disruption of LINC complexes in a “mechanical etiology” of some laminopathies. Top panel: Depiction of LINC complexes establishing a physical connection between the nuclear lamina and the cytoskeleton. Lower left panel: Disruption of LINC complexes, i.e. the displacement of endogenous Nesprins from the NE to the ER by overexpression of EGFP-KASH, results in a softening of the cytoskeleton. Lower right panel: Putative role of LINC complexes in human laminopathies. A-type lamin mutations that weaken/prevent the interaction of A-type lamins with Sun proteins would disrupt the structural integrity of LINC complexes and, as a result, induce a loss of mechanical stiffness of the cytoskeleton.
The use of recombinant proteins also allowed us to identify additional structural and post-translational modification of LINC complex proteins. We observed that the 20 C-terminal amino acids of the luminal domain of Sun2 are strictly required for interaction with EGFP-KASH constructs. This observation suggests that this stretch of amino acids is required for either binding to KASH domains or for a correct conformation of the SUN domain. The importance of this region is further reinforced by the strictly conserved identity of three amino acid residues within that region among all metazoan SUN domains. The strict requirement of this C-terminal stretch of the SUN domain (like the requirement for the SUN domain itself) contradicts observations of a two-hybrid interaction between a region of the luminal domain of Sun1 located upstream from the Sun domain and downstream from the coiled-coil region and the KASH domain. This observation, however, was not directly confirmed in pull-down assays [11]. Using a similar indirect method, McGee et al. also observed an interaction of Unc-83 with sequences located upstream from the SUN domain of Unc-84. However, from a functional point of view, they confirmed that the SUN domain of UNC-84 is necessary for the recruitment of ANC-1 and UNC-83 at the NE as well as in functional assays based on nuclear positioning in C. elegans [40]. Taken together, we conclude that it is the evolutionarily conserved SUN domain that mediates the interaction between Sun proteins and Nesprins and that the conservation of the SUN domain reflects the constraint to provide an interacting pocket for the KASH domain.
In this study, we also showed that Sun proteins are glycosylated. In Sun2, a unique glycosylation site is located in the middle of the SUN domain. Mutation of that glycosylation site did not affect the efficiency of the SUN/KASH interaction. However, as we have shown, glycosylation of Sun proteins provided a simple and efficient way to confirm the luminal localization of Sun proteins or their recombinant constructs.
LINC complexes, mechanical stiffness and laminopathies
Laminopathies include a wide array of human diseases characterized by mutations along the gene encoding A-type lamins [41]. The molecular etiology of these diseases is unknown. Mice lacking A-type lamins display phenotypes that are reminiscent of human muscular dystrophies and cardiomyopathies [42]. Embryonic fibroblasts derived from these mice (Lmna−/− MEFs) are characterized by a lack of mechanical stiffness measured using different biophysical approaches [27,33,34]. Furthermore, Lmna−/− MEFs also display a mislocalization of Nesprins [35]. We therefore wanted to address whether the mislocalization of Nesprins is directly correlated to a loss of cellular mechanical stiffness. Using BIN, we measured a significant loss of mechanical stiffness that is specifically associated to the disruption of endogenous LINC complexes by recombinant KASH constructs of Nesprins 1, 2 and 3. KASH2ext, which was used as a negative control since it neither interacts with Sun proteins nor disrupts endogenous LINC complexes, did not significantly alter cellular mechanical stiffness. Interestingly, we observed that the normalized decrease of elastic moduli of Swiss3T3 transfected with EGFP-KASH2 or of Lmna−/− MEFs was comparable. These data therefore strongly suggest the involvement of LINC complexes in cellular mechanical stiffness. Coupled with previous reports indicating the direct interaction of the nucleoplasmic region of Sun proteins with A-type lamins [8,9], a pathological mechanism involved in specific laminopathies can be postulated. In that model (Fig. 6), A-type lamins mutations that specifically weaken/prevent the interaction with Sun1 and/or Sun2 would also weaken physical connections between the nuclear lamina and the cytoskeleton, resulting in a loss of mechanical stiffness. Importantly, a loss and/or weakening of interaction of mutated A-type lamins with Sun proteins would not signify that Sun proteins would necessarily mislocalize from the NE since their retention in that location does not entirely rely on lamins [15]. A-type lamin mutations that disrupt the interaction with Sun proteins could, however, be responsible for phenotypic manifestations related to a mechanical etiology of human laminopathies that involve a structural impairment of LINC complexes (Fig. 6). Future studies dedicated to the identification of A-type lamin domains that interact with the nucleoplasmic region of Sun proteins as well as the development of appropriate animal models of LINC complexes disruption will be required to confirm these hypotheses.
Acknowledgments
The authors are grateful to Dr. Brian Burke (University of Florida, Gainesville) for the kind gift of HA-Sun1 and HA-Sun3 cDNAs, to Dr. C. Shanahan (University of Cambridge, UK) for the kind gift of KASH2ext, to Dr. A. Sonnenberg (The Netherlands Cancer Institute, Amsterdam, The Netherlands) for the anti-Nesprin 3 serum, to Drs. E. Gomes and G. Gundersen (Columbia University, New York) for the kind gift of the anti-Nesprin 2 giant serum, and to Drs. P. Stahl, P. Hanson and A. Charron (Washington University School of Medicine, St Louis) for helpful discussions and comment on the manuscript. This work was supported by grants from the NIH (R21#EB006890 to DW) and from the Muscular Dystrophy Association (to DH).
References
- 1.Burke B, Stewart CL. Life at the edge: the nuclear envelope and human disease. Nat Rev Mol Cell Biol. 2002;3:575–585. doi: 10.1038/nrm879. [DOI] [PubMed] [Google Scholar]
- 2.Worman HJ, Courvalin JC. The inner nuclear membrane. J Membr Biol. 2000;177:1–11. doi: 10.1007/s002320001096. [DOI] [PubMed] [Google Scholar]
- 3.Stewart CL, Roux KJ, Burke B. Blurring the boundary: the nuclear envelope extends its reach. Science. 2007;318:1408–1412. doi: 10.1126/science.1142034. [DOI] [PubMed] [Google Scholar]
- 4.Starr DA, Han M. ANChors away: an actin based mechanism of nuclear positioning. J Cell Sci. 2003;116:211–216. doi: 10.1242/jcs.00248. [DOI] [PubMed] [Google Scholar]
- 5.Starr DA, Fischer JA. KASH 'n Karry: the KASH domain family of cargo-specific cytoskeletal adaptor proteins. Bioessays. 2005;27:1136–1146. doi: 10.1002/bies.20312. [DOI] [PubMed] [Google Scholar]
- 6.Wilhelmsen K, Ketema M, Truong H, Sonnenberg A. KASH-domain proteins in nuclear migration, anchorage and other processes. J Cell Sci. 2006;119:5021–5029. doi: 10.1242/jcs.03295. [DOI] [PubMed] [Google Scholar]
- 7.Tzur YB, Wilson KL, Gruenbaum Y. SUN-domain proteins: ‘Velcro’ that links the nucleoskeleton to the cytoskeleton. Nat Rev Mol Cell Biol. 2006;7:782–788. doi: 10.1038/nrm2003. [DOI] [PubMed] [Google Scholar]
- 8.Haque F, Lloyd DJ, Smallwood DT, Dent CL, Shanahan CM, Fry AM, Trembath RC, Shackleton S. SUN1 interacts with nuclear lamin A and cytoplasmic Nesprins to provide a physical connection between the nuclear lamina and the cytoskeleton. Mol Cell Biol. 2006;26:3738–3751. doi: 10.1128/MCB.26.10.3738-3751.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, Stahl PD, Hodzic D. Coupling of the nucleus and cytoplasm: role of the LINC complex. J Cell Biol. 2006;172:41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hodzic DM, Yeater DB, Bengtsson L, Otto H, Stahl PD. Sun2 is a novel mammalian inner nuclear membrane protein. J Biol Chem. 2004;279:25805–25812. doi: 10.1074/jbc.M313157200. [DOI] [PubMed] [Google Scholar]
- 11.Padmakumar VC, Libotte T, Lu W, Zaim H, Abraham S, Noegel AA, Gotzmann J, Foisner R, Karakesisoglou I. The inner nuclear membrane protein Sun1 mediates the anchorage of Nesprin-2 to the nuclear envelope. J Cell Sci. 2005;118:3419–3430. doi: 10.1242/jcs.02471. [DOI] [PubMed] [Google Scholar]
- 12.Malone CJ, Fixsen WD, Horvitz HR, Han M. UNC-84 localizes to the nuclear envelope and is required for nuclear migration and anchoring during C. elegans development. Development. 1999;126:3171–3181. doi: 10.1242/dev.126.14.3171. [DOI] [PubMed] [Google Scholar]
- 13.Hagan I, Yanagida M. The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. J Cell Biol. 1995;129:1033–1047. doi: 10.1083/jcb.129.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee KK, Starr D, Cohen M, Liu J, Han M, Wilson KL, Gruenbaum Y. Lamin-dependent localization of UNC-84, a protein required for nuclear migration in Caenorhabditis elegans. Mol Biol Cell. 2002;13:892–901. doi: 10.1091/mbc.01-06-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hasan S, Guttinger S, Muhlhausser P, Anderegg F, Burgler S, Kutay U. Nuclear envelope localization of human UNC84A does not require nuclear lamins. FEBS Lett. 2006;580:1263–1268. doi: 10.1016/j.febslet.2006.01.039. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Q, Ragnauth G, Greener MJ, Shanahan CM, Roberts RG. The nesprins are giant actin-binding proteins, orthologous to Drosophila melanogaster muscle protein MSP-300. Genomics. 2002;80:473–481. [PubMed] [Google Scholar]
- 17.Zhang Q, Skepper JN, Yang F, Davies JD, Hegyi L, Roberts RG, Weissberg PL, Ellis JA, Shanahan CM. Nesprins: a novel family of spectrin-repeat-containing proteins that localize to the nuclear membrane in multiple tissues. J Cell Sci. 2001;114:4485–4498. doi: 10.1242/jcs.114.24.4485. [DOI] [PubMed] [Google Scholar]
- 18.Apel ED, Lewis RM, Grady RM, Sanes JR. Syne-1, a dystrophin- and Klarsicht-related protein associated with synaptic nuclei at the neuromuscular junction. J Biol Chem. 2000;275:31986–31995. doi: 10.1074/jbc.M004775200. [DOI] [PubMed] [Google Scholar]
- 19.Mislow JM, Kim MS, Davis DB, McNally EM. Myne-1, a spectrin repeat transmembrane protein of the myocyte inner nuclear membrane, interacts with lamin A/C. J Cell Sci. 2002;115:61–70. doi: 10.1242/jcs.115.1.61. [DOI] [PubMed] [Google Scholar]
- 20.Padmakumar VC, Abraham S, Braune S, Noegel AA, Tunggal B, Karakesisoglou I, Korenbaum E. Enaptin, a giant actin-binding protein, is an element of the nuclear membrane and the actin cytoskeleton. Exp Cell Res. 2004;295:330–339. doi: 10.1016/j.yexcr.2004.01.014. [DOI] [PubMed] [Google Scholar]
- 21.Zhen YY, Libotte T, Munck M, Noegel AA, Korenbaum E. NUANCE, a giant protein connecting the nucleus and actin cytoskeleton. J Cell Sci. 2002;115:3207–3222. doi: 10.1242/jcs.115.15.3207. [DOI] [PubMed] [Google Scholar]
- 22.Warren DT, Zhang Q, Weissberg PL, Shanahan CM. Nesprins: intracellular scaffolds that maintain cell architecture and coordinate cell function? Expert Rev Mol Med. 2005;7:1–15. doi: 10.1017/S1462399405009294. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Q, Ragnauth CD, Skepper JN, Worth NF, Warren DT, Roberts RG, Weissberg PL, Ellis JA, Shanahan CM. Nesprin-2 is a multi-isomeric protein that binds lamin and emerin at the nuclear envelope and forms a subcellular network in skeletal muscle. J Cell Sci. 2005;118:673–687. doi: 10.1242/jcs.01642. [DOI] [PubMed] [Google Scholar]
- 24.Wilhelmsen K, Litjens SH, Kuikman I, Tshimbalanga N, Janssen H, van den Bout I, Raymond K, Sonnenberg A. Nesprin-3, a novel outer nuclear membrane protein, associates with the cytoskeletal linker protein plectin. J Cell Biol. 2005;171:799–810. doi: 10.1083/jcb.200506083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang X, Xu R, Zhu B, Yang X, Ding X, Duan S, Xu T, Zhuang Y, Han M. Syne-1 and Syne-2 play crucial roles in myonuclear anchorage and motor neuron innervation. Development. 2007;134:901–908. doi: 10.1242/dev.02783. [DOI] [PubMed] [Google Scholar]
- 26.Grady RM, Starr DA, Ackerman GL, Sanes JR, Han M. Syne proteins anchor muscle nuclei at the neuromuscular junction. Proc Natl Acad Sci U S A. 2005;102:4359–4364. doi: 10.1073/pnas.0500711102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee JS, Hale CM, Panorchan P, Khatau SB, George JP, Tseng Y, Stewart CL, Hodzic D, Wirtz D. Nuclear lamin a/c deficiency induces defects in cell mechanics, polarization, and migration. Biophys J. 2007;93:2542–2552. doi: 10.1529/biophysj.106.102426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Panorchan P, Lee JS, Daniels BR, Kole TP, Tseng Y, Wirtz D. Probing cellular mechanical responses to stimuli using ballistic intracellular nanorheology. Methods Cell Biol. 2007;83:115–140. doi: 10.1016/S0091-679X(07)83006-8. [DOI] [PubMed] [Google Scholar]
- 29.Lee JS, Panorchan P, Hale CM, Khatau SB, Kole TP, Tseng Y, Wirtz D. Ballistic intracellular nanorheology reveals ROCK-hard cytoplasmic stiffening response to fluid flow. J Cell Sci. 2006;119:1760–1768. doi: 10.1242/jcs.02899. [DOI] [PubMed] [Google Scholar]
- 30.Tseng Y, Kole TP, Wirtz D. Micromechanical mapping of live cells by multiple-particle-tracking microrheology. Biophys J. 2002;83:3162–3176. doi: 10.1016/S0006-3495(02)75319-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hubbard TJ, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, Down T, Dyer SC, Fitzgerald S, Fernandez-Banet J, Graf S, Haider S, Hammond M, Herrero J, Holland R, Howe K, Howe K, Johnson N, Kahari A, Keefe D, Kokocinski F, Kulesha E, Lawson D, Longden I, Melsopp C, Megy K, Meidl P, Ouverdin B, Parker A, Prlic A, Rice S, Rios D, Schuster M, Sealy I, Severin J, Slater G, Smedley D, Spudich G, Trevanion S, Vilella A, Vogel J, White S, Wood M, Cox T, Curwen V, Durbin R, Fernandez-Suarez XM, Flicek P, Kasprzyk A, Proctor G, Searle S, Smith J, Ureta-Vidal A, Birney E. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–d617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blom N, Gammeltoft S, Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol. 1999;294:1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- 33.Lammerding J, Schulze PC, Takahashi T, Kozlov S, Sullivan T, Kamm RD, Stewart CL, Lee RT. Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J Clin Invest. 2004;113:370–378. doi: 10.1172/JCI19670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Broers JL, Peeters EA, Kuijpers HJ, Endert J, Bouten CV, Oomens CW, Baaijens FP, Ramaekers FC. Decreased mechanical stiffness in LMNA−/− cells is caused by defective nucleo-cytoskeletal integrity: implications for the development of laminopathies. Hum Mol Genet. 2004;13:2567–2580. doi: 10.1093/hmg/ddh295. [DOI] [PubMed] [Google Scholar]
- 35.Libotte T, Zaim H, Abraham S, Padmakumar VC, Schneider M, Lu W, Munck M, Hutchison C, Wehnert M, Fahrenkrog B, Sauder U, Aebi U, Noegel AA, Karakesisoglou I. Lamin A/C-dependent localization of Nesprin-2, a giant scaffolder at the nuclear envelope. Mol Biol Cell. 2005;16:3411–3424. doi: 10.1091/mbc.E04-11-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reinsch S, Gonczy P. Mechanisms of nuclear positioning. J Cell Sci. 1998;111(Pt 16):2283–2295. doi: 10.1242/jcs.111.16.2283. [DOI] [PubMed] [Google Scholar]
- 37.Tsai LH, Gleeson JG. Nucleokinesis in neuronal migration. Neuron. 2005;46:383–388. doi: 10.1016/j.neuron.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 38.Ketema M, Wilhelmsen K, Kuikman I, Janssen H, Hodzic D, Sonnenberg A. Requirements for the localization of nesprin-3 at the nuclear envelope and its interaction with plectin. J Cell Sci. 2007;120:3384–3394. doi: 10.1242/jcs.014191. [DOI] [PubMed] [Google Scholar]
- 39.Wang Q, Du X, Cai Z, Greene MI. Characterization of the structures involved in localization of the SUN proteins to the nuclear envelope and the centrosome. DNA Cell Biol. 2006;25:554–562. doi: 10.1089/dna.2006.25.554. [DOI] [PubMed] [Google Scholar]
- 40.McGee MD, Rillo R, Anderson AS, Starr DA. UNC-83 IS a KASH protein required for nuclear migration and is recruited to the outer nuclear membrane by a physical interaction with the SUN protein UNC-84. Mol Biol Cell. 2006;17:1790–1801. doi: 10.1091/mbc.E05-09-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Worman HJ, Bonne G. “Laminopathies”: a wide spectrum of human diseases. Exp Cell Res. 2007;313:2121–2133. doi: 10.1016/j.yexcr.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sullivan T, Escalante-Alcalde D, Bhatt H, Anver M, Bhat N, Nagashima K, Stewart CL, Burke B. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J Cell Biol. 1999;147:913–920. doi: 10.1083/jcb.147.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]


