Abstract
Branching processes such as nerves and vessels share molecular mechanisms of path determination. Our study focuses on unc5b, a member of the unc5 axon guidance gene family. Here, we have cloned the full-length zebrafish ortholog of unc5b, mapped its chromosome location in the zebrafish genome, and compared its expression patterns to robo4, another axon guidance family member. In situ show that unc5b is expressed predominantly in sensory structures such as the eye, ear, and brain. Both unc5b and robo4 show robust expression in all three compartments of the embryonic brain, namely forebrain, midbrain, and hindbrain. In particular, the hind-brain rhombomere expression displays interesting patterns in that robo4 is expressed in medial rhombomere cell clusters when compared to unc5b expressed in lateral rhombomere clusters. A similar medial–lateral theme is observed in other neural structures such as the neural tube. Our expression analysis provides a starting point for studying the role of axon guidance genes in embryonic hindbrain patterning.
Key words: unc5b, robo4, Hindbrain, Axon guidance, Zebrafish, Staining
INTRODUCTION
Neural and vascular patterning networks are intricate branching processes that are precisely controlled by cues from surrounding milieu (3). Four different classes of axon guidance molecules and their cognate receptors, namely the ephrins-Eph, semaphorinsplexin, netrin-unc5, and slit-roundabout (robo), are responsible for orchestrating and coordinating axon guidance to their target. unc5b and robo4 are members of the unc5 and robo family, respectively, and are expressed in both neural and vascular systems. This study focuses on the neuronal expression of unc5b, and compares its brain expression patterns to robo4. unc5b, like robo4, is a single pass transmembrane receptor and contains immunoglobulin domains in the extracellular region. In the intracellular region, unc5b contains unique cytoplasmic motifs such as zonula oc-cludens-1 (ZU-1) and death domains (1). Prior to this study, a partial unc5b zebrafish clone was reported (8), but the temporal and spatial expression patterns during embryonic development were not reported. Here, we have cloned the full-length ortholog of unc5b, characterized its genome location, performed in situ across developmental stages, and compared its neural expression patterns to robo4 in the developing vertebrate brain.
MATERIALS AND METHODS
Zebrafish Stocks and Reagents
Zebrafish were grown and maintained at 28.5°C (12) under MCW animal protocol number 312-06-2 guidelines. Mating was routinely carried out at 28.5°C and the embryos were staged according to established protocols (6).
RT-PCR and Antisense Probe Generation
Total RNA isolated from 24 hpf embryos was subjected to RT using oligo dT primer. Primers used to amplify were (a) CGGACTCGTTTCCATCAGCT CCT, (b) GCGTGCACATAATCCCTCAGTGC, (c) CAGGAAGTATTAGAGGTTGA, (d) GCAATCGC CATCTGTCGT, (e) GAATTCCAGAGTGAGCCC GAGGA, (f) CTGACATTCACGACTGCGCCAAT, using PCR parameters previously described (2). The PCR products were cloned into pCR4Topo vectors, sequenced from both ends, and antisense unc5b DIG RNA probe was made with SpeI linearized vector using T7 RNA polymerase. The robo4 probe was generated as described previously (2).
Radiation Hybrid (RH) Mapping
The Zebrafish/Hamster radiation hybrid panel was obtained from University of Ottawa. PCR amplification was performed using the following primer pairs: forward: CATGTCTGCCTTCGTCTCAA, reverse: CCAGCATTCTCCAGTCACAG. The 96-well PCR samples were analyzed on 3% agarose gel and stained with ethidium bromide. The score was submitted to the Zon Lab Web site (http://134.174.23.167/zonrh-mapper/instantMapping.htm) for instant mapping information.
In Situ Hybridization and Sections
WT embryos were grown in 0.003% phenylthiourea until the desired stage, fixed overnight in 4% paraformaldehyde/PBS at 4°C, dechorionated, and stored in 100% methanol until use. Single probe whole mount in situ hybridization was carried out as described previously (15). Serial transverse robo4 and unc5b sections were generated from Spurr’s epoxy resin embedded 24 hpf embryos. Embryos were washed three times in PBS, pH 7.4, dehydrated in ethanol and propylene oxide, embedded into Spurr’s epoxy resin, and cured in an oven for 18 h at 68 °C. Sections 1–2 μm thick were made on a Pyramitome with glass knives, floated on a drop of distilled water on a microscopic slide, and dried on a hot plate. The slides were mounted with coverslip and Cytoseal XYL medium (Richard-Allan Scientific). In situ whole mount pictures were captured with a Zeiss SVII Epi-Fluorescence stereomicroscope, and sections were captured with Carl Zeiss Axioskop2 plus upright microscope equipped with 40× or 63× differential interference contrast (DIC) objective lens and an Axiocam HRC camera. For flat mount, the yolk was removed in glycerol and the deyolked tissue was mounted on a slide containing mounting medium with a coverslip.
RESULTS AND DISCUSSION
Cloning and Mapping of unc5b
We cloned the zebrafish ortholog of human unc5b gene by extracting nucleotide sequences from Sanger zebrafish shotgun genome sequences that matched the amino acids of unc5b. Because members of the unc5 family are highly homologous to each other, we used synteny as another parameter of orthology determination. We used a two-prong approach where first, regions of unc5b that were least conserved between the members of the family or uniquely identifiable in unc5b were used for searching the public databases, and second, the hits were blasted to potential linkage groups. If the region was syntenic to human unc5b, then we proceeded with further analysis. Using the two-prong blast-mapping approach, we located two sequences that hit the extreme 5′ and 3′ end of unc5b, and amplified these fragments individually first (Fig. 1A, lanes 1 and 2) followed by nested PCR primer sets e and f, which confirmed the bands (Fig. 1A, lane 3). To determine linkage between the two sequences, PCR was performed with primers located at the 5′ end of the first sequence (primer a) and 3′ end of the second sequence (primer d), which generated a 2.8-kb band (Fig. 1A, lane 4). Subsequent blastn analysis of the 2.8-kb sequence mapped to linkage group 13 (data not shown), and the sequence was deposited to GenBank (accession #DQ365815). We verified the mapping data from SSAHA server by performing radiation hybrid mapping, which confirmed the location (data not shown). Linkage group 13 is syntenic to human chromosome 10 where human unc5b gene is located. Because unc5b was mapped to linkage group 13, where cloche mutant maps to, we checked by RT-PCR if unc5b was expressed in cloche m39 (11) deletion allele, and it was present (data not shown), suggesting that cloche is not unc5b.
Figure 1.
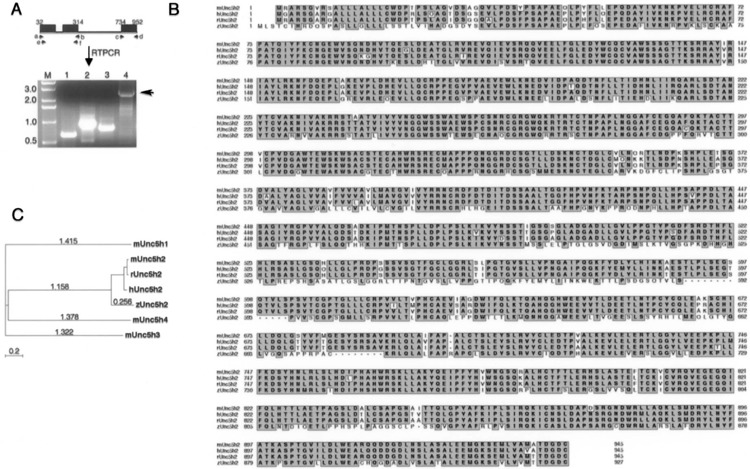
Cloning and phylogenetic analysis of unc5b.(A) The black boxes depicts the three exons that correspond to human unc5b amino acids, which hit putative zebrafish nucleotide sequence in Tblastn searches of Sanger zebrafish genome database. The six primer pairs (a to f) used for PCR are shown by a small black arrow. Lane M: marker (kb); lane 1: c and d; lane 2: a and b; lane 3: e and f: lane 4: a and d. Arrow indicates the 2.8-kb fragment of unc5b, which encompasses most of the unc5b gene except the first 35 amino acids. (B) Phylogenetic comparisons of zebrafish unc5b to unc5b members in other species and to mouse unc5 family (1, 3, and 4) are shown. The CLUSTALW program, as part of the MacVector 7.0 Software package was used for generating the tree and the accession numbers for sequences used here are zUnc5b (DQ365815), mUnc5h1 (NP_694771.1), mUnc5h3 (NP_033498), mUnc5H4 (NP_694775), mUnc5b (NP_084046), rUnc5b (NP_071543), and hUnc5b (NP_734465). Evolutionary tree was built using the “Neighbor joining” method, which does not assume constant divergent rates among sequences, and evolutionary distances were calculated using “Best tree” method, which estimates the number of substitutions per site under the assumption that the distribution follows a poisson distribution. (C) Sequence homology comparisons with different unc5b family members are shown. The human, mouse, and zebrafish unc5b amino acid sequences were aligned using CLUSTALW program in MacVector 7.0. Pairwise comparisons between human and mouse sequences showed 90% homology while between human or mouse to zebrafish unc5b was 65%. Unc5h1, Unc5h2, Unc5h3, and Unc5h4 is the old nomenclature for Unc5a, Unc5b, Unc5c, and Unc5d, respectively.
The zebrafish unc5b open reading frame encodes a protein of 929 amino acids (aa) with a single trans-membrane domain and shares 65% amino acid identity to the human or mouse protein (Fig. 1B). Phylogenetic tree analysis was performed with amino acid sequences of zebrafish, mouse, human, and rat unc5b to mouse unc5 family members. The cladogram depicts that zebrafish unc5b is closer to human unc5b than mouse unc5b (Fig. 1C). Based on amino acid identity, phylogenetic and synteny relationships, we have cloned the putative full-length ortholog of unc5b in zebrafish. These data also confirm the partial published unc5b sequence (8), which encodes 452 amino acids.
Whole Mount unc5b In Situ Hybridization in Embryonic Zebrafish Development
A 2.8-kb digoxigenin (DIG) RNA probe was synthesized, and whole mount in situ was performed as described previously (2). unc5b expression commences postgastrulation in 12 somite embryo, and is expressed in the head, tail, and future dorsal side of the embryo (Fig. 2A, B). In the dorsal head region (Fig. 2C), unc5b is expressed in three distinct regions of the neural plate (asterisks), which outlines the general location of sensory and motor neurons. The unc5b-expressing lateral sensory neural plate cells (Fig. 2C, black asterisk) are symmetrically aligned on either side of the central motor neuron region (Fig. 2C, white asterisk) of the neural plate. In 14 somite embryos, unc5b expression is ubiquitous with strong expression seen in epidermal cells that line the embryo architecture (Fig. 2D). Over the course of the next few hours in development unc5b expression is restricted primarily to the brain. At 25 hpf, unc5b expression is seen in the eye, forebrain, hindbrain, and otic vesicle (Fig. 2E). Dorsal view of the 25 hpf head region (Fig. 2F) shows prominent expression in eye, otic vesicle, and forebrain telencephalon regions. At 30 hpf (Fig. 2G), unc5b expression is robust in anterior regions with dorsal view (Fig. 2H) showing expression in all three regions of the brain and otic vesicle. The expression clearly crosses into dorsal hindbrain regions, and continues into the dorsal roof plate (Fig. 2G) in caudal regions of the embryo.
Figure 2.
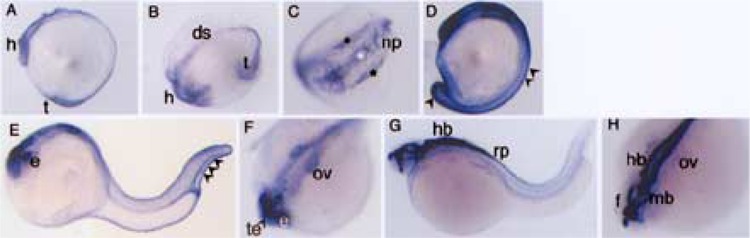
Expression patterns of unc5b in zebrafish embryonic development. A montage of unc5b whole mounts in situ stained embryos is shown. (A, B, C) 12 somite, (D) 14 somite, (E, F) 25 hpf, (G, H) 30 hpf. (A), (D), (E), and (G) are lateral view. (B), (C), (F), and (H) are dorsal views. The arrowheads in (D) and (E) indicate outer epidermal cells. The black asterisk and white asterisk in (C) indicate lateral sensory neural plate cells and central motor neuron region, respectively. Abbreviations used are ds: dorsal side, f: forebrain, h: head, e: eye, hb; hindbrain, mb: midbrain, np: neural plate, ov: otic vesicle, rp: roof plate, t: tail, te: telencephalon.
unc5b Expression Comparison to robo Family Member in Embryonic Zebrafish Brain Development
When performing whole mount unc5b in situ at 24 hpf we noticed that unc5b was expressed in lateral rhombomere cells in contrast to robo4, which we already knew was expressed in middle rhombomere cells in the same temporal window. To check whether this differential exclusivity in the gene expression patterns for unc5b and robo4 at 24 hpf is preserved in early and late embryos, we expanded our analysis to 16–18 hpf and 48 hpf time points. At 16 hpf (Fig. 3A′), robo4 expression is observed in telencephalon, diencephalon, and mesencephalon with clear absence in the midbrain–hindbrain boundary. Further, robo4 expression continues dorsally into the neural tube and in the notochord (Fig. 3A). unc5b expression is ubiquitous during this time period (Fig. 3D). At 24 hpf (Fig. 3B′, E′), a pattern of differential exclusive expression for the two genes is pronounced in the hind-brain regions anterior and posterior to the otic vesicle. In hindbrain, robo4 expression is restricted to the medial cluster of rhombomere cells (Fig. 3B′, white asterisk) while unc5b expression is restricted to the lateral clusters (Fig. 3E′, black asterisk). The rhom-bomere boundaries in the 24 hpf robo4 hindbrain also show disproportionate staining with more expression in the middle rhombomere boundaries (Fig. 3B′) when compared to the end ones. Both genes show expression in the otic vesicle (Figs. 3B′, E′). A consolidation of the 24 hpf unc5b and robo4 hindbrain rhombomere expression patterns at 24 hpf is depicted in Figure 3G. At 48 hpf, the robo4 brain expression pattern is remarkably defined with clear demarcated regions of expression (Fig. 3C′) in the optic tectum, rhomboencephalon, mesencephalon, and diencephalon and distinct lack of robo4 expression in the central region of the midbrain (Fig. 3C′). The telencephalon expression is also interrupted with regions of strong expression and no expression (Fig. 3C′, arrow). In the case of unc5b, brain expression is ubiquitous at 48 hpf and specific expression is noticed in the heart, cloaca, and posterior pronephric duct cells (Fig. 3F, black arrows). The expression patterns of netrins, ligands for unc5b, and slits, ligands for robos, have been previously reported. For netrins, only netrin-2 is observed in the fourth rhombomere during unc5b expression in rhombomere boundaries (14), and for slits, slit1a and slit1b are expressed in the bilateral cell clusters of hindbrain and floorplate (5) when robo4 is expressed.
Figure 3.
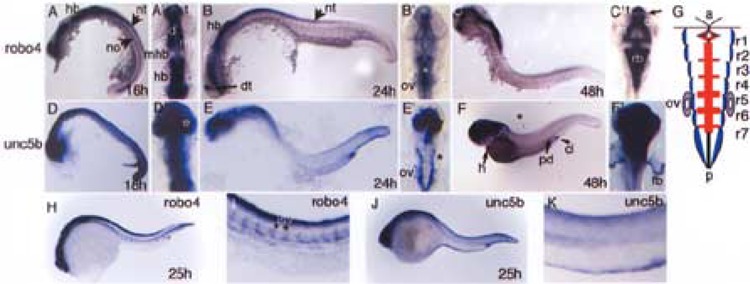
Comparisons of robo4 and unc5b brain expression patterns during zebrafish embryonic brain development. Wild-type embryos probed for robo4 and unc5b transcripts across three comparable developmental stages of brain development are shown. For each whole mount in situ, corresponding flat mount of the head region is depicted. (A), (B), and (C) show robo4 in situ embryos at 16, 24, and 48 hpf, respectively, along with (A′), (B′), and (C′) showing the corresponding flat mounts of the head region. Similarly, (D), (E), and (F) show unc5b in situ embryos at 18, 24, and 48 hpf along with their corresponding flat mounts of head region in (D′), (E′), and (F′). Asterisks in (B′) medial clusters of rhombomere cells and in (E′) unc5b expression in bilateral cluster of cells in rhombomeres. Arrows in (A) and (B) depict neural structures neural tube and notochord; arrow in (C′) shows separation of robo4 and non-robo4 expression domains in telencepha-lon. Short arrow in (F) shows cloaca and long arrows depict posterior pronephric duct region and heart. (G) A pictorial representation of blue regions (unc5b, E′), red regions (robo4, B′) in the rhombomeres (r1–r7) of a 24 hpf developing zebrafish hindbrain along with otic vesicle shown in purple to depict both transcript expression. (H–K) In situs for robo4 and unc5b genes at 25 hpf with emphasis on vascular expression in the trunk region (I, K). Abbreviations used include a: anterior, cl: cloaca, d: diencephalon, e: eye, fb: fin bud, h: heart, hb: hindbrain, isv: intersomitic vessels, m: mesencephalon, mb: midbrain, mhb: midbrain hindbrain boundary, no: notochord, nt: neural tube, ov: otic vesicle, p: posterior, pd: pronephric duct, t: telencephalon.
Sectional Analysis of unc5b and robo4 In Situ Embryos
To map the expression domains in greater detail, we compared serial sections of 24 hpf unc5b and robo4 in situ embryos. Serial sections of 24 hpf unc5b in situ embryos beginning with the most anterior (Fig. 4A) and moving posteriorly (Fig. 4F) show expression in various sensory organs. In embryonic brain development, unc5b and robo4 are expressed in forebrain, midbrain, and hindbrain regions. In some sections, robo4 expression is focused in the central regions and is enveloped by unc5b expression in lateral position. For example, in the midbrain section, robo4 is predominantly expressed in central midbrain regions (Fig. 4G) when compared to unc5b expression in more lateral regions (Fig. 4A). This theme continues into the hindbrain, where unc5b expression is focused on more dorsolateral rhombomere cells (Fig. 4C, D) and robo4 in central rhombomere cells (Fig. 4I, J). In posterior sections (Fig. 4F, L), robo4 expression is predominant in the central neural tube (Fig. 4L) when compared to unc5b expression, which is restricted to roof plate (Fig. 4F). The lateral–dorsal preference patterns for the two genes does not hold true throughout the embryo. For example, in fore-brain telencephalon, no preference is noted for the expression patterns for both genes (Fig. 4B, H). Sensory structures such as eye and ear show prominent unc5b expression. In the developing eye, unc5b is robustly expressed in the ventral temporal retina (Fig. 4B), and both genes are expressed in the optic cup (Figs. 4A, G). Similarly, both genes show robust expression in the developing ear (Fig. 4E, K).
Figure 4.
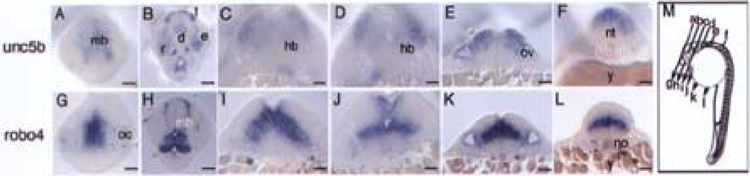
Comparative sectional analysis of embryonic brain regions of unc5b and robo4 in situ embryos. unc5b and robo4 24 hpf whole mount embryos were embedded in Spurr’s epoxy resin and sectioned as depicted in (M). All images were photographed at 40× magnification. Images were corrected for brightness, contrast, and sharpened in Adobe Photoshop. Scale bars: 50 μm. Abbreviations used include d: diencephalon, e: eye, hb: hindbrain, le: lens, mb: midbrain, no: notochord, nt: neural tube, oc: optic cup, ov: otic vesicle, r: retina, t: telencephalon, y: yolk.
Comparative Analysis of Mouse and Zebrafish unc5b Expression
Previously published mouse unc5b expression analysis (4) helps compare and contrast the zebrafish and mouse gene expression patterns. Spatially, the mouse and zebrafish genes show a high degree of similar expression profile in sensory organs but differences in other tissues. In the developing eye, both mouse and zebrafish genes show expression in the ventral temporal retina and lower towards the central retina. Further, lens expression is observed for zebra-fish unc5b (data not shown), but is absent for mouse unc5b (8). In the developing ear, unc5b expression is seen in both species with mouse unc5b expressed strongly in the semicircular canals (4) and zebrafish unc5b in the otic vesicles (Figs. 3E′ and 4E). In the vascular system, mouse unc5b is expressed in intersomitic vessels (ISVs) and CNS endothelial cells (4,8). In zebrafish, we failed to notice unc5b expression in developing ISVs using identical in situ conditions as that of age-matched robo4 in situ embryos (Fig. 3H, I). A recent study reported that unc5b knockdown embryos display ectopic ISVs sprouting defect (8), suggesting that unc5b + ISVs are guided to their target by cues from surrounding tissue. Another recent report (13) suggests that ISV sprouting defects in unc5b or netrins1a morphants are secondary to parachordal vessel defects. So it is unclear at this point whether ISV sprouting defects in unc5b morphants are primary or secondary. Interestingly, mouse robo4 is reported to be vascular specific (9) while the zebrafish robo4 is clearly not (2), suggesting that expression patterns for axon guidance genes differ across species.
Based on expression analysis in this study, unc5b’s original name derived from “uncoordinated” phenotype mutants in Caenorhabditis elegans correlates well with the expression in sensory tissues. In zebra-fish, unc5b is expressed in eye, ear, and brain, which corresponds to all relevant organ systems for coordination. Interestingly, netrin-1 mutant mice show severe semicircular canal malformations (10). unc5b is the only known unc5 member expressed in the semicircular canal (4). It is not known if ear defects were observed in unc5b knockout mice or unc5b morphant zebrafish embryos (8). Our expression analysis with unc5b and robo4 suggest that axon guidance genes may function during rhombomere specification in hindbrain development. Other axon guidance family members such as Ephrin-eph receptor–ligand signaling system have already been implicated in rhombomere formation by restricting cell intermingling and communication (7). Our studies not only extend these observations to other classes of axon guidance gene families such as unc5 and robos but also suggest that cooperative interactions between members of disparate axon guidance families may play a role in hindbrain development.
ACKNOWLEDGMENTS
The sequences that were used here were produced by the Danio Rerio Sequencing Group at the Sanger Institute and can be obtained from ftp://ftp.sanger.ac.uk/pub/zebrafish. Additional information regarding the Zebrafish sequencing project is available at http://www.sanger.ac.uk/Projects/D_rerio/. We thank Iain Drummond (MGH, HMS) and Ajay Chitnis (NICHD, NIH) for guidance on section interpretation, and Lyndsay Field of Charles River Laboratory for maintaining our fish stocks. R.R. is a recipient of NCI Scholar Award. The authors declare that they have no competing financial interests.
REFERENCES
- 1. Arakawa H. Netrin-1 and its receptors in tumorigenesis. Nat. Rev. Cancer 4:978–987; 2004. [DOI] [PubMed] [Google Scholar]
- 2. Bedell V. M.; Yeo S. Y.; Park K. W.; Chung J.; Seth P.; Shivalingappa V.; Zhao J.; Obara T.; Sukhatme V. P.; Drummond I. A.; Li D. Y.; Ramchandran R. roundabout4 is essential for angiogenesis in vivo. Proc. Natl. Acad. Sci. USA 102:6373–6378; 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Carmeliet P.; Tessier-Lavigne M. Common mechanisms of nerve and blood vessel wiring. Nature 436:193–200; 2005. [DOI] [PubMed] [Google Scholar]
- 4. Engelkamp D. Cloning of three mouse Unc5 genes and their expression patterns at mid-gestation. Mech. Dev. 118:191–197; 2002. [DOI] [PubMed] [Google Scholar]
- 5. Hutson L. D.; Jurynec M. J.; Yeo S. Y.; Okamoto H.; Chien C. B. Two divergent slit1 genes in zebra-fish. Dev. Dyn. 228:358–369; 2003. [DOI] [PubMed] [Google Scholar]
- 6. Kimmel C. B.; Ballard W. W.; Kimmel S. R.; Ullmann B.; Schilling T. F. Stages of embryonic development of the zebrafish. Dev. Dyn. 203:253–310; 1995. [DOI] [PubMed] [Google Scholar]
- 7. Klein R. Bidirectional signals establish boundaries. Curr. Biol. 9:R691–694; 1999. [DOI] [PubMed] [Google Scholar]
- 8. Lu X.; Le Noble F.; Yuan L.; Jiang Q.; De Lafarge B.; Sugiyama D.; Breant C.; Claes F.; De Smet F.; Thomas J. L.; Autiero M.; Carmeliet P.; Tessier-Lavigne M.; Eichmann A. The netrin receptor UNC5B mediates guidance events controlling morphogenesis of the vascular system. Nature 432:179–186; 2004. [DOI] [PubMed] [Google Scholar]
- 9. Park K. W.; Morrison C. M.; Sorensen L. K.; Jones C. A.; Rao Y.; Chien C. B.; Wu J. Y.; Urness L. D.; Li D. Y. Robo4 is a vascular-specific receptor that inhibits endothelial migration. Dev. Biol. 261:251–267; 2003. [DOI] [PubMed] [Google Scholar]
- 10. Salminen M.; Meyer B. I.; Bober E.; Gruss P. Netrin 1 is required for semicircular canal formation in the mouse inner ear. Development 127:13–22; 2000. [DOI] [PubMed] [Google Scholar]
- 11. Stainier D. Y.; Weinstein B. M.; Detrich H. W. 3rd; Zon L. I.; Fishman M. C. Cloche, an early acting zebrafish gene, is required by both the endothelial and hematopoietic lineages. Development 121:3141–3150; 1995. [DOI] [PubMed] [Google Scholar]
- 12. Westerfield M. The aebrafish book, 4th ed. Eugene: University of Oregon Press; 2000. [Google Scholar]
- 13. Wilson B. D.; Ii M.; Park K. W.; Suli A.; Sorensen L. K.; Larrieu-Lahargue F.; Urness L. D.; Suh W.; Asai J.; Kock G. A.; Thorne T.; Silver M.; Thomas K. R.; Chien C. B.; Losordo D. W.; Li D. Y. Netrins promote developmental and therapeutic angiogenesis. Science 313:640–644; 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Won Park K.; Urness L. D.; Senchuk M. M.; Colvin C. J.; Wythe J. D.; Chien C. B.; Li D. Y. Identification of new netrin family members in zebrafish: Developmental expression of netrin 2 and netrin 4. Dev. Dyn. 234:726–731; 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yeo S. Y.; Little M. H.; Yamada T.; Miyashita T.; Halloran M. C.; Kuwada J. Y.; Huh T. L.; Okamoto H. Overexpression of a slit homologue impairs convergent extension of the mesoderm and causes cyclopia in embryonic zebrafish. Dev. Biol. 230:1–17; 2001. [DOI] [PubMed] [Google Scholar]


