Abstract
The contribution of genes to the etiology of heroin dependence is greater than for any other illicit drug. The specific genes mediating this effect remain unknown, despite several candidate gene association studies of the condition. Here we report the results of a genome-wide search for heroin dependence susceptibility loci using multipoint linkage analysis. In phase I, we ascertained 207 independent affected sibling pairs from 202 Han Chinese families from Yunnan Province, China (near Asia's “Golden Triangle”). After data-cleaning, 194 fully independent sibling pairs (i.e., with no overlapping individuals) from 192 families were genotyped on 404 short tandem-repeat markers spaced at an average inter-marker distance of 9cM. Although none of our findings achieved genome-wide significance, we found two regions with nonparametric linkage (NPL) Z-scores greater than 2.0. An NPL Z-score of 2.19 (uncorrected p-value = 0.014) was observed at D4S1644, located at 143.3 cM on chromosomal region 4q31.21. The highest NPL Z-score of 2.36 (uncorrected p-value = 0.009) was observed at 53.4 cM on chromosomal region 17q11.2 at marker D17S1880. This is among the first published reports of a genome-wide linkage analysis of heroin dependence. Forthcoming results from other groups and from two additional waves of ascertainment (one planned, one currently ongoing) for our own study should be able to support or refute the putative susceptibility loci we have identified, after which positional candidate genes can be further evaluated as risk factors for the illness.
Keywords: affected sibling-pairs, chromosome 17, genome-wide linkage analysis, heroin dependence, opioid dependence
Introduction
Heroin dependence is a particularly disabling psychiatric disorder, characterized by severe psychological and physical dependence and withdrawal, which often prevents cessation and sustains usage. The severity of the recent global heroin epidemic is reflected in some statistics from the United States, where heroin dependence is at its highest levels since the late 1970s (Substance Abuse and Mental Health Services Administration - Office of Applied Studies 2003b). In 2002, approximately 166,000 individuals in the U.S. were current heroin users, and the lifetime prevalence of heroin use has been increasing since the mid-1990s among the youngest segments of the population. For example, from 1995 to 2002, the rate of heroin use among youths between the ages of 12 and 17 years increased from 0.1 to 0.4%; among young adults between the ages of 18 and 25 years, this rate rose from 0.8 to 1.6% during the same period. The latest surge in heroin usage has been accompanied by an increase in medical emergencies and mortality directly precipitated by the use of the drug (Substance Abuse and Mental Health Services Administration - Office of Applied Studies 2003a; Substance Abuse and Mental Health Services Administration - Office of Applied Studies 2004). Thus, heroin dependence is a major threat to global public health, and efforts to identify avenues for its prevention are sorely needed. Unfortunately, the specific factors that predispose individuals to abuse heroin are not yet well enough understood to target these for intervention and prevention efforts.
Evidence from several twin studies (Karkowski and others 2000; Kendler and others 2000; Tsuang and others 2001; Tsuang and others 1996; van den Bree and others 1998) suggests that genes account for between 23 and 54% of the total liability toward heroin dependence, with the remainder of variance in liability attributable to environmental factors. In contrast to other substance use disorders, a large portion of the genetic risk for heroin dependence is conferred by genes that specifically increase risk for dependence on this substance alone, rather than by genes that predispose toward substance dependence more generally (Tsuang and others 1998). The search for risk genes for heroin dependence is therefore well warranted.
Several attempts have been made to relate particular allelic variants of functional candidate genes with heroin dependence through association mapping. The majority of genes that historically have been selected for examination of association with the illness can be roughly classified into two groups based on the functions of their proteins: 1) monoamine pathway genes, due to the known role of serotonin and dopamine neurotransmission in mediating the effects of several drugs of abuse; and 2) opioid pathway genes, due to their expression in or direct interaction with the opioid system, which is the primary transmitter system targeted by heroin. The collective evidence for an association between heroin dependence and any of these functional candidate genes is quite limited (Kreek and others 2005), suggesting that the selection of positional candidate genes based on the results of genome-wide linkage analysis might be a beneficial alternate strategy.
Toward that end, we collected DNA and clinical data from Han Chinese sibling-pairs (both of whom were affected by heroin dependence), and collected DNA from their parents and additional family members from Yunnan Province, China, which borders an area of Myanmar and Laos known as the “Golden Triangle”. This region is the source of more than 20% of the world's heroin (Cai 1998), and increases in heroin smuggling through Yunnan Province during the last two decades (Beyrer and others 2000; Stimson 1993; Stimson 1994) have produced a concomitant rise in heroin dependence among the Province's 37 million residents (Beyrer and others 2000; McCoy and others 2001), especially the region's youth (Kulsudjarit 2004). These high levels of exposure to a necessary environmental factor (i.e., heroin) make this geographical region ideal for investigating the genetic bases of heroin dependence. The first phase of data collection for this project has been completed, and this report documents the results of the initial genome-wide linkage analysis of these families.
Materials and Methods
Ascertainment and Clinical Assessment
Heroin-dependent probands were recruited from the Yunnan Institute of Drug Abuse (YIDA), Yunnan Province, China. An initial screening was performed to determine if the proband had any affected siblings; if so, the proband was enrolled and asked for permission to contact their family members, who were then individually evaluated for inclusion in the study. The final sample for the present analyses included 604 individuals (including 194 affected sibling-pairs) from 192 families. The average number of genotyped people per family was 2.4. (Table 1).
Table 1. Descriptive Statistics of the Sample.
The table provides frequencies of individuals, sibling-pairs, and families used in the present analyses.
| Statistic | N (% of sample) |
|---|---|
| Genotyped individuals | 453 |
| Genotyped individuals by sex | |
| male | 332 (0.73) |
| female | 121 (0.27) |
| Genotyped individuals by affection status | |
| affected | 386 (0.67) |
| unaffected | 67 (0.33) |
| Families with the indicated number of genotyped individuals | |
| 2 | 132 (0.70) |
| 3 | 37 (0.20) |
| 4 | 17 (0.09) |
| 5 | 2 (0.01) |
| Independent Sibling-pairs | 194 |
| Affected full sibling-pairs | 180 |
| Affected half-sibling-pairs | 14 |
Each proband and their affected sibling underwent a confirmatory diagnostic screen using supplemental medical records and a semi-structured interview that was based on the Diagnostic and Statistical Manual, Fourth Edition (DSM-IV) (American Psychiatric Association 1994). Following this screen, the Mandarin Chinese version of the Diagnostic Interview for Genetic Studies (DIGS) (Faraone and others 1996; Nurnberger and others 1994; Wang and others 2004) was administered. Interviewers underwent rigorous training on the DIGS to ensure an accurate diagnostic assessment. Test-retest reliabilities of diagnoses based on the English, French, Korean, and Colombian versions of the DIGS have previously been shown to be excellent (Faraone and others 1996; Joo and others 2004; Nurnberger and others 1994; Palacio and others 2004; Preisig and others 1999); test-retest reliability of the Chinese version may be similarly excellent, however this issue has not yet been empirically determined. We supplemented structured interview data with information extracted from medical records. Best-estimate final diagnoses were made by two board-certified psychiatrists independently based on all the clinical information that was collected. When these psychiatrists disagreed, a third diagnostician was used as the tiebreaker. All procedures used in this study were approved by the Institutional Review Boards of the participating institutions, including YIDA, Harvard Medical School, and the University of California, San Diego.
Genotyping
Approximately 10ml of blood was drawn from each subject and immediately shipped to the NIDA Center for Genetic Studies at the Rutgers University Cell and DNA Repository, where cells were immortalized via transformation with Epstein-Barr virus. DNA was extracted from these cell lines and sent to the Center for Inherited Disease Research (CIDR) for genotyping. CIDR genotyped all samples on 386 microsatellite markers spaced at an average inter-marker distance of 9 cM, following their standard procedures, which are published on the web (http://www.cidr.jhmi.edu/protocol.html). No gap was greater than 20 cM between any two markers. Marker distances were generated using the sex-averaged Marshfield genetic map (Broman and others 1998). Pedigree inconsistencies were evaluated using RELCHCK (Boehnke and Cox 1997) and GRR (Abecasis and others 2001). Once the pedigree errors were identified, each error was examined manually and corrected accordingly or, if the source of the discrepancy could not be identified, the pedigree was eliminated from the analyses. Mendelian inconsistencies were checked and removed using PEDCHECK (O'Connell and Weeks 1998). Additional unlikely genotypes were removed using pedwipe, a command available in Merlin (Abecasis and others 2001).
Linkage Analysis
Multipoint nonparametric linkage analysis was performed across all autosomes and the X chromosome using the computer program Merlin (Abecasis and others 2001). Although all relative-pairs were included in the data analyses, only data from affected sibling-pairs contributed to the test statistics. This was due to unavoidable limitations on the data collection protocol, which prohibited the collection of detailed phenotypic information from family members other than probands and their affected siblings. Results are reported as nonparametric linkage (NPL) Z-scores calculated from Merlin, the method of which is described in detail by Whittemore and Halpern (1994). Allele frequencies were calculated in Merlin using all available individuals, and NPL Z-scores were calculated at each marker throughout the genome. The statistical significance of these Z-scores was determined by performing 1000 simulations of the data under the assumption of no linkage, and then performing linkage analyses in Merlin using that simulated data. Through this process, we determined that an NPL score of 2.18 (LOD score of 1.51) or greater would be expected to occur once by change in a genomewide linkage scan and, thus, would meet traditional criteria for “suggestive” evidence for linkage, while an NPL score of 3.03 (LOD score of 2.85) would occur only 0.5 times per linkage scan and would qualify as “significant” evidence for linkage (Lander and Kruglyak 1995). In the analysis reported here, individuals with either heroin abuse or dependence were considered affected. This study only collected data on nuclear families and therefore there were no extended relatives or three-generation families included in the analysis. Power analyses were conducted using the Genetic Power Calculator (Purcell and others 2003).
Results
To date, we have collected clinical and genetic marker data on 207 independent affected sibling-pairs from 202 Han Chinese families. Of these, 192 families were retained for genetic linkage analyses after applying the various data-cleaning procedures resulting in a total of 194 independent sibling-pairs in the linkage analysis. Table 1 contains descriptive information on the final sample used in the genetic linkage analysis presented here.
We found two regions with nonparametric linkage (NPL) Z-scores greater than 2.0 (Figure 1). A peak NPL Z-score of 2.19 was observed at marker D4S1644 (uncorrected p-value = 0.014), located at 143.3 cM on chromosomal region 4q31.21 (Figure 2). The corresponding LOD-score for this peak is approximately 1.22, and the 1-LOD drop-down region surrounding this peak extended from 122-172 cM on chromosome 4. The highest NPL Z-score of 2.36 (uncorrected p-value = 0.009) was observed at 53.4 cM on chromosomal region 17q11.2 at marker D17S1880 (Figure 3). This peak NPL Z-score corresponded to a LOD score of 1.77, and the 1-LOD drop-down region surrounding this peak extended from 30-82 cM. The evidence for linkage at both markers (on chromosomes 4 and 17) exceeded the threshold for “suggestive” evidence for linkage, but neither attained the level of “significant” evidence according to traditional criteria.
Figure 1.
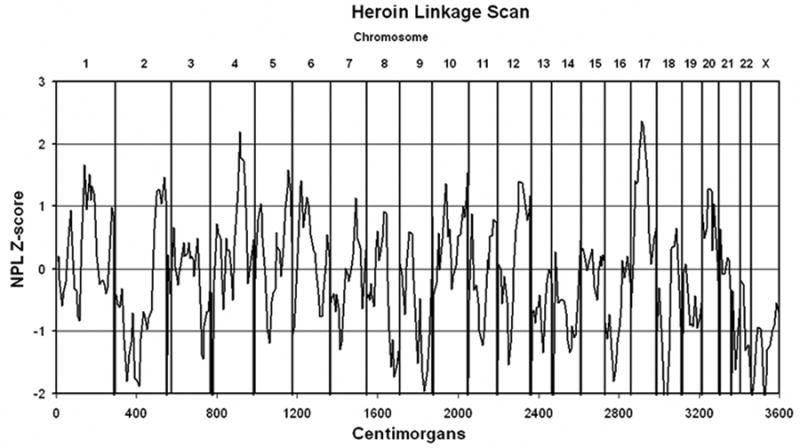
The plot shows a curve of NPL Z-scores observed at markers mapping to the indicated centiMorgan positions throughout the genome.
Figure 2.
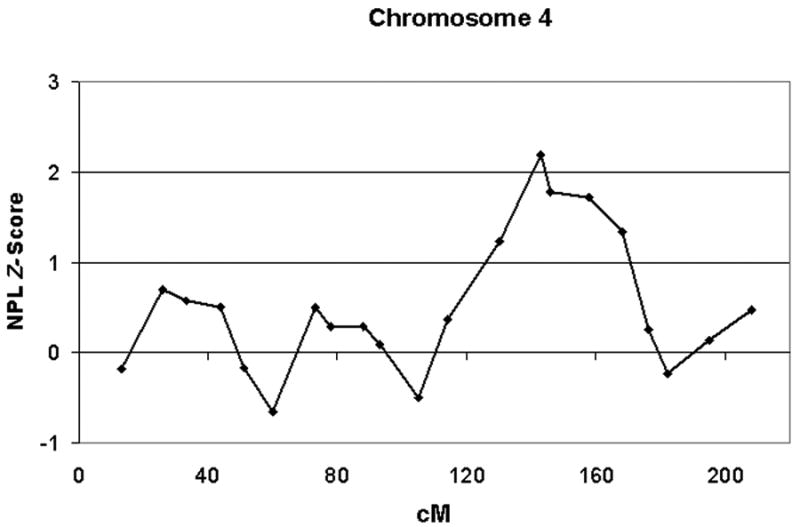
The plot shows a curve of NPL Z-scores observed at markers mapping to the indicated centiMorgan positions on chromosome 4, including a peak NPL Z-score of 2.19 at marker D4S1644 (143.3 cM).
Figure 3.
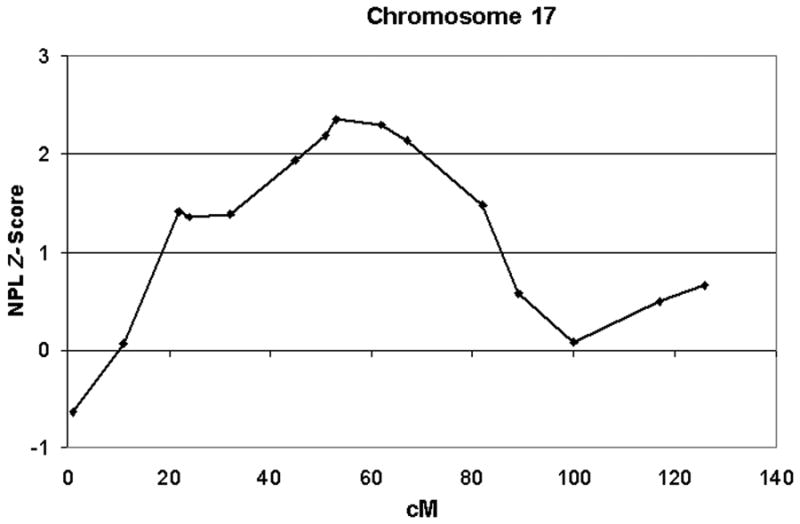
The plot shows a curve of NPL Z-scores observed at markers mapping to the indicated centiMorgan positions on chromosome 17, including a peak NPL Z-score of 2.36 at marker D17S1880 (53.4 cM).
Given the observed marker heterozygosity, spacing, and density, many regions of the genome had relatively low information content. For example, less than twenty-five percent of the genome had information content greater than 0.55, half the genome had information content less than 0.52, and no place along the genome had an information content greater than 0.64. These values may be lower than those observed in other samples for which the CIDR marker set was optimized (e.g., European-American samples), which may cause a reduction in power in analyzing our samples with the same markers. However, this suggests that the evidence for linkage that we have observed presently could be strengthened by further fine-mapping or by the evaluation of additional carefully selected markers that show high heterozygosity in the Han Chinese population.
Discussion
The present report documents some of the first known results of genome-wide linkage analysis of sibling-pairs affected by heroin dependence. Loci on chromosomes 4 and 17 showed some promise for harboring risk genes for the illness. Of potentially major importance, we note that the only marker to be significantly linked (LOD = 3.46) to non-opioid drug dependence in the sample of European-American families studied by Gelernter et al. (2006) is within 25 cM and the 1-LOD drop-down interval of the most strongly linked marker observed in our sample of Han Chinese heroin-dependent sibling-pairs (D17S1880 at 53.4 cM on chromosome 17q). However, the strongest evidence for linkage to opioid dependence reported by Gelernter et al. was observed at a more distal marker at 103.5 cM on 17q, beyond the 1-LOD drop-down interval around our most strongly linked marker. In addition, the evidence for linkage we have observed on chromosome 17 attained the level of “suggestive” evidence for linkage, but did not surpass criteria for genome-wide “significance” when accounting for multiple testing across the genome. Because of this, caution must be exercised when interpreting these findings.
Our immediate plans for following up on these results include ascertainment and analysis of additional affected sibling-pairs and families, as well as increasing the density of informative markers in the region of the observed linkage signals. Yet, even at this early stage, some of the genes under these preliminary linkage peaks might warrant attention as positional candidate genes for heroin abuse or dependence. For example, CDK5R1 maps to chromosome 17q11.2 and codes for regulatory subunit 1 of cyclin-dependent kinase 5 (cdk5), which has been implicated in the psychological dependence on and behavioral sensitization to morphine in rodents (Narita and others 2005; Wang and others 2004). Cdk5 protein was also found to be down-regulated in neuronal tissue from opioid addicts and opiate-treated rats (Ferrer-Alcon and others 2003). Thus, CDK5R1 may now be considered a reasonable candidate gene for heroin dependence on both positional and functional grounds. In addition, ARRB2, which codes for arrestin beta 2, might be considered a candidate gene worth pursuing since mu-opioid receptor agonism usually involves recruitment of this molecule, and Arrb2 has been shown to directly regulate opioid-induced synaptic release and presynaptic inhibition (Bradaia and others 2005); however, ARRB2 maps to chromosome 17p13, which is quite distal to the region on chromosome 17q11.2 that was the most tightly linked in our study. These and other candidate genes under the observed linkage peaks are now being evaluated for potential functional relevance to the condition, as well as their suitability for allelic association analysis. However, we must reiterate that the evidence for linkage at even our most strongly linked candidate regions is preliminary in nature and these regions are very broad, encompassing hundreds of genes; thus, any pursuit of candidate genes in these regions must proceed with due caution.
Of note, none of the “traditional” functional candidate genes for heroin dependence (e.g., monoamine and opioid pathway genes) code to either of the two regions implicated in this study. This result suggests that functional candidate gene association studies of complex conditions such as heroin dependence may not be the only—or even the best—strategy for identifying responsible polymorphisms. Such approaches may work better when they are complemented by systematic genome-wide linkage and association screens that are not limited by the constraints of existing disease models. Indeed, the full compendium of risk factors for heroin dependence may include some genes that would have been identified as functional candidates (e.g., opioid receptors), some genes that are positional candidates, and some which are both (e.g., CDK5R)
The results reported here are from the first phase of data collection for an ongoing study, and therefore must be considered preliminary. With the addition of more families and markers, the evidence for linkage at the loci on chromosomes 4 and 17 may become stronger or disappear altogether. On the other hand, loci with NPL Z-scores greater than 1.0 but less than 2.0 (such as those on chromosomes 1, 5, and 10) may yield additional evidence to strengthen their position as linked loci. In addition to the preliminary nature of this study, another related limitation is the relatively small sample size, which only afforded us adequate power (β > 0.80) to detect loci with relatively large effects (i.e., accounting for ≥ 10% of the variance) on risk for heroin dependence. The genetic architecture of heroin dependence is unknown but is likely to be quite complex, with multiple genes and environmental factors each making small contributions to the overall liability toward dependence. Therefore, our sample (to date) is probably too small to detect the majority of loci that have small but reliable influences on risk for heroin dependence.
This study also has several strengths which may offset these limitations to some degree. Three of these strengths relate to three distinct aspects of the sampled population. First, the rate of exposure to heroin among individuals in Yunnan Province is much higher than in most other places in the world, and exposure to heroin is of course a necessary step in becoming dependent. In the presence of such high levels of exposure, our ability to detect risk genes for heroin dependence may be heightened because more individuals are put in a position where the risk-conferring genes will have the opportunity to manifest their influence. On the other hand, the prevalence of phenocopies in our sample may be heightened. Second, families in China (even those with heroin-dependent members) tend to remain intact, thus increasing our continued ability to ascertain families that are useful for linkage analysis and, ultimately, family-based association analyses. Third, the Han Chinese population is genetically more homogeneous than other populations (Oota and others 2002), especially that of the United States. This greater homogeneity may increase our ability to detect risk genes by decreasing the background “noise” against which risk genes generate a small but observable linkage “signal”.
In summary, we have identified two loci that may contain genes that influence risk for heroin dependence. The results of this first-pass positional cloning effort should provide useful information beyond that already attained using candidate gene association methods, and provide the foundation for future analyses of positional candidate genes that also have physiological plausibility. The evidence linking these loci to heroin dependence will either be strengthened or refuted by additional samples collected in our study, as well as the results of other ongoing linkage and association studies of opioid and heroin dependence.
Acknowledgments
This project is a joint effort between the American and Chinese project sites and research team members. The authors would like to thank Ma Kejian, Ji Hongrui, Wu Li, Wang Hua, Liu Xianling, Li Yu, Yang Liping, Wang Jing, Gong Xuemei, Shi Huaihai, Wang Haibin, Fu Li, Li Peikai, Shen Jiucheng, Xu Yan, Duan Chunmei, and Deng Yuan from the Yunnan Institute of Drug Abuse for their valuable contributions to the successful completion of this study. In addition, we thank the Human Genetics Resources Administration of China (HGRAC) for their support of this project. This work was supported by a grant (R01DA012846) from the National Institutes of Health to Ming T. Tsuang.
References
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. GRR: graphical representation of relationship errors. Bioinformatics. 2001;17(8):742–3. doi: 10.1093/bioinformatics/17.8.742. [DOI] [PubMed] [Google Scholar]
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders (DSM-IV) Washington, DC: American Psychiatric Association; 1994. [Google Scholar]
- Beyrer C, Razak MH, Lisam K, Chen J, Lui W, Yu XF. Overland heroin trafficking routes and HIV-1 spread in south and south-east Asia. Aids. 2000;14(1):75–83. doi: 10.1097/00002030-200001070-00009. [DOI] [PubMed] [Google Scholar]
- Boehnke M, Cox NJ. Accurate inference of relationships in sib-pair linkage studies. American Journal of Human Genetics. 1997;61(2):423–9. doi: 10.1086/514862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradaia A, Berton F, Ferrari S, Luscher C. beta-Arrestin2, interacting with phosphodiesterase 4, regulates synaptic release probability and presynaptic inhibition by opioids. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(8):3034–3039. doi: 10.1073/pnas.0406632102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman KW, Murray JC, Sheffield VC, White RL, Weber JL. Comprehensive human genetic maps: individual and sex-specific variation in recombination. American Journal of Human Genetics. 1998;63(3):861–9. doi: 10.1086/302011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai ZJ. Research on drug dependence and epidemiological investigation of drug abuse in China. Journal of Toxicological Sciences. 1998;23 Suppl 2:191–3. doi: 10.2131/jts.23.supplementii_191. [DOI] [PubMed] [Google Scholar]
- Faraone SV, Blehar M, Pepple J, Moldin S, Norton J, Tsuang MT, Nurnberger JI, Malaspina D, Kaufmann CA, Reich T, et al. Diagnostic accuracy and confusability analyses: An application to the diagnostic interview for genetic studies. Psychological Medicine. 1996;26:401–410. doi: 10.1017/s0033291700034796. [DOI] [PubMed] [Google Scholar]
- Ferrer-Alcon M, La Harpe R, Guimon J, Garcia-Sevilla JA. Downregulation of neuronal cdk5/p35 in opioid addicts and opiate-treated rats: relation to neurofilament phosphorylation. Neuropsychopharmacology. 2003;28(5):947–955. doi: 10.1038/sj.npp.1300095. [DOI] [PubMed] [Google Scholar]
- Gelernter J, Panhuysen C, Wilcox M, Hesselbrock V, Rounsaville B, Poling J, Weiss R, Sonne S, Zhao H, Farrer L, et al. Genomewide linkage scan for opioid dependence and related traits. American Journal of Human Genetics. 2006;78:759–769. doi: 10.1086/503631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo EJ, Joo YH, Hong JP, Hwang S, Maeng SJ, Han JH, Yang BH, Lee YS, Kim YS. Korean version of the diagnostic interview for genetic studies: Validity and reliability. Comprehensive Psychiatry. 2004;45(3):225–229. doi: 10.1016/j.comppsych.2004.02.007. [DOI] [PubMed] [Google Scholar]
- Karkowski LM, Prescott CA, Kendler KS. Multivariate assessment of factors influencing illicit substance use in twins from female-female pairs. American Journal of Medical Genetics (Neuropsychiatric Genetics) 2000;96(5):665–70. [PubMed] [Google Scholar]
- Kendler KS, Karkowski LM, Neale MC, Prescott CA. Illicit psychoactive substance use, heavy use, abuse, and dependence in a US population-based sample of male twins. Archives of General Psychiatry. 2000;57(3):261–269. doi: 10.1001/archpsyc.57.3.261. [DOI] [PubMed] [Google Scholar]
- Kreek MJ, Bart G, Lilly C, LaForge KS, Nielsen DA. Pharmacogenetics and human molecular genetics of opiate and cocaine addictions and their treatments. Pharmacological Reviews. 2005;57(1):1–26. doi: 10.1124/pr.57.1.1. [DOI] [PubMed] [Google Scholar]
- Kulsudjarit K. Drug problem in southeast and southwest Asia. Annals of the New York Academy of Sciences. 2004;1025:446–457. doi: 10.1196/annals.1316.055. [DOI] [PubMed] [Google Scholar]
- Lander E, Kruglyak L. Genetic dissection of complex traits: Guidelines for interpreting and reporting linkage results. Nature Genetics. 1995;11(3):241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- McCoy CB, McCoy HV, Lai S, Yu Z, Wang X, Meng J. Reawakening the dragon: changing patterns of opiate use in Asia, with particular emphasis on China's Yunnan province. Substance Use and Misuse. 2001;36(1-2):49–69. doi: 10.1081/ja-100000228. [DOI] [PubMed] [Google Scholar]
- Narita M, Shibasaki M, Nagumo Y, Narita M, Yajima Y, Suzuki T. Implication of cyclin-dependent kinase 5 in the development of psychological dependence on and behavioral sensitization to morphine. Journal of Neurochemistry. 2005;93(6):1463–1468. doi: 10.1111/j.1471-4159.2005.03136.x. [DOI] [PubMed] [Google Scholar]
- Nurnberger JI, Jr, Blehar MC, Kaufmann CA, York-Cooler C, Simpson SG, Harkavy-Friedman J, Severe JB, Malaspina D, Reich T, Miller M, et al. Diagnostic interview for genetic studies. Rationale, unique features, and training. Archives of General Psychiatry. 1994;51:849–859. doi: 10.1001/archpsyc.1994.03950110009002. [DOI] [PubMed] [Google Scholar]
- O'Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. American Journal of Human Genetics. 1998;63(1):259–66. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oota H, Kitano T, Jin F, Yuasa I, Wang L, Ueda S, Saitou N, Stoneking M. Extreme mtDNA homogeneity in continental Asian populations. American Journal of Physical Anthropology. 2002;118(2):146–153. doi: 10.1002/ajpa.10056. [DOI] [PubMed] [Google Scholar]
- Palacio CA, Garcia J, Arbelaez MP, Sanchez R, Aguirre B, Garces IC, Montoya GJ, Gomez J, Agudelo A, Lopez CA, et al. Validation of the Diagnostic Interview for Genetic Studies (DIGS) in Colombia. Biomedica. 2004;24(1):56–62. [PubMed] [Google Scholar]
- Preisig M, Fenton BT, Matthey ML, Berney A, Ferrero F. Diagnostic interview for genetic studies (DIGS): inter-rater and test-retest reliability of the French version. European Archives of Psychiatry and Clinical Neuroscience. 1999;249(4):174–179. doi: 10.1007/s004060050084. [DOI] [PubMed] [Google Scholar]
- Purcell S, Cherny SS, Sham PC. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19(1):149–50. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- Stimson GV. The global diffusion of injecting drug use: implications for human immunodeficiency virus infection. Bulletin on Narcotics. 1993;45(1):3–17. [PubMed] [Google Scholar]
- Stimson GV. Reconstruction of subregional diffusion of HIV infection among injecting drug users in Southeast Asia: implications for early intervention. Aids. 1994;8(11):1630–2. doi: 10.1097/00002030-199411000-00023. [DOI] [PubMed] [Google Scholar]
- Substance Abuse and Mental Health Services Administration - Office of Applied Studies. Emergency Department Trends From the Drug Abuse Warning Network, Final Estimates 1995–2002. Report nr DAWN Series: D-24, DHHS Publication No. (SMA) 03-3780. Rockville, MD: 2003a. [Google Scholar]
- Substance Abuse and Mental Health Services Administration - Office of Applied Studies. Results from the 2002 National Survey on Drug Use and Health: National Findings. Report nr NHSDA Series H-22, DHHS Publication No. SMA 03–3836. Rockville, MD: 2003b. [Google Scholar]
- Substance Abuse and Mental Health Services Administration - Office of Applied Studies. Mortality Data From the Drug Abuse Warning Network, 2002. Report nr DAWN Series D25, DHHS Publication No. (SMA) 043875. Rockville, MD: 2004. [Google Scholar]
- Tsuang MT, Bar JL, Harley RM, Lyons MJ. The Harvard Twin Study of Substance Abuse: what we have learned. Harvard Review of Psychiatry. 2001;9(6):267–79. [PubMed] [Google Scholar]
- Tsuang MT, Lyons MJ, Eisen SA, Goldberg J, True W, Lin N, Meyer JM, Toomey R, Faraone SV, Eaves L. Genetic influences on DSM-III-R drug abuse and dependence: A study of 3,372 twin pairs. American Journal of Medical Genetics (Neuropsychiatric Genetics) 1996;67:473–477. doi: 10.1002/(SICI)1096-8628(19960920)67:5<473::AID-AJMG6>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Tsuang MT, Lyons MJ, Meyer JM, Doyle T, Eisen SA, Goldberg J, True W, Lin N, Toomey R, Eaves L. Co-occurrence of abuse of different drugs in men. The role of drug-specific and shared vulnerabilities. Archives of General Psychiatry. 1998;55:967–972. doi: 10.1001/archpsyc.55.11.967. [DOI] [PubMed] [Google Scholar]
- van den Bree MB, Johnson EO, Neale MC, Pickens RW. Genetic and environmental influences on drug use and abuse/dependence in male and female twins. Drug and Alcohol Dependence. 1998;52(3):231–241. doi: 10.1016/s0376-8716(98)00101-x. [DOI] [PubMed] [Google Scholar]
- Wang CH, Lee TH, Tsai YJ, Liu JK, Chen YJ, Yang LC, Lu CY. Intrathecal cdk5 inhibitor, roscovitine, attenuates morphine antinociceptive tolerance in rats. Acta Pharmacologica Sinica. 2004;25(8):1027–1030. [PubMed] [Google Scholar]
- Whittemore AS, Halpern J. A class of tests for linkage using affected pedigree members. Biometrics. 1994;50(1):118–27. [PubMed] [Google Scholar]


