Abstract
Helicobacter hepaticus, a causative agent of chronic hepatitis and hepatocellular carcinoma in mice, possesses a hydrogenase and a urease, both of which are nickel-containing enzymes. Analysis of the genome sequence of H. hepaticus revealed a full set of accessory genes which are required for the nickel maturation of each enzyme in other micro-organisms. Erythromycin-resistant mutants were constructed in four of these genes, hypA, hypB, ureE and ureG. Controls for polar effect were provided for hypA or hypB mutants by disrupting each gene located immediately downstream, i.e. hp0809 or hypC, respectively. Urease and hydrogenase activities were determined for each strain with or without supplemented nickel in the medium. As expected, the ureE and the ureG mutants had negligible urease activity, but they retained normal levels of hydrogenase activity. Urease levels could not be increased by the addition of nickel to the medium. The H. hepaticus hypA and hypB strains were deficient in both urease and hydrogenase activities, suggesting that both gene products act in a similar fashion as their counterparts in H. pylori. However, in contrast with the analogous mutants of H. pylori, the addition of nickel into the growth medium failed to restore either urease or hydrogenase enzyme levels in the H. hepaticus hypA or hypB mutants, indicating a probably unique role for these genes in the mouse liver pathogen.
INTRODUCTION
Helicobacter hepaticus is a spiral-shaped Gram-negative microaerophilic bacterium (Fox et al., 1994). It has been shown to be a causative agent of hepatic and enteric inflammatory diseases and malignancy in mice (Solnick & Schauer, 2001). Therefore, it is an excellent model for studying mechanisms of bacterium-associated liver carcinogenesis. The best known member of the genus is Helicobacter pylori, which causes peptic ulcers and gastric cancers in humans (Blaser, 1995; Sipponen et al., 1998). In this well studied bacterium, two nickel-containing enzymes, the hydrogen uptake hydrogenase and urease, have been shown to be required for early colonization and persistence in the stomach (Eaton et al., 1991; Olson & Maier, 2002). H. hepaticus also possesses both enzymes (Beckwith et al., 2001; Mehta et al., 2005; Suerbaum et al., 2003). While it is known that H. hepaticus hydrogenase mutants are deficient in hydrogen-supported amino acid uptake and cannot cause liver lesions in mice (Mehta et al., 2005), the role of urease in H. hepaticus survival in the host remains to be demonstrated.
In every bacterium studied so far, the maturation of hydrogenase or urease is a complex process which requires several accessory proteins. In H. pylori, hydrogenase maturation is achieved with the help of Hyp proteins (HypABCDEF) (Benoit et al., 2004; Olson et al., 2001), while urease maturation requires a set of accessory Ure proteins (UreEFGH) (Mehta et al., 2003a; Voland et al., 2003). Interestingly, in the gastric pathogen two hydrogenase accessory proteins, HypA and HypB, have been shown to be involved in urease maturation as well (Olson et al., 2001). H. pylori HypA is known to bind nickel (Mehta et al., 2003b), as well as zinc (Kennedy et al., 2007). The involvement of HypA in the nickel activation of urease is mediated through an identified interaction between this accessory protein and UreE, another nickel-binding protein likely to be the preferred donor to the urease apoenzyme (Benoit et al., 2007). Studies on HypB support its role as a GTPase enzyme (Mehta et al., 2003b) and the same role has been assigned to the UreG accessory protein. In both the H. pylori hypA and hypB mutants, addition of excess nickel to the medium can fully or partially restore enzyme activities to wild-type hydrogenase or urease levels, respectively (Olson et al., 2001). In contrast, the urease activity is severely affected in both the H. pylori ureE and ureG mutants and nickel supplementation cannot restore it to wild-type levels (Benoit & Maier, 2003; Benoit et al., 2007; Mehta et al., 2003a); the hydrogenase activity is not affected in these mutants.
Analysis of the H. hepaticus genome sequence (Suerbaum et al., 2003) has revealed the presence of hypA and hypB gene homologues, which prompted us to check whether inactivation of either gene would have a pleiotropic effect on both the urease and hydrogenase activity, as observed in H. pylori. Therefore, both genes were disrupted by insertion of an erythromycin resistance cassette and the effect on nickel enzyme activities was determined. The same procedure was followed for the ureE and ureG genes. To ensure that the observed phenotype was not due to polar effects on the genes transcribed downstream of each targeted gene, genes located immediately downstream of either hypA or hypB (that is, hp0809 or hypC, respectively) were individually disrupted as controls. The effect of nickel supplementation in the medium on nickel-related enzyme activities (hydrogenase or urease) was investigated in the mutant strains as well as the control strains. Taken together, our results suggest that while H. hepaticus ureE and ureG (and hypC) have similar functions as their counterparts in H. pylori, hypA and hypB seem to play different mechanistic roles when compared to their respective homologues in H. pylori.
METHODS
Bacterial strains and growth conditions
Helicobacter hepaticus and Escherichia coli strains used in this study are listed in Table 1. H. hepaticus strain ATCC 51449 was the parent strain. H. hepaticus cells were routinely grown at 37 °C on Brucella agar (Difco) plates supplemented with 10% defibrinated sheep blood (BA plates) under microaerophilic conditions (1–2% O2, 5% CO2, N2). The effect of nickel supplementation on urease and hydrogenase activity was studied by adding 1, 5, 10 or 50 µg NiCl2 ml−1 into the medium. E. coli was grown aerobically in Luria–Bertani (LB) or Brain–Heart infusion (BHI) medium or on plates at 37 °C. When required, the following antibiotics were added into the medium: erythromycin (150 µg ml−1 in BHI or 5 µg ml−1 in BA), ampicillin (100 µg ml−1 in LB) or chloramphenicol (30 µg ml−1 in LB or 20 µg ml−1 in BA).
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics* | Source or reference |
|---|---|---|
| H. hepaticus | ||
| 51449 | Parent strain for all H. hepaticus strains | ATCC† |
| hypA mutant | hypA::ery; Err | This study |
| hypB mutant | hypB::ery; Err | This study |
| hypC mutant | hypC::ery; Err | This study |
| ureE mutant | ureE::ery; Err | This study |
| ureG mutant | ureG::ery; Err | This study |
| ΔureG mutant | ΔDureG::ery; Err | This study |
| hh0809 mutant | hh0809::ery; Err | This study |
| E. coli | ||
| TOP10 | Cloning strain | Invitrogen |
| Plasmids | ||
| pGEM-T | Cloning vector; Apr | Promega |
| pBC-KS | Cloning vector; Cmr | Stratagene |
| pBluescript-KS | Cloning vector; Apr | Stratagene |
| pSLB167 | pBC-KS with ery (EcoRV); Cmr Err; source of ery cassette | Mehta et al. (2007) |
| pGEM-hypA | pGEM with a 760 bp sequence containing H. hepaticus hypA and part of hh0809 | This study |
| pSLB164 | pGEM-hypA with ery gene inserted within NruI of hypA | This study |
| pGEM-hypB | pGEM with a 814 bp sequence containing H. hepaticus hypB | This study |
| pSLB165 | pGEM-hypB with ery gene inserted within HindIII of hypB | This study |
| pGEM-ureE | pGEM with a 734 bp sequence containing H. hepaticus ureE | This study |
| pSLB166 | pGEM-ureE with ery gene inserted within HindIII of ureE | This study |
| pBC-ureG | pBC-KS with a 904 bp sequence containing H. hepaticus ureG | This study |
| pSLB173 | pBC-ureG with ery gene inserted within AvaII of ureG | This study |
| pGEM-ureG | pGEM with a 904 bp H. hepaticus ureG with an engineered HindIII | This study |
| pSLB175 | pGEM-ureG with ery gene inserted within HindIII of ureG | This study |
| pSLB290 | pGEM-hypA deleted for MluI (PstI/NsiI-digested; religated) | This study |
| pSLB293 | pSLB290 with ery inserted within MluI of hh0809 | This study |
Apr, Ampicillin resistance; Cmr, chloramphenicol resistance; Err, erythromycin resistance.
ATCC, American Type Culture Collection, Manassas, VA, USA.
Construction of site-directed H. hepaticus mutants
Genomic DNA from strain 51449 was used as a template to amplify hypA and part of the hp0809 gene, hypB, hypC, ureE or ureG. Primers (Table 2) were designed to amplify each gene together with some flanking sequences in order to have at least 200 bp on each side of the restriction site used for the marker insertion. PCR products of 760, 814, 734 or 904 bp were obtained for hypA and part of hh0809, hypB, ureE or ureG, respectively, and cloned into either pGEM-T or pBC-KS vector (Table 1), before being transformed into E. coli. To generate allelic exchange vectors for construction of hypA, hypB or ureE mutants, an 1140 bp long ery cassette conferring resistance to erythromycin (Mehta et al., 2007; Stabb & Ruby, 2002) was inserted into a unique restriction site within each gene (Table 1 and Fig. 1). Since there was no convenient unique site within ureG, two different approaches were followed. First, a new HindIII restriction site was generated in this gene using the overlapping PCR approach; briefly, two PCR products were obtained by using either primers UpureG with UreGmut2 or RevureG with UreGmut1, gel-purified, mixed together and a final PCR product containing a new HindIII site was obtained after amplification with UpureG and RevureG primers. This PCR product was cloned into pGEM-T vector and the new restriction site was used to insert the ery gene, generating plasmid pSLB175 (Table 1). For the second approach, a ΔureG::ery vector was constructed by cloning the 904 bp PCR product containing ureG into pBC-KS; two AvaII sites present within ureG were used to insert the ery gene, generating plasmid pSLB173 (Table 1).
Table 2.
Primers used in this study
| Name | Sequence (5′–3′)* | Usage |
|---|---|---|
| UphypA | ataggtgcatcaccgcctactg | Construction of hypA and hh0809 mutant |
| RevhypA | tttgctcaatggcgcgaagg | Construction of hypA and hh0809 mutant |
| UphypB | gccaaacacattcttgctcc | Construction of hypB mutant |
| Revhyp B | cgcgagtattagtttcgccc | Construction of hypB mutant |
| UphypC | ttgtgagcaaggcagatatg | Construction of hypC mutant |
| Revhyp C | cttcaagcactataatagg | Construction of hypC mutant |
| HypC-ery1 | GCGATACCGTCGAGATtacaactgccatattattg | Construction of hypC mutant |
| HypC-ery2 | CTAGCGATAAGCTTgatacattaggcgtgag | Construction of hypC mutant |
| UpureE | ggcttacaggctggattgaaag | Construction of ureE mutant |
| RevureE | tgcacatagctttctagccc | Construction of ureE mutant |
| UpureG | ctcaaagcgatgggcaagag | Construction of ureG mutant |
| RevureG | atcctcatcttctagtgggc | Construction of ureG mutant |
| UreGmut1 | AGAGAAGAAgcttctatgaatcttg | Creation of HindIII within ureG |
| UreGmut2 | TCATAGAAGCTtcttctctgattgctg | Creation of HindIII within ureG |
Lower-case letters indicate H. hepaticus-derived sequences and bold letters indicate newly generated restriction sites. ery-specific sequences are underlined. All primers were purchased from Integrated DNA Technology (IDT).
Fig. 1.
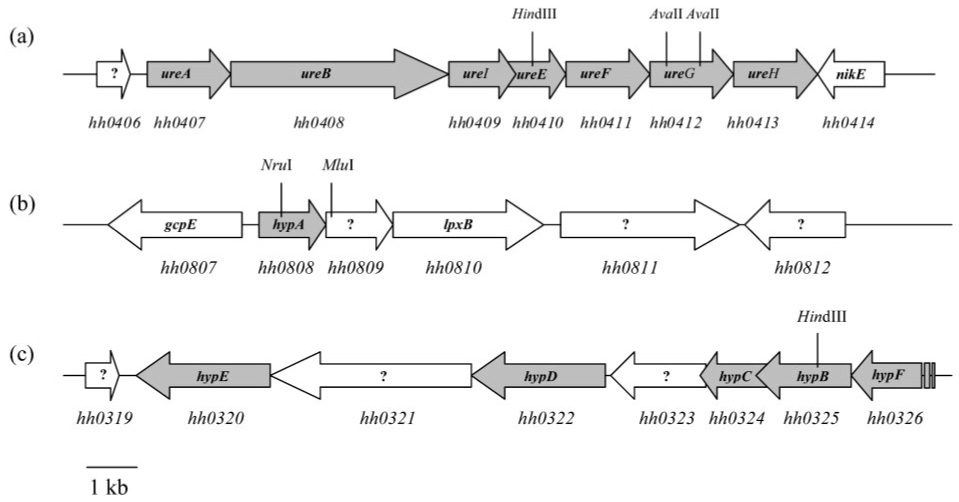
Genome location and organization of the ure and hyp genes in H. hepaticus. Gene annotations are according to the TIGR website (www.tigr.org). The ure and hyp genes are shown in grey. Restriction sites used to disrupt each of the genes studied are indicated. An approximate scale is shown bottom left.
Site-directed mutants in hh0809 or hypC, the respective controls (for polar effect) of hypA or hypB mutants, were constructed as follows. Since part of hh0809 (242 bp) is also present in the pGEM-hypA plasmid, a unique MluI restriction site located at the beginning of hh0809 was used to insert the ery cassette. However, prior to use of this site, another MluI site (from pGEM-T) had to be removed by digesting pGEM-hypA with PstI and NsiI and religating. The newly generated plasmid, pSLB290, was then digested with MluI and ligated with ery to yield plasmid pSLB293 (Table 1). All plasmids were checked by restriction analysis. To construct the H. hepaticus hypC mutant, an all-PCR overlapping approach was used. Briefly, two sets of primers, UphypC with hypC-ery1 and RevhypC with hypC-ery2, were used in independent PCR reactions to amplify hypC upstream and hypC downstream fragments of 280 and 505 bp, respectively. Primers hypC-ery1 and hypC-ery2 were designed to introduce ery priming sequences into each PCR product. A final round of PCR was carried out with both (purified) PCR products, along with the whole ery cassette as well as primers UphypC and RevhypC. Finally, an 1885 bp PCR product was obtained, purified and directly introduced into wild-type strain 51449 to generate a hypC::ery mutant.
Each of the six allelic exchange suicide vectors designed to generate either hypA::ery, hypB::ery, hh0809::ery, ureE::ery, ureG::ery or ΔureG::ery was introduced into H. hepaticus strain 51449 by electroporation (a pulse of 2.5 kV in a Transporator Plus; BTX). Cells were plated on non-selective BA medium and transferred after 24 h onto BA plates supplemented with 5 µg erythromycin ml−1. Isolated clones resulting from homologous recombination appeared after 3–5 days incubation. The disruption of each gene was confirmed by PCR using genomic DNA from each mutant as a template and specific primers for each gene.
Urease assays
The urease activity of fresh lysates was determined by measuring ammonia production from urea hydrolysis with the phenol/hypochlorite assay as described by Weatherburn (1967). Briefly, cells were grown for 48–72 h on BA plates supplemented with various NiCl2 concentrations, harvested and resuspended in 50 mM HEPES-NaOH (pH 7.5), and then washed once and resuspended in the same buffer. After sonication, the protein concentration was determined using the BCA assay (Pierce). Lysates were incubated for 20 min in presence of 25 mM urea, and the amount of ammonia released was then assayed with the phenol/hypochlorite assay. A standard ammonium chloride concentration curve was used to convert the absorbance at 625 nm to nmol ammonia. Urease specific activity is measured in units of µmol ammonia produced min−1 (mg protein) −1.
Hydrogenase assays
Hydrogenase assays were carried out on intact whole cells (grown for 48–72 h) using a previously described amperometric method (Maier et al., 1996). Hydrogenase specific activity is measured in units of nmol H2 used min−1 per 109 cells.
Immunoblotting
Whole-cell extracts (10 µg total protein) from cells grown without added nickel in the medium were subjected to SDS-PAGE (12.5 %) with a mini-protean II apparatus (Bio-Rad), according to the method of Laemmli (1970). Prestained SDS-PAGE standards were purchased from Bio-Rad. Proteins were transferred to nitrocellulose as described by Towbin et al. (1992) using a Bio-Rad Trans Blotter. Immunoblotting was performed using anti-H. pylori UreA or UreB rabbit antisera (a gift from H. L. Mobley, University of Michigan Medical School, USA), diluted 1 : 2000 to 1 : 5000, and goat anti-rabbit immunoglobulin G-alkaline phosphatase (Bio-Rad), diluted 1 : 2000. Bound antibodies were detected by the addition of the chromogenic reagents nitro blue tetrazolium (0.25 mg ml−1) and 5′-bromo-4-chloro-3-indolyl phosphate (0.125 mg ml−1) (Sigma).
RESULTS
Location and homology of the hydrogenase and urease maturation genes
Analysis of the sequenced genome of H. hepaticus strain 51449 revealed the presence of homologues of genes needed for the maturation of both the urease (ure) and hydrogenase (hyp) enzymes in the related species H. pylori (Suerbaum et al., 2003). The H. hepaticus genes are located and arranged in a similar fashion as their H. pylori counterparts: all the ure accessory genes, ureE (annotated as hh0410 in the TIGR database), ureF (hh0411), ureG (hh0412) and ureH (hh0413) are clustered in an operon that also contains the urease structural genes ureA (hh0407) and ureB (hh0408), as well as a homologue of ureI (hh0409), encoding the urea trans-porter in H. pylori (Fig. 1). The respective location of each ure gene within the ureABIEFGH operon is the same as in H. pylori. The hyp genes are located on two different loci. The hypA gene (annotated as hh0808 in the TIGR database) is part of a four-gene operon which also includes hh0809, a homologue of the hp0868 gene whose role in H. pylori is still unknown despite attempts to characterize it (Mehta et al., 2003b), hh0810, encoding a putative lipid A disaccharide synthetase, and hh0811, which encodes a conserved hypothetical protein. The hypB gene (hh0325) is part of a seven-gene operon which also includes hypC (hh0324), hypD (hh0322), hypE (hh0320) and hypF (hh0326) as well as two other genes whose roles have yet to be determined (hh0321 and hh0323).
The corresponding proteins share somewhat significant or even high degrees of similarity or identity with their homologues in H. pylori. For the four proteins of interest in this study, the respective percentage of similarity and identity with homologues in the sequenced H. pylori strain 26695 are as follows: HypA, 71.9 and 52.6 %; HypB, 83.1 and 62.8 %; UreE, 60.8 and 30.8 %; UreG, 93 and 82.4 %. Further analysis of the protein sequence of H. hepaticus HypA reveals the presence of a conserved histidine residue (His2) shown to be involved in nickel binding in H. pylori (Mehta et al., 2003b) (Fig. 2); likewise, H. hepaticus UreE possesses a His residue (His111), conserved among the UreE proteins and shown to participate in the nickel-binding process in the well studied Klebsiella aerogenes (His96) and Bacillus pasteurii (His100) (Colpas et al., 1999; Remaut et al., 2001). Both the H. hepaticus HypB and UreG proteins possess a nucleotide-binding domain, GSGKT, shown to be essential for the maturation of either hydrogenase or urease in H. pylori (Mehta et al., 2003a); indeed, substitution of the conserved lysine residue with an alanine in either HypB or UreG led to inactivation of the urease (in both hypB and ureG mutants) and hydrogenase (only in the hypB mutant). Therefore, the presence of these conserved amino acids and the overall strong protein homology between these four H. hepaticus proteins and their H. pylori counterparts was in agreement with an anticipated role for HypA, HypB, UreE and UreG in urease and/or hydrogenase maturation.
Fig. 2.

Partial protein sequences of H. hepaticus HypA, HypB, UreE and UreG. Conserved residues whose roles have been demonstrated in homologous proteins from H. pylori (HypA, HypB, UreG), K. aerogenes (UreE) or B. pasteurii (UreE) are shown in bold. A conserved p-loop involved in GTPase activity is highlighted for HypB or UreG.
Construction of mutant and control strains
Primers were designed to amplify a DNA sequence specific for each gene with at least 200 bp flanking each side of the restriction site used for the marker insertion (Table 1 and Table 2). Erythromycin resistance was used to inactivate each of the four genes, as this marker has proved to be the best available so far to generate mutants in H. hepaticus (Mehta et al., 2007). The cassette was introduced within a unique restriction site (for hypA, hypB or ureE) or between two naturally existing AvaII restriction sites (for ureG) (Fig. 1). Another ureG mutant was also created by inserting the same cassette into a unique, PCR-generated, HindIII restriction site. After introduction of each suicide vector into H. hepaticus by electroporation, cells were allowed to recover for 24 h on non-selective medium plates before being transferred onto selective plates containing erythromycin. For each experiment, dozens of recombinant H. hepaticus clones appeared after 3–5 days. Several colonies were picked and their genomic DNA was extracted. The presence of the erythromycin resistance cassette in each gene was confirmed by PCR, using genomic DNA from each mutant as a template and specific primers for each gene (Fig. 3). Indeed, PCR products corresponding to the size of each original PCR product (760 bp for hypA, 814 bp for hypB, 734 bp for ureE and 904 bp for ureG) plus the size of the erythromycin resistance cassette (1140 bp) were obtained. A 1669 bp PCR product was obtained when genomic DNA from the ΔureG::ery strain was used as a template, in good agreement with the expected 375 bp deletion generated within ureG in this mutant.
Fig. 3.
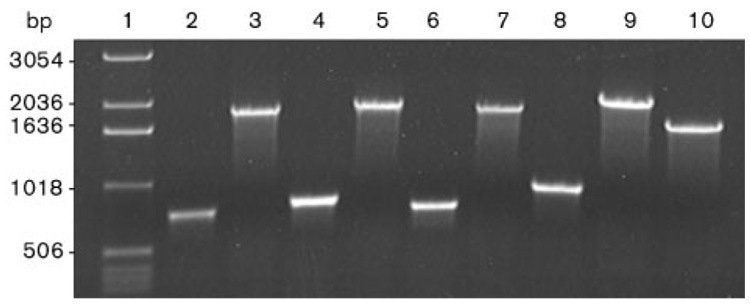
Agarose gel of PCR products used to verify cassette insertions. Lane 1 contains a 1 kb DNA ladder, with sizes indicated on the left. Lanes 2–10 contain fragments amplified from H. hepaticus genomic DNA from the following strains: 2, 4, 6 or 8, wild-type strain 51449; 3, hypA::ery; 5, hypB::ery; 7, ureE::ery; 9, ureG::ery; 10: ΔureG::ery. The following primers were used: lanes 1–2, UphypA and RevhypA; 3–4, UphypB and RevhypB; 5–6, UpureE and RevureE; 7–10, UpureG and RevureG.
Control strains were provided by disrupting the gene immediately downstream of hypA (i.e. hh0809) or hypB (i.e. hypC) (Fig. 1). Both mutants were generated by following the method previously used to generate other mutants (insertion of ery into the targeted gene). However, it is worth noting that purified PCR product (instead of plasmid DNA) was used to generate the hypC::ery mutant. To our knowledge, it is the first time a linear PCR product has been used for this purpose in H. hepaticus. The presence of the 1140 bp erythromycin resistance gene within hh0809 or hypC was verified by PCR using genomic DNA from several individual colonies and primers specific for each gene (data not shown).
Hydrogenase and urease activity in presence of various amounts of nickel
All five mutant strains (hypA, hypB, ureE, ureG and ΔureG), as well as the control hh0809 and hypC strains and the wild-type strain were grown on BA medium without supplemented nickel or with increasing amounts of NiCl2 (up to 50 mM). Cells were harvested after 2 days, washed and used the same day for hydrogenase and urease assays.
Hydrogenase activity was amperometrically determined with whole cells (Table 3). The hydrogenase activity in the wild-type strain was enhanced when nickel was added to the medium, as described previously (Mehta et al., 2005). The activity was the same as wild-type in the H. hepaticus ureE or the ureG mutants, suggesting that neither gene is involved in the maturation of hydrogenase in the mouse pathogen. Similar results have been reported for H. pylori ureE and ureG mutants (Benoit & Maier, 2003; Mehta et al., 2003a). In contrast, in the hypA or hypB mutant, the hydrogenase activity was abolished, indicating that both genes are involved in the maturation of the respiratory enzyme, as expected. Addition of up to 10 µM nickel to the medium could not restore the activity, although some hydrogenase activity was detected in both hypA and hypB mutant strains when 50 µM nickel was added to the medium; this represented only about 2% of the wild-type level (Table 3). This is a significant difference from the results for H. pylori hypA or hypB mutants, which can be partially complemented even with low amounts of nickel (5 µM) (Olson et al., 2001). The hydrogenase activity in the control strain hh0809::ery was comparable to the wild-type level, suggesting that this gene is not required for hydrogenase activity in H. hepaticus, at least under the conditions tested in this study; this result also confirmed that the hydrogenase-negative phenotype observed for the hypA mutant can be assigned directly to hypA rather than to a polar effect on hh0809. Hydrogenase activity was totally abolished in the hypC::ery control strain, indicating that the hypC gene is required for hydrogenase activity in H. hepaticus (Table 3). The result obtained with this control strain was expected, as HypC is known to be required for hydrogenase maturation in various microorganisms, including H. pylori and E. coli (Benoit et al., 2004; Blokesch & Bock, 2002).
Table 3.
Hydrogenase activity of H. hepaticus parental and mutant strains as a function of nickel added to the growth medium
| Strain | Hydrogenase activity (nmol H2 min−1 per 109 cells) |
|||||
|---|---|---|---|---|---|---|
| Nickel added (µM): | 0 | 1 | 5 | 10 | 50 | |
| WT 51449 | 0.77±0.04 | 1.15±0.08 | 1.90±0.15 | 3.64±0.50 | 9.18±1.20 | |
| hypA | <0.01 | <0.01 | <0.01 | <0.01 | 0.20±0.12 | |
| hh0809 | 0.83±0.10 | 0.95±0.20 | 1.33±0.23 | 2.50±0.18 | 4.09±0.85 | |
| hypB | <0.01 | <0.01 | <0.01 | <0.01 | 0.16±0.05 | |
| hypC | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | |
| ureE | 0.91±0.15 | 1.12±0.06 | 1.50±0.22 | 3.01±0.21 | 5.32±0.90 | |
| ureG | 0.87±0.19 | 0.92±0.25 | 1.24±0.10 | 5.64±0.07 | 9.81±1.05 | |
| ΔureG | 0.67±0.05 | 0.85±0.12 | ND | ND | 8.21±1.32 | |
Cells were harvested from Brucella agar plates supplemented with various amounts of NiCl2. Hydrogen uptake activity was determined amperometrically from whole cells, with O2 provided as the final electron acceptor. Results shown are means of two to four independent cultures±SD, with each assay made in duplicate or triplicate. ND, Not determined.
For urease assays, cells were broken, cell-free extracts were isolated and the protein concentration was determined. The ammonia produced from urea hydrolysis was measured by using the phenol/hypochlorite assay (Table 4). Addition of NiCl2 to the medium dramatically increased the urease activity in the wild-type (from 0.06±0.02 units of specific activity in unsupplemented medium to 5.44±1.42 units in medium supplemented with 50 µM NiCl2), as reported by Belzer et al. (2005). Compared to the wild-type, urease activities were severely affected in all four mutants in non-nickel-supplemented medium, suggesting that hypA, hypB, ureE and ureG play a role in urease activity, as do their counterparts in H. pylori (Table 4). The urease activity was not restored to wild-type level in H. hepaticus ureE::ery, ureG::ery or the ΔureG::ery mutant, even when the medium was supplemented with up to 50 µM nickel. The same effect has been previously reported for H. pylori ureE and ureG mutants (Benoit et al., 2007). Interestingly, the urease activity of H. hepaticus hypA or hypB mutants could not be restored to wild-type level by nickel supplementation either; this is in contrast with the urease activity of H. pylori hypA or hypB mutants, which has been shown to be complemented by adding 5 µM NiCl2 to the medium (Olson et al., 2001). Results obtained with the hh0809::ery and hypC::ery control strains confirmed the non-polarity effect of the ery cassette and the definitive involvement of both HypA and HypB in the urease maturation process. These results also suggested a possible role for the hh0809 gene product and more probably for the HypC protein. Indeed, urease activity levels in the hh0809::ery control strain were in general slightly lower than that of wild-type when nickel was added at between 1 and 10 µM to the medium. In contrast, the H. hepaticus hypC mutant consistently displayed higher urease activity than the wild-type when grown under the same conditions. Both genes are currently being investigated for their respective roles in nickel enzyme maturation.
Table 4.
Urease activity of H. hepaticus parental and mutant strains as a function of nickel added to the growth media
| Strain | Urease activity [µmol NH3 min−1 (mg total protein)−1] |
|||||
|---|---|---|---|---|---|---|
| Nickel added (µM): | 0 | 1 | 5 | 10 | 50 | |
| WT 51449 | 0.06±0.02 | 0.26±0.05 | 0.54±0.12 | 1.97±0.23 | 5.44±1.42 | |
| hypA | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | |
| hh0809 | 0.08±0.03 | 0.13±0.04 | 0.34±0.08 | 0.58±0.13 | 3.30±0.54 | |
| hypB | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | |
| hypC | 0.16±0.01 | 0.57±0.03 | 1.52±0.12 | 2.85±0.34 | 14.96±1.83 | |
| ureE | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | |
| ureG | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | |
| ΔureG | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | |
Cells were harvested from Brucella agar plates supplemented with various amounts of nickel chloride, washed and lysed by sonication. Protein concentration was determined by using the BCA method (Pierce). Urease activity was determined by measuring ammonia production from urea hydrolysis by using the phenol/hypochlorite assay. Results shown are means of two to five independent cultures±SD, with each assay made in duplicate or triplicate.
The amount of urease synthesized is similar in the mutants and the wild-type
To determine whether the decreased urease activity in the various mutants was linked to a decreased amount of urease apoenzyme, immunoblotting was performed on cell-free extracts from each mutant and the wild-type grown on unsupplemented BA medium (Fig. 4). The significant degree of identity between H. pylori and H. hepaticus UreA and UreB (65 and 69 %, respectively), allowed us to use antisera raised against H. pylori UreA or UreB. As shown in Fig. 4, the amount of the structural UreA subunit (expected molecular mass 25.2 kDa) was similar in every lane, indicating that the difference in urease activity between the wild-type and the hypA, hypB, ureE or ureG strains was not due to decreased synthesis of UreA. This was also observed in the ΔureG::ery mutant (data not shown). A similar result was also observed with all these mutants and the wild-type when the anti-UreB antiserum was used (data not shown). Therefore, it does not seem that a transcriptional repression mechanism of the urease structural genes accounts for the decreased urease activity in these four mutants. Rather, this phenotype is probably due to a deficient post-translational nickel maturation of the urease enzyme.
Fig. 4.
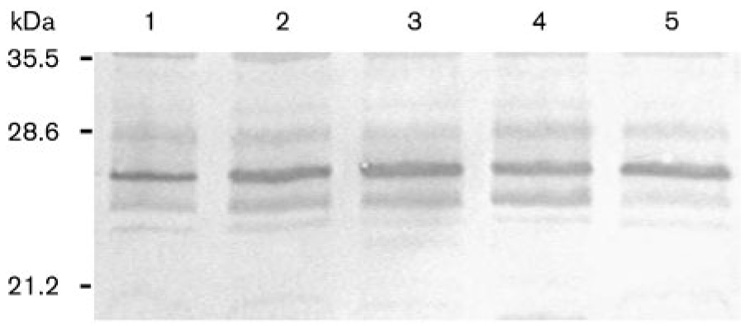
Immunoblotting analysis of H. hepaticus parental and mutant strain cell extracts with anti-H. pylori UreA. Ten micrograms of cell-free extract from each strain was subjected to SDS-PAGE, transferred to nitrocellulose and blotted with anti-UreA antibodies. Lanes: 1, wild-type strain 51449; 2, hypA::ery; 3, hypB::ery; 4, ureE::ery; 5, ureG::ery. Sizes corresponding to prestained molecular mass markers are indicated on the left.
DISCUSSION
Although H. hepaticus and H. pylori share a similar battery of accessory proteins involved in hydrogenase and/or urease maturation (i.e. UreEFGH and HypABCDEF), our work suggests that HypA, HypB and HypC have a different mechanistic role in each bacterium. While mutants in hypA or hypB are urease-negative and hydrogenase-negative, respectively, in both Helicobacter species, partial or total complementation of both enzyme activities by nickel supplementation only seems to happen in the human gastric pathogen, H. pylori. Hence, addition of up to 50 µM NiCl2 in the growth medium of H. hepaticus did not restore urease activity and could only restore hydrogenase activity up to 2% of the wild-type level in either the hypA or the hypB mutant. This unexpected absence of complementation by nickel in both mutants indicates that both proteins are required at all times for urease activity as well as hydrogenase activity in this bacterium, independently of the nickel content of the cell (the latter is assumed to increase with increasing amounts of nickel added to the medium). These data further suggest that both proteins could be involved in the final stages of maturation of either of the nickel-containing proteins in the mouse liver pathogen, but they are probably part of early stages of the maturation processes in H. pylori.
Although protein sequences of H. hepaticus and H. pylori HypA share only 52.6% identity, some key residues are well conserved; for instance, H. hepaticus HypA possesses a conserved histidine residue (His2) known to be involved in nickel binding and essential for urease activity in H. pylori (Mehta et al., 2003b). Likewise, the HypB protein seems well conserved between H. hepaticus and H. pylori, with the same GTPase P-loop motif (GSGKT) located at the exact same position in the sequence. Nevertheless, despite all these similarities between species, HypA and HypB appear to play a different role in H. hepaticus compared to H. pylori. Differences in actual protein complexes might account for these discrepancies. Direct interactions between H. pylori HypA and HypB have been shown by cross-linking and immunoblotting using antibodies against HypA or HypB (Mehta et al., 2003b); however, interactions between HypA/HypB and either the urease or the hydrogenase apoenzyme complexes have never been reported in H. pylori. It is very much anticipated that H. hepaticus HypA and HypB proteins will be able to interact with each other, although it remains to be demonstrated; most of all, future studies will be aimed at isolating complexes between HypA/HypB and HydA-HydB or UreA-UreB heterodimers. The HypC protein is also likely to play an interesting role with respect to these putative protein complexes, if one looks at the phenotype of the H. hepaticus hypC mutant (hydrogenase-negative and urease hyperpositive). It is not clear whether the HH0809 hypothetical protein plays a role in the urease maturation process in H. hepaticus; indeed, the hh0809 mutant showed slightly or more pronounced decreased urease activity when the cells when grown on medium supplemented with 1–10 mM nickel. We do not have any explanation for this phenotype. HH0809 is a homologue of the HP0868 hypothetical protein in H. pylori, and an H. pylori hp0868 mutant has been shown to have a wild-type-like phenotype for both urease and hydrogenase activities.
Our study also showed that H. hepaticus ureE and ureG mutants are totally devoid of urease activity. These results were expected, because UreE and UreG have been shown to be required for urease activity in several micro-organisms, including H. pylori (Mehta et al., 2003a; Voland et al., 2003), Proteus mirabilis (Island & Mobley, 1995) and K. aerogenes (Mulrooney & Hausinger, 1990). As shown previously for H. pylori (Benoit et al., 2007), nickel supplementation of the growth medium did not restore urease activity in either H. hepaticus mutant. This seems to favour UreE and UreG being involved in the later, irreversible steps of insertion of nickel within the urease apoenzyme.
A recent study in our lab showed interactions between H. pylori HypA and UreE; these interactions are believed to be needed when nickel is scarce in the medium and the bacterial cell (Benoit et al., 2007). Under these conditions, it seems that H. pylori cannot rely on UreE alone to provide urease with nickel because the UreE protein has a poor nickel-binding capacity, especially when compared to other histidine-rich UreE proteins, like that of P. mirabilis (Jones & Mobley, 1989) or K. aerogenes (Mulrooney & Hausinger, 1990). In H. hepaticus, given that UreE does not possess histidine-rich sequences either, and HypA appears to be required for urease activity, it is likely that a HypA-UreE transient association is also needed for transfer of nickel from HypA to the apourease.
While the addition of nickel did not restore urease activity in any of the four H. hepaticus mutants of interest in this study (i.e. hypA, hypB, ureE, ureG), it increased the activity in the wild-type strain 51449. Analysis of the genome sequence of the mouse liver pathogen reveals the presence of a gene (hh0352) encoding a homologue of NikR, a nickel-responsive regulatory protein involved in urease regulation in H. pylori. However, Belzer et al. (2005) have shown that the surge in urease activity in the wild-type was the consequence of a post-translational nickel activation of the apourease rather than a transcriptional mechanism. This result is in good agreement with the finding that the four nickel-related mutants have similar amounts of urease, as shown by immunoblotting against UreA or UreB. This confirms that the dramatic decrease of urease activity in these mutants is due to deficient post-translational nickel maturation, not to transcriptional repression of the urease structural genes.
ACKNOWLEDGEMENTS
This work was supported by the Georgia Research Foundation and by National Institute of Health Grant RO1DK062852. Antisera against H. pylori UreA or UreB were kindly provided by H. L. Mobley, University of Michigan Medical School, Ann Arbor, MI, USA.
REFERENCES
- Beckwith CS, McGee DJ, Mobley HL, Riley LK. Cloning, expression, and catalytic activity of Helicobacter hepaticus urease. Infect Immun. 2001;69:5914–5920. doi: 10.1128/IAI.69.9.5914-5920.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belzer C, Stoof J, Beckwith CS, Kuipers EJ, Kusters JG, van Vliet AH. Differential regulation of urease activity in Helicobacter hepaticus and Helicobacter pylori. Microbiology. 2005;151:3989–3995. doi: 10.1099/mic.0.28188-0. [DOI] [PubMed] [Google Scholar]
- Benoit S, Maier RJ. Dependence of Helicobacter pylori urease activity on the nickel-sequestering ability of the UreE accessory protein. J Bacteriol. 2003;185:4787–4795. doi: 10.1128/JB.185.16.4787-4795.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit S, Mehta N, Wang G, Gatlin M, Maier RJ. Requirement of hydD, hydE, hypC and hypE genes for hydrogenase activity in Helicobacter pylori. Microb Pathog. 2004;36:153–157. doi: 10.1016/j.micpath.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Benoit SL, Mehta N, Weinberg MV, Maier C, Maier RJ. Interaction between the Helicobacter pylori accessory proteins HypA and UreE is needed for urease maturation. Microbiology. 2007;153:1474–1482. doi: 10.1099/mic.0.2006/003228-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaser MJ. The role of Helicobacter pylori in gastritis and its progression to peptic ulcer disease. Aliment Pharmacol Ther. 1995;9(Suppl 1):27–30. doi: 10.1111/j.1365-2036.1995.tb00780.x. [DOI] [PubMed] [Google Scholar]
- Blokesch M, Bock A. Maturation of [NiFe]-hydrogenases in Escherichia coli: the HypC cycle. J Mol Biol. 2002;324:287–296. doi: 10.1016/s0022-2836(02)01070-7. [DOI] [PubMed] [Google Scholar]
- Colpas GJ, Brayman TG, Ming LJ, Hausinger RP. Identification of metal-binding residues in the Klebsiella aerogenes urease nickel metallochaperone, UreE. Biochemistry. 1999;38:4078–4088. doi: 10.1021/bi982435t. [DOI] [PubMed] [Google Scholar]
- Eaton KA, Brooks CL, Morgan DR, Krakowka S. Essential role of urease in pathogenesis of gastritis induced by Helicobacter pylori in gnotobiotic piglets. Infect Immun. 1991;59:2470–2475. doi: 10.1128/iai.59.7.2470-2475.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox JG, Dewhirst FE, Tully JG, Paster BJ, Yan L, Taylor NS, Collins MJ, Jr, Gorelick PL, Ward JM. Helicobacter hepaticus sp. nov., a microaerophilic bacterium isolated from livers and intestinal mucosal scrapings from mice. J Clin Microbiol. 1994;32:1238–1245. doi: 10.1128/jcm.32.5.1238-1245.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Island MD, Mobley HL. Proteus mirabilis urease: operon fusion and linker insertion analysis of ure gene organization, regulation, and function. J Bacteriol. 1995;177:5653–5660. doi: 10.1128/jb.177.19.5653-5660.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones BD, Mobley HL. Proteus mirabilis urease: nucleotide sequence determination and comparison with jack bean urease. J Bacteriol. 1989;171:6414–6422. doi: 10.1128/jb.171.12.6414-6422.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy DC, Herbst RW, Iwig JS, Chivers PT, Maroney MJ. A dynamic Zn site in Helicobacter pylori HypA: a potential mechanism for metal-specific protein activity. J Am Chem Soc. 2007;129:16–17. doi: 10.1021/ja066958x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Maier RJ, Fu C, Gilbert J, Moshiri F, Olson J, Plaut AG. Hydrogen uptake hydrogenase in Helicobacter pylori. FEMS Microbiol Lett. 1996;141:71–76. doi: 10.1111/j.1574-6968.1996.tb08365.x. [DOI] [PubMed] [Google Scholar]
- Mehta N, Benoit S, Maier RJ. Roles of conserved nucleotide-binding domains in accessory proteins, HypB and UreG, in the maturation of nickel-enzymes required for efficient Helicobacter pylori colonization. Microb Pathog. 2003a;35:229–234. doi: 10.1016/s0882-4010(03)00151-7. [DOI] [PubMed] [Google Scholar]
- Mehta N, Olson JW, Maier RJ. Characterization of Helicobacter pylori nickel metabolism accessory proteins needed for maturation of both urease and hydrogenase. J Bacteriol. 2003b;185:726–734. doi: 10.1128/JB.185.3.726-734.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta NS, Benoit S, Mysore JV, Sousa RS, Maier RJ. Helicobacter hepaticus hydrogenase mutants are deficient in hydrogen-supported amino acid uptake and in causing liver lesions in A/J mice. Infect Immun. 2005;73:5311–5318. doi: 10.1128/IAI.73.9.5311-5318.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta NS, Benoit SL, Mysore J, Maier RJ. In vitro and in vivo characterization of alkyl hydroperoxide reductase mutant strains of Helicobacter hepaticus. Biochim Biophys Acta. 2007;1770:257–265. doi: 10.1016/j.bbagen.2006.09.022. [DOI] [PubMed] [Google Scholar]
- Mulrooney SB, Hausinger RP. Sequence of the Klebsiella aerogenes urease genes and evidence for accessory proteins facilitating nickel incorporation. J Bacteriol. 1990;172:5837–5843. doi: 10.1128/jb.172.10.5837-5843.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson JW, Maier RJ. Molecular hydrogen as an energy source for Helicobacter pylori. Science. 2002;298:1788–1790. doi: 10.1126/science.1077123. [DOI] [PubMed] [Google Scholar]
- Olson JW, Mehta NS, Maier RJ. Requirement of nickel metabolism proteins HypA and HypB for full activity of both hydrogenase and urease in Helicobacter pylori. Mol Microbiol. 2001;39:176–182. doi: 10.1046/j.1365-2958.2001.02244.x. [DOI] [PubMed] [Google Scholar]
- Remaut H, Safarov N, Ciurli S, Van Beeumen J. Structural basis for Ni2+ transport and assembly of the urease active site by the metallochaperone UreE from Bacillus pasteurii. J Biol Chem. 2001;276:49365–49370. doi: 10.1074/jbc.M108304200. [DOI] [PubMed] [Google Scholar]
- Sipponen P, Hyvarinen H, Seppala K, Blaser MJ. Review article: pathogenesis of the transformation from gastritis to malignancy. Aliment Pharmacol Ther. 1998;12(Suppl. 1):61–71. doi: 10.1111/j.1365-2036.1998.00005.x. [DOI] [PubMed] [Google Scholar]
- Solnick JV, Schauer DB. Emergence of diverse Helicobacter species in the pathogenesis of gastric and enterohepatic diseases. Clin Microbiol Rev. 2001;14:59–97. doi: 10.1128/CMR.14.1.59-97.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabb EV, Ruby EG. RP4-based plasmids for conjugation between Escherichia coli and members of the Vibrionaceae. Methods Enzymol. 2002;358:413–426. doi: 10.1016/s0076-6879(02)58106-4. [DOI] [PubMed] [Google Scholar]
- Suerbaum S, Josenhans C, Sterzenbach T, Drescher B, Brandt P, Bell M, Droge M, Fartmann B, Fischer HP, et al. The complete genome sequence of the carcinogenic bacterium Helicobacter hepaticus. Proc Natl Acad Sci U S A. 2003;100:7901–7906. doi: 10.1073/pnas.1332093100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. 1979. Biotechnology. 1992;24:145–149. [PubMed] [Google Scholar]
- Voland P, Weeks DL, Marcus EA, Prinz C, Sachs G, Scott D. Interactions among the seven Helicobacter pylori proteins encoded by the urease gene cluster. Am J Physiol Gastrointest Liver Physiol. 2003;284:G96–G106. doi: 10.1152/ajpgi.00160.2002. [DOI] [PubMed] [Google Scholar]
- Weatherburn MW. Phenol-hypochlorite reaction for determination of ammonia. Anal Chem. 1967;39:971–974. [Google Scholar]


