Abstract
We report characterization of the nanostructures of complexes formed between the redoxactive lipid bis(n-ferrocenylundecyl)dimethylammonium bromide (BFDMA) and DNA using small-angle neutron scattering (SANS) and cryogenic transmission electron microscopy (cryo-TEM). A particular focus was directed to the influence of lipid oxidation state (where reduced BFDMA has a net charge of + 1 and oxidized BFDMA has a charge of +3) on the nanostructures of the solution aggregates formed. Complexes were characterized over a range of charge ratios of reduced BFDMA to DNA (1.1:1, 2.75:1 and 4:1) in solutions of 1mM Li2SO4. For these complexes, a single peak in the SANS data at 1.2nm-1 indicated that a nanostructure with a periodicity of 5.2nm was present, similar to that observed with complexes of the classical lipids DODAB:DOPE and DNA (multilamellar spacing of 7.0nm). The absence of additional Bragg peaks in all the SANS data indicated that the periodicity did not extend over large distances. Both Inverse Fourier Transform analysis and form factor fitting suggested formation of a multilamellar vesicle. These results were confirmed by cryo-TEM images in which multilamellar complexes with diameters between 50 and 150nm were observed with no more than seven lamellae per aggregate. In contrast to complexes of reduced BFDMA and DNA, Bragg peaks were absent in SANS spectra of complexes formed by oxidized BFDMA and DNA at all charge ratios investigated. The low q behavior of the SANS data obtained using oxidized BFDMA and DNA complexes suggested that large, loose aggregates were formed, consistent with complementary cryo-TEM images showing predominantly loose disordered aggregates. Some highly ordered sponge-like and cubic phase nanostructures were also detected in cryo-TEM images. We conclude that control of BFDMA oxidation state can be used to manipulate the nanostructures of lipid-DNA complexes formed using BFDMA.
Keywords: nanostructure, cationic lipid, lipoplex, small-angle neutron scattering, cryo-TEM
Introduction
DNA assumes extended conformations in aqueous solution due to its anionic polyelectrolyte character. Cationic lipids such as dimethyldioctadecylammonium bromide (DODAB), which form vesicle-like aggregates in aqueous solution, have been shown to complex strongly with DNA to form lipid-DNA complexes called lipoplexes.1,2 Within these complexes, the DNA is highly compacted as compared to cationic lipid-free solutions of DNA in the absence of cationic lipids. A number of past studies have demonstrated that DNA, when compacted by certain cationic lipids, is able to pass across cell membranes and transfect cells.3-6 Recently, we demonstrated that changing the oxidation state of a double-tailed cationic lipid that incorporates the redox-active group ferrocene, bis(n-ferrocenylundecyl)dimethylammonium bromide or BFDMA (Figure 1), dramatically influences the efficiency of delivery of DNA to cells.7,8 Oxidation of the ferrocene groups of reduced BFDMA to ferrocenium ions leads to a change in molecular charge from +1 to +3 and a change in the amphiphilic character of BFDMA. Reduced BFDMA was shown to be an effective agent for transfection, whereas under the same conditions oxidized BFDMA did not promote high levels of transfection.8 The capability to control transfection by manipulation of the oxidation state of BFDMA may find application in the development of new non-viral gene delivery agents. As discussed in greater detail below, past studies have established that the nanostructures of lipoplexes can influence the efficiency with which cationic lipids can promote cell transfection. In the study reported in this paper, we seek to understand how the oxidation state of BFDMA influences the nanostructures of BFDMA and DNA complexes and provide possible insight into results arising from our past transfection studies.
Figure 1.
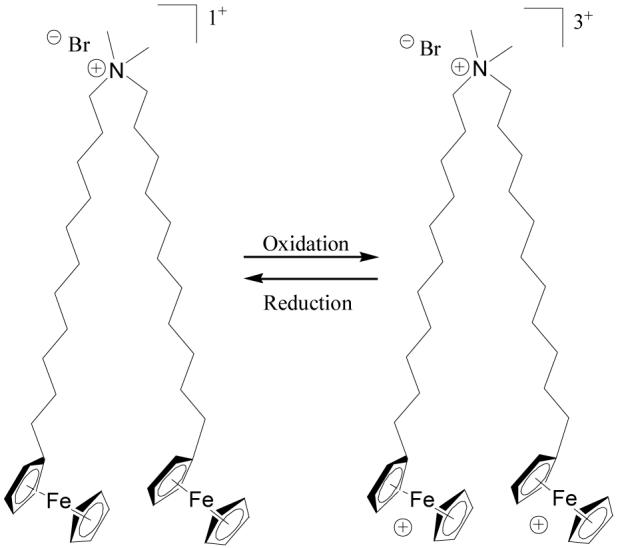
Molecular structure of BFDMA.
Our previous characterization of complexes formed upon the addition of DNA to reduced or oxidized BFDMA used dynamic light scattering and zeta potential measurements at concentrations relevant to our past transfection experiments (2.4μg/mL DNA and 2-100μM BFDMA).9 Our past studies revealed that the hydrodynamic sizes of lipoplexes of reduced BFDMA and DNA passed through a maximum, and that the zeta potentials changed sign from negative to positive, as a function of increasing ratio of reduced BFDMA to DNA,9 a behavior similar to that observed using complexes of DNA and conventional vesicle-forming cationic lipids.1,2,10 In contrast, complexes of DNA and oxidized BFDMA exhibited a minimum in hydrodynamic size as a function of increasing oxidized BFDMA concentration, and negative zeta potentials were observed at all concentrations of BFDMA investigated, similar to complexes formed between DNA and non-self-associating molecules with multiple positive charges.11,12 In addition, past light scattering measurements revealed that the DNA molecules had substantial conformational freedom when complexed with oxidized BFDMA, as compared to reduced BFDMA.9 These results, when combined, suggest that differences in the properties of complexes formed by BFDMA and DNA with oxidation state can be understood in terms of a change in amphiphilicity of BFDMA with oxidation state. These past studies also hint at the possibility that the nanostructures of the complexes formed by BFDMA and DNA may depend on the oxidation state of BFDMA.
In the study reported in this paper, we used small-angle neutron scattering (SANS) and cryogenic transmission electron microscopy (cryo-TEM) to build from these size and charge measurements to investigate the nanostructures of these complexes. SANS and cryo-TEM are complementary characterization techniques for investigations of the nanostructures of complexes of lipid and DNA. Cryo-TEM probes local regions of a solution and SANS provides access to statistically averaged data describing populations of aggregates present throughout an entire solution. The combination of these two techniques thus allows one to compare detailed nanostructural information obtained from individual complexes to the statistically averaged solution nanostructure. Previous studies using small-angle neutron13 and x-ray5,14-17 scattering, cryo-TEM,18-20 freeze-fracture-replication electron microscopy6,21 and atomic force microscopy5 have shown that the nanostructures of cationic lipid-DNA aggregates can be complex, often involving multilamellar 5,6,14,16,17 or hexagonal phases.5,6,14 Some studies employing cryo-TEM and AFM have revealed evidence for the coexistence of hexagonal and lamellar nanostructures in solution.5,6,18 Although ongoing research seeks to understand how the nanostructures of lipoplexes formed between cationic lipids and DNA influence the ability of the lipoplexes to transfect cells, a number of studies have reported that cationic lipid-DNA complexes with hexagonal nanostructures can lead to higher transfection efficiencies than those lipoplexes with lamellar nanostructures.5,22 Complementary studies using light microscopy have shown that anionic vesicles, a primitive model for a cellular membrane, can interact with hexagonal cationic lipid-DNA complexes, resulting in rapid fusion and subsequent DNA release.14 This rapid fusion process did not occur when a lamellar cationic lipid-DNA complex was used (lipid transfer occurred over several hours).14 These past studies suggest that the development of a full understanding of how the oxidation state of BFDMA influences transfection efficiency will require knowledge of the nanostructures of BFDMA-DNA complexes as a function of the oxidation state of BFDMA.
This paper is organized as follows: First, we describe small-angle neutron scattering from complexes formed by the addition of DNA to a classical lipid system comprising DODAB and DOPE. This classical lipid-DNA system was investigated so as to validate our methodology and to serve as a reference system for comparison to BFDMA-DNA systems. Second, we discuss results obtained from SANS and cryo-TEM nanostructural analysis of complexes formed by aqueous solutions of BFDMA in the presence of plasmid DNA. The oxidation state of the BFDMA is shown to influence the nanostructures of the complexes that it forms with DNA. We note that in addition to our past studies of the BFDMA-DNA systems described above,7-9 there exist past studies of the interactions of other ferrocene-containing surfactants with either DNA23 or cellulose-derived polymers.24 These past studies do not provide SANS or cryo-TEM characterization of the nanostructures of the complexes formed between these species.
Materials and Methods
Materials
BFDMA was synthesized according to methods published elsewhere.25 Salmon sperm DNA (2000 ± 500 base pairs) was purchased from Invitrogen (Carlsbad, CA) and plasmid DNA encoding enhanced green fluorescent protein (pEGFP-N1 (4.7kbps) >95% supercoiled) was purchased from the Waisman Clinical Biomanufacturing Facility at the University of Wisconsin-Madison. Dioctadecyldimethylammonium bromide (DODAB, Sigma Aldrich) and dioleoylphosphatidylethanolamine (DOPE, Avanti Polar Lipids) were used as received. All other chemicals were purchased from Sigma Aldrich (St. Louis, MO), each with a purity of at least 98%, and used without further purification unless otherwise stated.
Electrochemical Oxidation
Electrochemical oxidation of BFDMA (4ml, 1mM Li2SO4, pH5) was performed as described previously.7,8 The extent of oxidation of BFDMA was followed by monitoring the current passed at the working electrode and by using UV-visible spectrophotometry, as the lipid was converted from the reduced form (yellow, peak absorbance at 430nm) to the oxidized state (blue, peak absorbance at 625nm). UV-visible absorbance values were recorded on a Beckman Coulter DU520 UV-Vis Spectrophotometer (Fullerton, CA). For UV-visible spectrophotometry of solutions containing reduced BFDMA only, 40mM dodecyltrimethylammonium bromide (DTAB) was added to the lipid solutions prior to measurement of UV-visible spectra to disrupt large aggregates that caused the solutions to be turbid (DTAB does not absorb light in the visible region of interest, 400-800nm). As the oxidation reaction progressed, the peak in the spectrum at 430nm disappeared and a well defined peak appeared at 625nm. The fully oxidized lipid was stable in solution for at least 14 days at room temperature as determined by UV-visible spectroscopy. All lipid solutions (both reduced and oxidized) were prepared in 1mM Li2SO4 at pH 5 unless otherwise stated (alkaline conditions were avoided due to instability of oxidized ferrocene under alkaline conditions).26
Preparation of Samples for Characterization by SANS and cryo-TEM
DODAB:DOPE (1:1) extruded lipid mixtures and complexes formed by the subsequent addition of salmon sperm DNA at a charge ratio of 2:1 were prepared according to the method published by Barreleiro et al.13 We used salmon sperm DNA in these experiments to permit direct comparison with the previous studies by Barreleiro et al., and to validate our experimental methodology.
All other samples characterized by SANS and cryo-TEM contained BFDMA and plasmid DNA (pEGFP-N1). Stock solutions of BFDMA (1mM) were prepared in aqueous Li2SO4 (1mM) solutions. The poor solubility of the lipid limited the concentration of BFDMA in solution to 1mM. The stock solution of BFDMA (1mM in Li2SO4) was diluted with a DNA stock solution (with vortexing for 5 seconds) to give the desired charge ratios of BFDMA and DNA in solution (where the charge ratio is defined as the molar ratio of the positive charges in the cationic lipid to negatively charged DNA phosphate groups). We note that complexes formed from reduced BFDMA and DNA at a charge ratio of 1.1:1 contain the same molar concentrations of BFDMA and DNA as complexes formed by oxidized BFDMA and DNA at a charge ratio of 3.3:1. The charge ratios investigated in our study were similar to those used in previously reported transfection experiments.7,8 Although the charge ratios match previous transfection experiments, the absolute concentrations of BFDMA used in the SANS experiments (1mM) were substantially higher than the transfection experiments (10-40μM). The high concentrations of BFDMA in the SANS experiments were necessary to obtain a sufficient intensity of scattered neutrons.
Small-Angle Neutron Scattering
SANS measurements were performed using the NG3 instrument at the NIST Center for Neutron Research in Gaithersburg, MD. The incident neutron wavelength was on average 0.6nm, with a spread in wavelength, Δλ/λ, of 14%. Data were recorded at three different sample-to-detector distances (1.9m, 4.5m and 13.1m), giving q ranges from 0.03nm-1 - 0.45nm-1, 0.76nm-1 - 1.44nm-1 and 0.18nm-1 - 3.38nm-1, respectively. To ensure statistically relevant data, at least 500,000 counts were collected for each sample at each detector distance. The samples were contained in quartz cells with a 2mm path length, and placed in a sample chamber held at 25.0 ± 0.1 °C. The data were corrected for detector efficiency, background radiation, empty cell scattering and incoherent scattering to determine the intensity on an absolute scale.27 The background scattering from the solvent was subtracted unless otherwise stated. The processing of data was performed using Igor Pro (WaveMetrics, Lake Oswego, Oregon) with a program provided by NIST. Guinier analysis, Indirect Fourier Transform (IFT) analysis and form factor fitting were used to interpret the scattering. These techniques are described in detail elsewhere,28-34 and are only summarized briefly here.
The intensity of neutrons scattered from an isotropic sample, I(q) may be described as the product of a form factor P(q), giving the intrinsic particle shape, and a structure factor S(q), describing inter-particle interactions, as given below
| (1) |
where np is the number density of scattering objects. Guinier analysis relates I(q) to the form factor P(q) for a range of geometries such as cylinders and bilayers (vesicles), and predicts that a plot of ln[I(q)] versus ln(q) should be linear with slopes of -1 and -2 for cylindrical and bilayer aggregates, respectively.35 This approximation holds true for qRx < 1, where Rx is the radius of gyration of the aggregate.
We also used Indirect Fourier transform (IFT) analysis to calculate the pair distance distribution function (PDDF) or p(r) from I(q) according to the following relationship:
| (2) |
When the system is dilute (the structure factor S(q) = 1) and the aggregates are monodisperse, the calculated PDDF allows direct identification of the particle structure.36 We performed the IFT analysis using software provided by Glatter.37 The only input parameter required for the IFT analysis is an estimate of Dmax, the maximum distance between two points in the particle, which imposes the constraint in the PDDF that p(r) = 0 when r > Dmax.
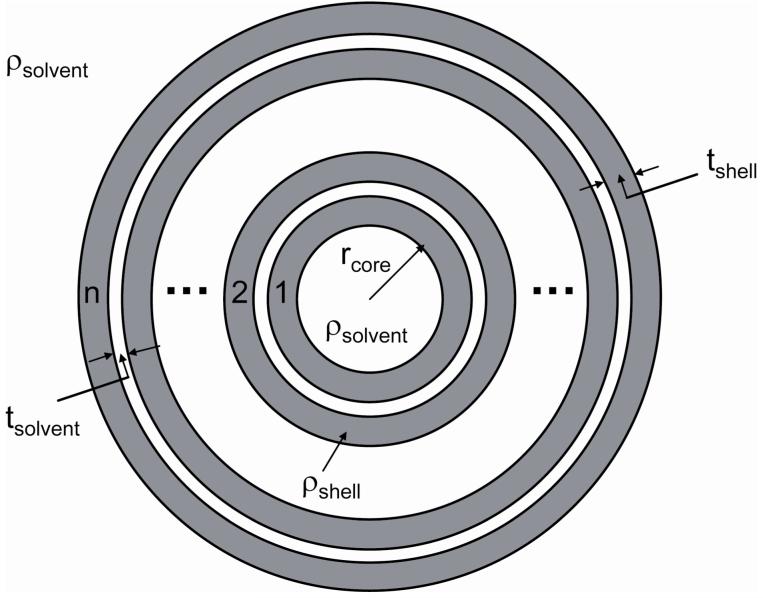
We used the calculation of the PDDF to suggest models for form factor fitting (models provided by NIST for use with Igor Pro).27,38 The model used here allows calculation of the scattering from a system of multi-shell spherical particles (Figure 2). The scattering length densities of the shells (ρshell) and solvent (ρsolvent) were determined using the NIST scattering length density calculator39 leaving four parameters to optimize in the fitting process; the core radius (rcore), the shell thickness (tshell), the solvent thickness (tsolvent) and the number of shells (n). Polydispersity in the number of shells was included in the model using a Gaussian distribution about the mean number of shells.
Figure 2.
Schematic illustration of multi-shell sphere model used for form factor fitting to experimental scattering data.
Cryogenic Transmission Electron Microscopy
Samples were prepared in a controlled environment vitrification (CEVS) system at 25°C and 100% relative humidity, as previously described.40,41 Samples were examined using a Philips CM120 microscope (Eindhoven, The Netherlands) operated at 120kV, using an Oxford CT-3500 cooling holder and transfer-station (Abingdon, England). Specimens were equilibrated in the microscope below -178°C, then examined in the low-dose imaging mode to minimize electron beam radiation damage, and recorded at a nominal focus of 1-2μm to enhance phase-contrast. Images were acquired digitally by a Gatan MultiScan 791 cooled charge coupled device camera (Pleasanton, CA) using the Digital Micrograph software package.
Results
Characterization of Complexes of DODAB:DOPE and DNA
Prior to performing SANS measurements with BFDMA and plasmid DNA, we validated our methodology using vesicles of dioctadecyldimethylammonium bromide (DODAB) and dioleoylphosphatidylethanolamine (DOPE), in a 1:1 ratio, in the presence of salmon sperm DNA (Figure 3). We note here that the DODAB:DOPE system has been the subject of a previous stopped-flow SANS study by Barreleiro et al.,13 using the D22 instrument at the Institute Laue-Langevin, Grenoble, France. These investigators examined the behavior of the system on very short (millisecond to minute) time scales. The extremely high flux at the D22 beamline allows time-resolved measurements to be performed. It was not clear at the outset of our study that investigation of the nanostructures of lipid-DNA complexes was possible using the lower flux SANS NG3 instrument at NIST, Gaithersburg, MD. Our experiments with the “model” DODAB/DOPE system revealed that the scattering intensity at the NG3 beamline was sufficiently low that (i) it was necessary to use pure D2O as the solvent, rather than the 70:30 D2O:H2O mixture employed by Barreleiro et al. to contrast match the scattering density of salmon sperm DNA and the solvent, and (ii) it was necessary to collect data for 2-4 hours per sample. Our scattering measurements using DNA in pure D2O showed negligible scattering in the q range of interest, suggesting that even in 100% D2O the DNA does not contribute to the scattering, and that any nanostructure detected results from the organization of the lipid alone.
Figure 3.
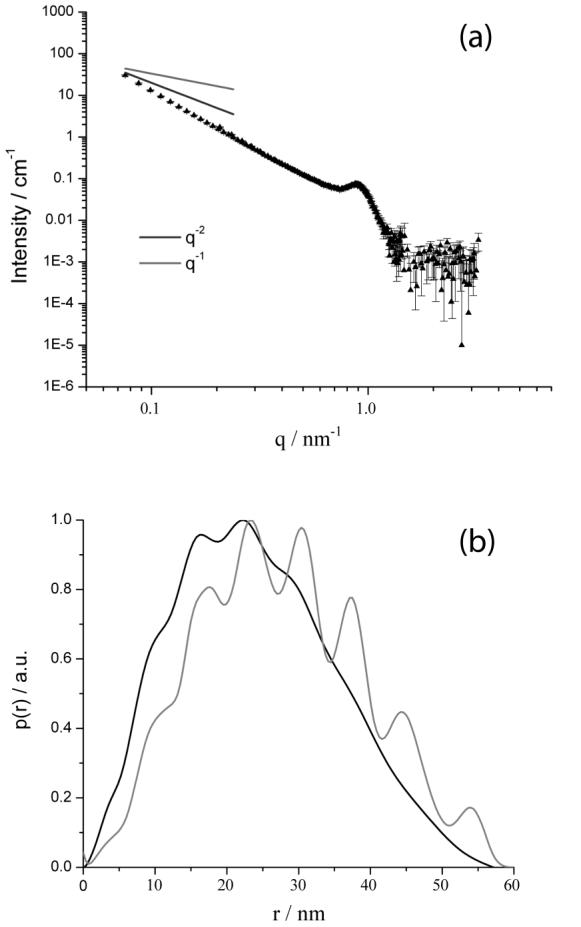
(a) SANS spectrum measured using complexes formed by DODAB:DOPE (1:1) and salmon sperm DNA at a charge ratio of 2:1. (b) PDDF calculated from the indirect Fourier transform of the data in (a) (dark line). For comparison, the gray line shows the PDDF calculated for a multilamellar vesicle model with a core radius of 5.0nm and on average 6 shells, where each shell has a thickness 4.5nm and is separated by a water layer of thickness 2.5nm.
Figure 3a shows the scattering from complexes formed between DNA and the DODAB:DOPE mixture at a lipid:DNA charge ratio of 2:1. The presence of the peak at 0.9nm-1 indicates a periodic structure with a repeat distance of 7.0nm, and is in good agreement with the spacing observed from scattering data reported in the Barreleiro study.13 This spacing was proposed to correspond to a lamellar nanostructure and was determined to be the sum of the thicknesses of the lipid bilayer, the DNA strand and one water layer. The presence of a single peak in our measurements indicates that the ordered lamellar structure does not extend over large distances. The scattering at low q follows a q-2.7 dependence, indicative of the presence of complex large-scale aggregates with a fractal or network-like structure. A similar q-2.7 dependence was also observed in other past studies of the scattering from complexes formed by salmon sperm DNA and hexadecyltrimethylammonium bromide.42 This q-2.7 dependence deviates from the Barreleiro study, performed on short time scales (0-100s), where the scattering followed a slope of -2 at low q. This suggests that large scale aggregation processes may occur over the long time periods (several hours) required to perform our measurements.
The pair distance distribution function (PDDF) was calculated from the scattering data shown in Figure 3a and compared to the PDDF predicted using a multilamellar vesicle with polydispersity in the number of shells per aggregate (Figure 3b). The polydisperse multilamellar vesicles (with a mean number of shells of 6) were assumed to have core radii of 5.0nm and shell thicknesses of 4.5nm. Each shell was separated by a water layer of thickness 2.5nm. Figure 3b shows the presence of fluctuations in the PDDFs from both the experiment and the model, confirming the suggestion of a periodic structure in the DODAB/DOPE and DNA complexes. The periodicity of 6.5nm in the experimental PDDF for the lipid-DNA complex (Figure 3b) is similar to the spacing (7.0nm) obtained from the broad Bragg peak at approximately 0.9nm-1 (Figure 3a). We conclude that the complex formed by DODAB:DOPE and DNA is an aggregated multilamellar structure, similar to that observed by Barreleiro et al.13 The weak nature of the fluctuations in the PDDF for D > Dmax/2 indicates some polydispersity in the number of shells per aggregate.
Characterization of Complexes of Reduced BFDMA and DNA
Having determined that the NG3 beamline could be used to identify nanostructures of lipoplexes, we proceeded to characterize the nanostructures of complexes of BFDMA and plasmid DNA as a function of the oxidation state of BFDMA. Here we used plasmid DNA, rather than salmon sperm DNA, to allow more direct connection between these studies and previous light scattering and transfection studies of the BFDMA/plasmid DNA system.7-9
Our past studies established that complexes of reduced BFDMA and DNA formed at a charge ratio of 1.1:1 (lipid:DNA) result in high transfection efficiency.7,8 As mentioned above, this result guided the charge ratios investigated in the current study. When DNA was added to a solution of reduced BFDMA (1mM) to form a solution with a charge ratio of 1.1:1, visible yellow aggregates were observed to precipitate, resulting in a partial loss of color in the supernatant (faint yellow) and suggesting a decrease in the concentration of reduced BFDMA in solution. Although some sedimentation was observed in all solutions, the majority of the BFDMA remained in solution throughout the measurements (as determined by UV-visible spectrophotometry). Because the neutron beam passed through the center of the sample cell, the nanostructure of the precipitates was not characterized by our SANS measurements. Precipitate particles, hundreds of nanometers in size were, however, characterized by our direct imaging cryo-TEM measurements (see, for example, Figure 5C below). Furthermore, larger aggregates were examined by freeze-fracture-replication cryo-TEM, as shown in Figure S3C for oxidized BFDMA and DNA. The combined use of SANS and cryo-TEM characterization techniques, therefore, allowed analysis of all nanostructures in the BFDMA/DNA system. We note that sedimentation has previously been observed with complexes of cationic lipids and DNA, sometimes associated with the formation of hexagonal structures from solution.43
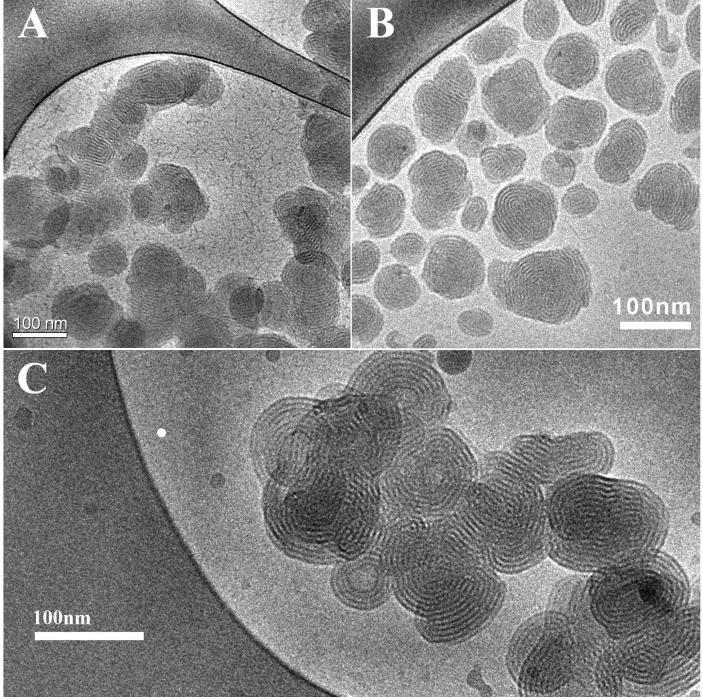
Figure 5.
Cryo-TEM images of lipoplexes of reduced BFDMA and DNA. A) 8 μM reduced BFDMA + 2.4 μg/ml DNA (charge ratio 1.1:1); B) 1mM reduced BFDMA + 2.9 mg/ml DNA (charge ratio 1.1:1); C) 1 mM reduced BFDMA + 2.9 μg/ml DNA (charge ratio 4.0: 1).
The SANS spectrum obtained using solutions of reduced BFDMA and DNA at a charge ratio of 1.1:1 is shown in Figure 4a. A peak observed at q = 1.2nm-1 corresponds to a periodic structure with a wavelength of 5.2nm, a dimension that is qualitatively similar to that observed with the DODAB/DOPE:DNA complexes at q = 0.9nm-1 (7.0nm, Figure 3a). The difference of 1.8nm in the periodicity observed between the two lipid systems (7.0nm and 5.2nm) may result from differences in the relative lengths of the lipids, the relative thicknesses of the water layers and the strength of binding of the DNA to the interface of the lipid assembly. We note that a spacing of 5.2nm is consistent with a model in which DNA is intercalated between lipid bilayers, where the double strand of DNA and a D2O layer have a combined thickness of 2.5nm 13 and the lipid has a thickness of 2.7nm. The latter thickness is close to that expected for a bilayer of reduced BFDMA, based on a molecular length of BFDMA of approximately 1.5nm in an all trans configuration.
Figure 4.
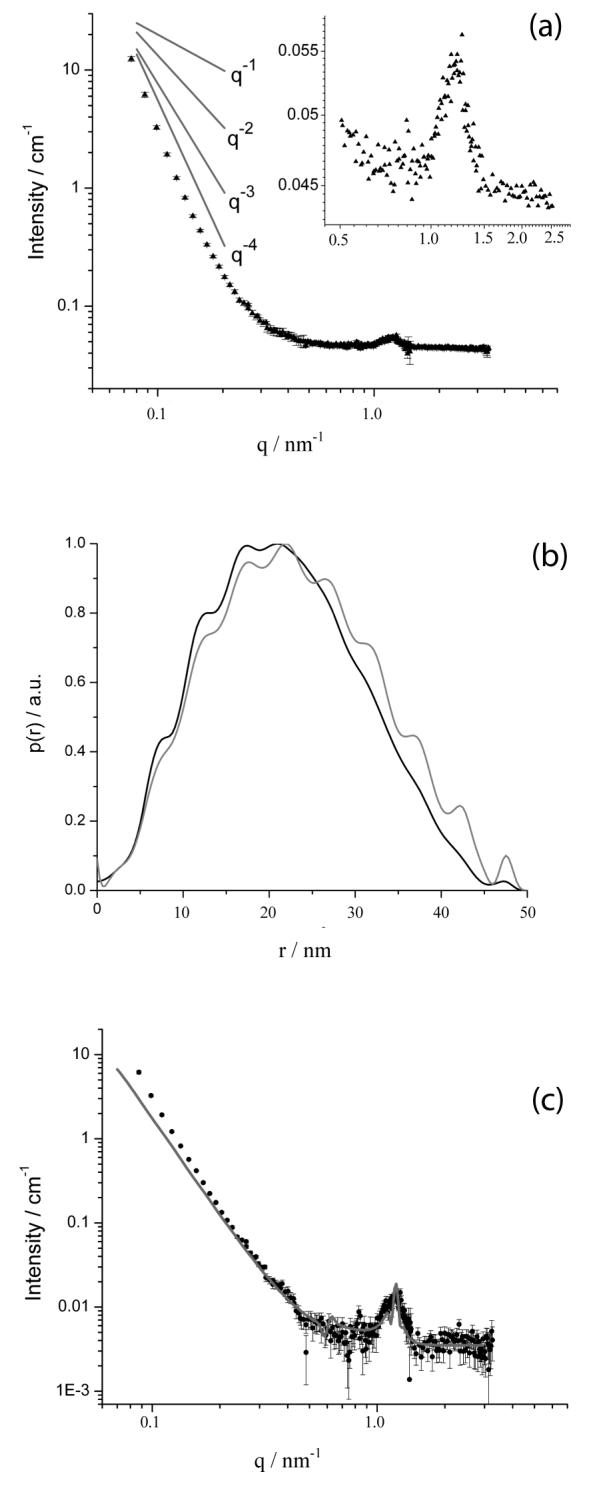
(a) SANS spectrum measured using an aqueous solution of reduced BFDMA (1mM) in the presence of DNA (2.9mg/ml) at a charge ratio of 1.1:1. The inset shows the peak observed at 1.2nm-1. (b) The PDDF calculated from IFT analysis of the data in (a) (dark line). For comparison, the gray line shows the PDDF calculated from a multilamellar vesicle model with a core radius of 5.0nm and 10 shells, where each shell has a thickness of 2.7nm and is spaced by a water layer of thickness 2.5nm. (c) Comparison of SANS spectrum shown in (a) to a form factor fit using on a multi-shell sphere model which includes polydispersity in the number of shells (gray line). The fit yields a core radius of 1.0nm surrounded by, on average, 6 shells. Each shell, of thickness 2.7nm, is separated by a 2.5nm water layer.
The absence of a secondary peak in the spectrum obtained from the reduced BFDMA:DNA complexes (Figure 4a) suggests that the periodicity associated with these nanostructures does not extend over large distances. The absence of multiple peaks (with spacing expected from a lamellar or hexagonal phase) also prevents the use of SANS to differentiate between different types of periodic nanostructures. However, the results of cryo-TEM reported below support the interpretation of the SANS data in terms of aggregates having a lamellar nanostructure. The similarity of the nature of the SANS spectrum obtained from lipoplexes prepared with reduced BFDMA with that obtained using DODAB/DOPE is consistent with our past study that noted similarities between these systems on the basis of dynamic light scattering and zeta potential measurements.9 SANS spectra obtained using complexes formed from reduced BFDMA and DNA at different charge ratios (1.1:1, 2.75:1 and 4:1) also showed the existence of a peak at 1.2nm-1, suggesting that the periodic structure formed does not change over this range of solution compositions (see Supporting Information).
The PDDF determined by IFT of the data presented in Figure 4a is shown in Figure 4b. The PDDF calculated from a model of a multilamellar vesicle (where the mean number of shells is 6) of core radius 5.0nm, shell thickness 2.7nm and water layer thickness 2.5nm has been included for comparison. As in the case of DODAB/DOPE:DNA complexes, the overall shape of the PDDF obtained from the scattering from the reduced BFDMA and DNA complexes is similar to the PDDF of the model multilamellar structure. The fluctuations in the PDDF obtained from the experimental data correspond to the presence of a structure with a periodicity of approximately 5.0nm, consistent with the spacing (5.2nm) obtained from the peak at q = 1.2 nm-1. Dmax, the maximum aggregate diameter, is a parameter required for the IFT analysis; Dmax of 50nm produced the best fit to the experimental data when performing the IFT analysis. As discussed below, cryo-TEM images suggest that a Dmax of 50nm corresponds approximately to the diameters of the lipoplexes (Figure 5). Similar to the lipoplexes formed using DOPE/DODAB, the weak nature of the fluctuations in the PDDF for D>Dmax/2 indicates polydispersity.
Figure 4c shows the form factor fit to the SANS data calculated using the multi-shell sphere model described above, where the multi-shell sphere model was evaluated by assuming that the scattering length density of the D2O and DNA layer is similar to that of D2O. The multi-shell spherical structure used as a model has a core radius of 5.0nm and each shell, of thickness 2.7nm, is separated by a water layer 2.5nm thick. The shell periodicity of 5.2nm is fixed, although the fit was insensitive to variations as large as ± 0.3nm in the thickness of the shell relative to the thickness of the water layer. Polydispersity in the number of shells was included in the model by assuming a Gaussian distribution about the mean number of six shells, as described above.
Inspection of Figure 4a reveals the scattering from reduced BFDMA and DNA complexes in the low q region to follow a q dependence that is close to q-4. The q-4 scattering behavior is predicted for the polydisperse multilamellar vesicle model (Figure 4c). Inspection of Figure 4c reveals that the measured scattering in the low q region deviates slightly from the form factor fit. This deviation likely results from the presence of large scale aggregation (for example, where two or more individual complexes associate to form a larger non-spherical multilamellar aggregate; see below for additional comments based on cryo-TEM imaging).
Cryo-TEM imaging provided additional insight into the nature of the periodic nanostructure formed by BFDMA and DNA, as well as the overall sizes of the lipoplexes and their state of aggregation. The cryo-TEM imaging was performed using samples having BFDMA concentrations comparable to those used in the previous transfection7,8 and dynamic light scattering8,9 experiments, as well as using samples formed at the higher BFDMA concentrations required for the SANS measurements. The nanostructures of the complexes of BFDMA and DNA were not observed to depend on the concentration of the BFDMA. The micrographs shown in Figure 5 (discussed below) are representative of all images obtained by cryo-TEM using lipoplexes formed using reduced BFDMA. Additional images are included as Supporting Information.
Figure 5 shows cryo-TEM images of complexes formed by reduced BFDMA at concentrations of BFDMA of 8 μM (Figure 5A) and 1 mM (Figure 5B and Figure 5C) in the presence of DNA at charge ratios of 1.1:1 (Figure 5A and Figure 5B) and 4:1 (Figure 5C). Inspection of these images reveals that the complexes are roughly spherical in shape, multilamellar and possess a compact core. Aggregation of the complexes (as clearly seen in Figure 5A) was a general characteristic of samples containing reduced BFDMA and DNA. The aggregates are polydisperse with diameters ranging from approximately 50 to 150nm. Smaller aggregates have fewer layers, but the periodicity measured from the images of 5.2nm ± 0.2nm is conserved between all aggregates, regardless of diameter. This periodicity is consistent with the position of the peak observed by SANS at 1.2nm-1 (Figure 4a). A value of Dmax of 50nm was used in the IFT analysis and is in good agreement with the diameters of the lipoplexes observed in the cryo-TEM images. The aggregates in the cryo-TEM images are polydisperse in nature, with a varying number of shells per aggregate. The maximum number of layers per aggregate observed from the cryo-TEM images is seven, consistent with the absence of a secondary peak in the SANS spectra. This latter finding was interpreted to indicate that the periodic structure does not persist over large length scales. The cryo-TEM images are also supportive of the form factor fit, which suggests polydispersity in the number of shells per aggregate. Although the form factor model of the multilamellar vesicle has a hollow core, the fit is not sensitive to core radii below 15nm. This core is not observed in cryo-TEM images. Here we also note that DNA is evident as thread-like nanostructures in the background of the micrographs shown in Figure 5A.
Characterization of Complexes of Oxidized BFDMA and DNA
Next, we sought to determine whether the nanostructures of complexes formed from oxidized BFDMA and DNA differed from those formed by reduced BFDMA and DNA. These measurements were performed at the same molar concentrations of BFDMA and plasmid DNA. As noted above, however, the oxidation of BFDMA results in a three-fold increase in the charge ratio (e.g. complexes formed by reduced BFDMA and DNA at a charge ratio of 1.1:1 contain the same molar concentrations of BFDMA and DNA as complexes formed by oxidized BFDMA and DNA at a charge ratio of 3.3:1). Prior to the addition of the DNA, the solutions of oxidized BFDMA were blue in color. Similar to complexes formed by reduced BFDMA and DNA, immediate aggregation and some precipitation was observed upon the addition of DNA to solutions of oxidized BFDMA. The solutions of oxidized BFDMA and DNA, however, remained blue in color. A white aggregate with a loose open floc-type structure was formed, which sedimented slowly with time.
For the purposes of comparison, Figure 6 shows the neutron scattering from complexes formed between reduced BFDMA and DNA (CR 1.1:1) and oxidized BFDMA in the presence of DNA (CR 3.3:1). Whereas complexes of reduced BFDMA and DNA show a Bragg peak at q = 1.2nm-1, no equivalent Bragg peak is observed (see Figure 6 inset plot) for complexes of oxidized BFDMA and DNA. This indicates that no measurable periodic nanostructure exists in the complexes formed by oxidized BFDMA and DNA. The marked difference in the scattering suggests that the interactions between DNA and BFDMA are strongly influenced by the lipid oxidation state.
Figure 6.
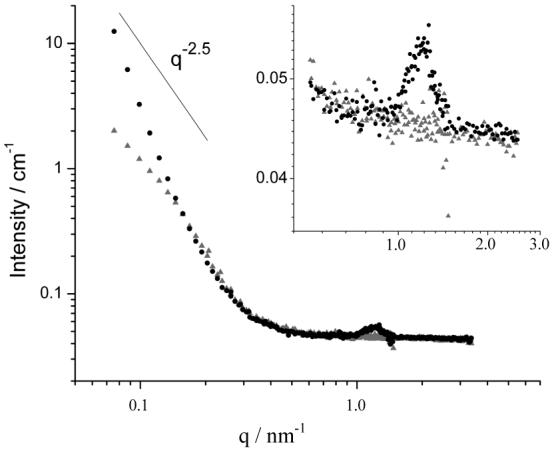
SANS spectra obtained using solutions of reduced (dark symbols, charge ratio 1.1:1) and oxidized (light symbols, charge ratio 3.3:1) BFDMA (1mM) in the presence of DNA (2.9mg/ml). A line with a slope of -2.5 is included for comparison.
The intensity of scattered neutrons measured at the low q detector setting was substantially lower for oxidized BFDMA and DNA complexes (112cps) than for the reduced BFDMA in the presence of DNA (342cps). This indicated that the extent of aggregation observed in samples containing oxidized lipid and DNA was much lower compared with the extent of aggregation observed in samples containing reduced lipid and DNA. No other peaks or features characteristic of a particular type of aggregate structure could be observed in the SANS data obtained with oxidized BFDMA and DNA.
The behavior of the SANS spectra in the low q region in Figure 6 also indicates that complexes formed by reduced BFDMA and DNA are very different in nanostructure from complexes formed by oxidized BFDMA and DNA. The scattering from reduced BFDMA and DNA complexes in the low q region showed a q dependence of q-4, similar to that predicted for the polydisperse multilamellar vesicle model. In the case of oxidized BFDMA, however, the low q scattering followed a slope of q-2.5. A q dependence of -2.5 in the low q region is consistent with the presence of a loose network structure similar to that observed in loose polymer networks or aggregate structures.44 This interpretation is supported by the cryo-TEM images shown in Figure 7A and 7B (oxidized BFDMA concentration of 1mM and a charge ratio of 3.3:1). Inspection of these images reveals the presence of either large, loose non-lamellar aggregates (Figure 7A; diameters between 10nm and 200nm) or a network of lamellae (Figure 7B). Both of these types of aggregate nanostructures could be described by a q dependence of - 2.5. The loose aggregate structure seen in Figure 7A is consistent with our previous measurements of dynamic light scattering and zeta potential, which indicated that oxidized BFDMA acts to condense DNA into loosely packed complexes with coronas enriched with negatively charged DNA.8,9 The formation of this type of nanostructure appears to result from ionic interactions between the oxidized BFDMA and DNA, similar to other non-self associating molecules with three positive charges (such as the interactions between DNA and spermidine).45-47
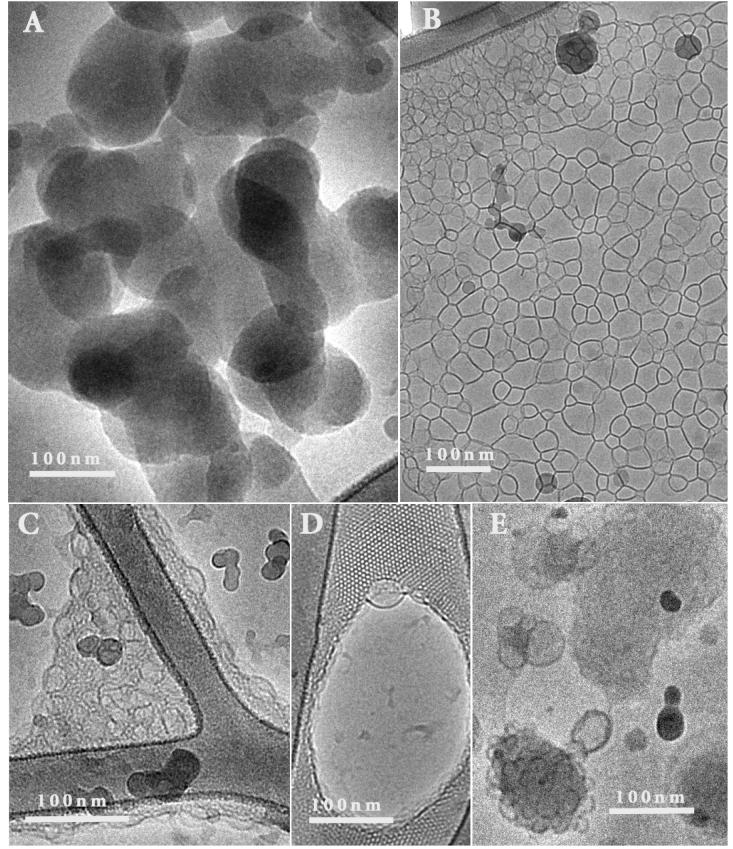
Figure 7.
Cryo-TEM images of complexes formed by oxidized BFDMA (1mM) and DNA (2.9mg/ml), at a charge ratio of 3.3. A) Amorphous, non-lamellar structures seen in most experiments; B) Network of lamellae; C) Dense sponge-like phase; D) Ordered cubic phase with vesicles emerging; E) Fused vesicles.
Interestingly, while the images in Figure 7A and 7B are representative of the nanostructures observed in the majority of samples, we also observed other nanostructures to be present in some samples (Figure 7C - Figure 7E). Figure 7C shows the presence of a dense sponge-like phase, Figure 7D shows an ordered cubic phase with vesicles emerging from the phase boundary, and Figure 7E shows fused vesicles. The factors that govern the formation of these complex and ordered nanostructures is not yet fully understood, and will be the subject of future study. It is possible that some of these nanostructures correspond to non-equilibrium states of the system.
Conclusions
In summary, the results above demonstrate that the nanostructures of complexes formed by the redox-active lipid BFDMA and DNA depend upon the oxidation state of the BFDMA. Upon addition of DNA to reduced BFDMA, complexes with multilamellar nanostructures are formed, as evidenced by cryo-TEM images and an analysis of SANS data by both the IFT technique and form-factor fitting. The presence of a single Bragg peak in the SANS measurement indicates a fixed periodic nanostructure of 5.2nm comprising two layers: a water/DNA layer and a lipid bilayer. The absence of additional Bragg peaks in the SANS data suggests that this multilamellar structure does not extend over large distances. These results are in good agreement with cryo-TEM images of the complexes, which demonstrate the presence of complexes between 50 and 150nm in diameter. The nanostructures formed by reduced BFDMA and DNA are similar to the nanostructures observed with complexes of DNA and a classical lipid system. In comparison to complexes formed using reduced BFDMA, the nanostructures of complexes of oxidized BFDMA and DNA are markedly different. The absence of peaks in the SANS data suggests that a periodic nanostructure does not exist within these complexes. The scattering in the low q region follows a slope that is greater than -2, indicating the presence of a loose, large scale aggregate structure. This result is consistent with the results of our cryo-TEM studies involving oxidized BFDMA and DNA. In the majority of these experiments, cryo-TEM images revealed predominantly loose, disordered aggregates of oxidized BFDMA and DNA. Some highly ordered sponge-like and cubic phase nanostructures were also detected in cryo-TEM images.
The key finding of the study reported in this paper is that the oxidation state of BFDMA impacts the nanostructure of complexes of BFDMA and DNA. This finding is significant as it suggests a nanostructural origin to previously reported changes in the properties of these complexes that are known to generally influence the efficiency of transfection of cells. In particular, we note that we previously reported that the oxidation state of BFDMA impacts the zeta-potentials of complexes formed by DNA and BFDMA.9 Because uptake of lipid-DNA complexes into cells is known to be influenced by zeta-potential, the oxidation-state-dependent nanostructure of BFDMA-DNA complexes reported in this paper could influence transfection via its impact on zeta-potential. More broadly, our results, when combined, suggest that a full understanding of the influence of BFDMA oxidation state on transfection will require consideration of the dependence of BFDMA/DNA nanostructure on lipid oxidation state.
Supplementary Material
ACKNOWLEDGMENT
Financial support was provided by the NSF (CTS-0327489 and CTS-0553760), the NIH (EB-002746 and EB-006168), and the Arnold and Mabel Beckman Foundation. We acknowledge the support of the National Institute of Standards and Technology, U.S. Department of Commerce, in providing the neutron facilities used in this work. The cryo-TEM work was carried out in the Hannah and George Krumholz Laboratory for Advanced Microscopy, Part of the Technion Project on Complex Fluids, Microstructure and Macromolecules. Partial support for that work was provided by the Technion Russell Berrie Nanotechnology Institute (RBNI) and the Israel Science Foundation.
REFERENCES
- (1).Birchall JC, Kellaway IW, Mills SN. International Journal of Pharmaceutics. 1999;183:195. doi: 10.1016/s0378-5173(99)00117-9. [DOI] [PubMed] [Google Scholar]
- (2).Choosakoonkriang S, Wiethoff CM, Anchordoquy TJ, Koe GS, Smith JG, Middaugh CR. Journal of Biological Chemistry. 2001;276:8037. doi: 10.1074/jbc.M010592200. [DOI] [PubMed] [Google Scholar]
- (3).Felgner PL, Tsai YJ, Sukhu L, Wheeler CJ, Manthorpe M, Marshall J, Cheng SH. Annals of the New York Academy of Sciences. 1995;772:126. doi: 10.1111/j.1749-6632.1995.tb44738.x. [DOI] [PubMed] [Google Scholar]
- (4).Miller AD. Angewandte Chemie International Edition. 1998;37:1768. [Google Scholar]
- (5).Safinya CR. Current Opinion in Structural Biology. 2001;11:440. doi: 10.1016/s0959-440x(00)00230-x. [DOI] [PubMed] [Google Scholar]
- (6).Nicolazzi C, Garinot M, Mignet N, Scherman D, Bessodes M. Current Medicinal Chemistry. 2003;10:1263. doi: 10.2174/0929867033457467. [DOI] [PubMed] [Google Scholar]
- (7).Abbott NL, Jewell CM, Hays ME, Kondo Y, Lynn DM. Journal of the American Chemical Society. 2005;127:11576. doi: 10.1021/ja054038t. [DOI] [PubMed] [Google Scholar]
- (8).Jewell CM, Hays ME, Kondo Y, Abbott NL, Lynn DM. Journal of Controlled Release. 2006;112:129. doi: 10.1016/j.jconrel.2006.01.014. [DOI] [PubMed] [Google Scholar]
- (9).Hays ME, Jewell CM, Kondo Y, Lynn DM, Abbott NL. Biophys J. 2007;93:4414. doi: 10.1529/biophysj.107.107094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Almofti MR, Harashima H, Shinohara Y, Almofti A, Baba Y, Kiwada H. Archives of Biochemistry and Biophysics. 2003;410:246. doi: 10.1016/s0003-9861(02)00725-7. [DOI] [PubMed] [Google Scholar]
- (11).Widom J, Baldwin RL. Journal of Molecular Biology. 1980;144:431. doi: 10.1016/0022-2836(80)90330-7. [DOI] [PubMed] [Google Scholar]
- (12).Raspaud E, Chaperon I, Leforestier A, Livolant F. Biophysical Journal. 1999;77:1547. doi: 10.1016/S0006-3495(99)77002-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Barreleiro PCA, May RP, Lindman B. Faraday Discussions. 2002;122:191. doi: 10.1039/b200796g. [DOI] [PubMed] [Google Scholar]
- (14).Koltover I, Salditt T, Radler JO, Safinya CR. Science (Washington, D. C.) 1998;281:78. doi: 10.1126/science.281.5373.78. [DOI] [PubMed] [Google Scholar]
- (15).Mel’nikova YS, Mel’nikov SM, Lofroth J-E. Biophysical Chemistry. 1999;81:125. doi: 10.1016/s0301-4622(99)00088-5. [DOI] [PubMed] [Google Scholar]
- (16).Pitard B. Biotechnology Intelligence Unit. 2003;7:1. [Google Scholar]
- (17).Caracciolo G, Caminiti R. Chemical Physics Letters. 2005;411:327. [Google Scholar]
- (18).Alfredsson V. Current Opinion in Colloid & Interface Science. 2005;10:269. [Google Scholar]
- (19).Simberg D, Weisman S, Talmon Y, Barenholz Y. Critical Reviews in Therapeutic Drug Carrier Systems. 2004;21:257. doi: 10.1615/critrevtherdrugcarriersyst.v21.i4.10. [DOI] [PubMed] [Google Scholar]
- (20).Weisman S, Hirsch-Lerner D, Barenholz Y, Talmon Y. Biophys J. 2004;87:609. doi: 10.1529/biophysj.103.033480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Sternberg B, Sorgi F, Huang L. FEBS Letters. 1997;356:361. doi: 10.1016/0014-5793(94)01315-2. [DOI] [PubMed] [Google Scholar]
- (22).Natali F, Castellano C, Pozzi D, Castellano AC. Biophysical Journal. 2005;88:1081. doi: 10.1529/biophysj.104.042788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Hays ME, Jewell CM, Lynn DM, Abbott NL. Langmuir. 2007;23:5609. doi: 10.1021/la0700319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Hays ME, Abbott NL. Langmuir. 2005;21:12007. doi: 10.1021/la051640o. [DOI] [PubMed] [Google Scholar]
- (25).Yoshino N, Shoji H, Kondo Y, Kakizawa Y, Sakai H, Abe M. Journal of the Japanese Oil Chemistry Society. 1996;45:769. [Google Scholar]
- (26).Bennett DE, Gallardo BS, Abbott NL. Journal of the American Chemical Society. 1996;118:6499. [Google Scholar]
- (27).NIST SANS Analysis; Vol. http://www.ncnr.nist.gov/programs/sans/data/data_anal.html.
- (28).Guinier A, Fournet G. Small Angle X-ray Scattering. John Wiley & Sons; New York, NY: 1955. [Google Scholar]
- (29).Glatter O. Journal of Applied Crystallography. 1977;10:415. [Google Scholar]
- (30).Porod G. Small Angle Scattering. London; Academic Press: 1982. [Google Scholar]
- (31).Brasher LL, Kaler EW. Langmuir. 1996;12:6270. [Google Scholar]
- (32).Fritz G, Bergmann A, Glatter O. Journal of Chemical Physics. 2000;113:9733. [Google Scholar]
- (33).Hassan P, Fritz G, Kaler EW. Journal of Colloid and Interface Science. 2003;257:154. doi: 10.1016/s0021-9797(02)00020-6. [DOI] [PubMed] [Google Scholar]
- (34).Gonzalez YI, Stjerndahl M, Danino D, Kaler EW. Langmuir. 2004;20:7053. doi: 10.1021/la0493464. [DOI] [PubMed] [Google Scholar]
- (35).Guinier A, Fournet G. Small Angle X-ray Scattering. John Wiley & Sons; New York: 1955. [Google Scholar]
- (36).Svergun DI, Koch MHJ. Reports on Progress in Physics. 2003;66:1735. [Google Scholar]
- (37).Glatter O, Bergman A, Brunner-Popela J, Fritz G, Mittelbach R, Weyerich B. Graz, Austria: 2000. GIFT for Windows; 1.01.02 ed. [Google Scholar]
- (38).Kline SR. Journal of Applied Crystallography. 2006;39:895. [Google Scholar]
- (39).NIST Scattering Length Density Calculator; Vol. http://www.ncnr.nist.gov/resources/sldcalc.html.
- (40).Bellare JR, Davis HT, Scriven LE, Talmon Y. Journal of Electron Microscopy Technique. 1988;10:87. doi: 10.1002/jemt.1060100111. [DOI] [PubMed] [Google Scholar]
- (41).Danino D, Bernheim-Groswasser A, Talmon Y. Colloids and Surfaces a-Physicochemical and Engineering Aspects. 2001;183:113. [Google Scholar]
- (42).Cardenas M, Dreiss CA, Nylander T, Chan CP, Cosgrove T, Lindman B. Langmuir. 2005;21:3578. doi: 10.1021/la047251w. [DOI] [PubMed] [Google Scholar]
- (43).Zuhorn IS, Hoekstra D. Journal of Membrane Biology. 2002;189:167. doi: 10.1007/s00232-002-1015-7. [DOI] [PubMed] [Google Scholar]
- (44).Beaucage G. Journal of Applied Crystallography. 1996;29:134. [Google Scholar]
- (45).de Frutos M, Raspaud E, Leforestier A, Livolant F. Biophysical Journal. 2001;81:1127. doi: 10.1016/S0006-3495(01)75769-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).He SQ, Arscott PG, Bloomfield VA. Biopolymers. 2000;53:329. doi: 10.1002/(SICI)1097-0282(20000405)53:4<329::AID-BIP5>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- (47).Raspaud E, Chaperon I, Leforestier A, Livolant F. Biophysical Journal. 1999;77:1547. doi: 10.1016/S0006-3495(99)77002-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.