Abstract
We report the chromosomal localization, mutant gene identification, ophthalmic appearance, histology, and functional analysis of two new hereditary mouse models of retinal degeneration not having the Pde6brd1 (“r”, “rd”, or “rodless”) mutation. One strain harbors an autosomal recessive mutation that maps to mouse chromosome 5. Sequence analysis showed that the retinal degeneration is caused by a missense point mutation in exon 13 of the beta-subunit of the rod cGMP phosphodiesterase (β-PDE) gene (Pde6b). The gene symbol for this strain was set as Pde6brd10, abbreviated rd10 hereafter. Mice homozygous for the rd10 mutation showed histological changes at postnatal day 16 (P16) of age and sclerotic retinal vessels at four weeks of age, consistent with retinal degeneration. Retinal sections were highly positive for TUNEL and activated caspase-3 immunoreactivity, specifically in the outer nuclear layer (ONL). ERGs were never normal, but rod and cone ERG a- and b-waves were easily measured at P18 and steadily declined over 90% by two months of age. Protein extracts from rd10 retinas were positive for β-PDE immunoreactivity starting at about the same time as wild type (P10), though signal averaged less than 40% of wild type. Interestingly, rearing rd10 mice in total darkness delayed degeneration for at least a week, after which morphological and functional loss progressed irregularly. With the second strain, a complementation test with rd1 mice revealed that the retinal degeneration phenotype observed represents a possible new allele of Pde6b. Sequencing demonstrated a missense point mutation in exon 16 of the beta-subunit of rod phosphodiesterase gene, different from the point mutations in rd1 and rd10. The gene symbol for this strain was set as Pde6bnmf137, abbreviated nmf137 hereafter. Mice homozygous for this mutation showed retinal degeneration with a mottled retina and white retinal vessels at three weeks of age. The exon 13 missense mutation (rd10) is the first known occurrence of a second mutant allele spontaneously arising in the Pde6b gene in mice and may provide a model for studying the pathogenesis of autosomal recessive retinitis pigmentosa (arRP) in humans. It may also provide a better model for experimental pharmaceutical-based therapy for RP because of its later onset and milder retinal degeneration than rd1 and nmf137.
Keywords: mouse model, retinal degeneration, retinitis pigmentosa, rd1, rd10, nmf137, PDE6b, betaphosphodiesterase, rod photoreceptor, cGMP-PDE, beta-subunit of rod cGMP phosphodiesterase gene
Introduction
One of the earliest reported animal models of retinal degeneration is the “rodless retina” mouse, described by Keeler as being caused by an autosomal recessive mutation (gene symbol, r; (Keeler 1924)). In affected animals, the rod photoreceptor cells begin degenerating at about postnatal day 8 (P8), and by four weeks no photoreceptors are left (Keeler 1924; Keeler 1966; Farber and Lolley 1974; LaVail and Sidman 1974; Pittler and Baehr 1991). The retinal degeneration is preceded by accumulation of cyclic guanosine monophosphate (cGMP) in the retina and is correlated with deficient activity of rod photoreceptor cGMP-phosphodiesterase type 6 (cGMP-PDE) (Farber and Lolley 1974; Farber and Lolley 1976; Beavo, Conti, Heaslip, 1994). cGMP is a key messenger molecule that links absorption of photons of light energy to neural signaling in the vertebrate photoreceptor. When a photon is absorbed by rhodopsin, cGMP-PDE and transducin are activated. cGMP-PDE catalyzes the degradation of cGMP to guanosine-5′-monophosphate (5′-GMP), closing ionic channels gated by cGMP and eventually generating a visual signal. Thus, cGMP-PDE is pivotal in photoreceptor conversion of light to neural impulse (Yau and Baylor 1989). The retinal degeneration is caused by a defective Pde6b, the gene that encodes the β subunit of rod photoreceptor cGMP phosphodiesterase type 6, which contains both an intronic MLV virus insertion and a nonsense point mutation (Pittler and Baehr 1991; Bowes, Li et al. 1993) (gene symbol Pde6brd1; abbreviated rd1 hereafter (Beavo, Conti, Heaslip, 1994). This pair of anomalies is widely distributed in inbred and wild-derived mouse strains from different parts of the world (Pittler and Baehr 1991; Bowes, Li et al. 1993; Clapcote et al., 2005). That the 'rodless retina' mutation was proven to carry the same sequence anomalies suggests an ancient history for this mutation prior to the development of laboratory strains (Pittler, Keeler et al. 1993).
Numerous mutations in the catalytic domain of the human gene, PDE6B, have been found in patients suffering from autosomal recessive retinitis pigmentosa (arRP; OMIM 180072) (McLaughlin, Sandberg et al. 1993; McLaughlin, Ehrhart et al. 1995). Hence, the rd1 mouse and other mouse strains harboring Pde6b mutant alleles are considered animal models of RP. Interest remains strong in using these as a research tools. Indeed, beyond those naturally occurring, new strains are being made. Hart et al. recently published data correlating the genotypes and phenotypes of several novel Pde6b mutants produced by N-ethyl-N-nitrosourea (ENU) mutation induction (Hart, McKie, et al., 2005). Similarly, we recently identified two new mouse strains that exhibit retinal degeneration and sequence anomalies in the Pde6b gene, Pde6brd10 (abbreviated rd10 hereafter) (Chang, Hawes et al. 2002) and Pde6bnmf137 (abbreviated nmf137 hereafter). Here we describe their genotypes and phenotypes in detail. The nmf137 mutant degenerates as rapidly as rd1, but the onset of degeneration in the rd10 mouse is later and can be delayed by rearing in darkness, suggesting that it may be a useful research tool.
Materials and Methods
Animals
The mice in this study were initially bred and maintained in standardized conditions in the Research Animal Facility at The Jackson Laboratory, Bar Harbor, ME and subsequently at Emory University and the Veteran’s Administration Hospital, both in Atlanta, GA. They were maintained on NIH31 6% fat chow and acidified water, with a 14-hour light/10-hour dark cycle in conventional facilities that are monitored regularly to maintain a pathogen-free environment. Light intensity in the cages was measured at 50 to 200 lux. A subset of rd10 mice was raised in total darkness. All experiments were approved by the respective Institutional Animal Care and Use Committees and conducted in accordance with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research.
Origin
Pde6brd10 was discovered in the CXB-1 recombinant inbred line with white retinal vessels at weaning age (Fig. 1). Subsequently, the rd10 stock has been maintained by repeated backcrossing to C57BL/6J to make a congenic inbred strain, hereafter referred to as B6-rd10, or simply the rd10 strain. Major aspects of the phenotype do not appear to differ on the two genetic backgrounds. Pde6bnmf137 was identified in a mutation screen where B6 mice were mutagenized by ENU. Both first-generation (G1) and third generation (G3) mice obtained through a three-generation breeding scheme were tested to identify dominant and recessive mutations, respectively.
Figure 1.
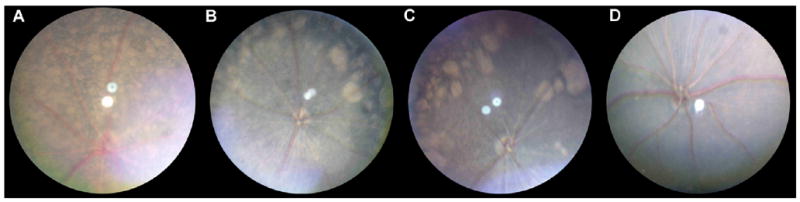
Fundus appearance of various mouse strains at two months of age: rd1 (A), rd10 (B), nmf137 (C), and C57BL/6J wild type (D). The retinal degeneration in the rd10 and nmf137 strains was easily distinguished from wild type and rd1 retinal appearance at two months of age by fundoscopy.
Clinical retinal evaluation
All mice in the characterization studies and linkage crosses had pupils dilated with 1% atropine ophthalmic drops (Bausch and Lomb Pharmaceuticals Inc., Tampa, FL) and were evaluated by indirect ophthalmoscopy with a 78 diopter lens. Signs of retinal degeneration, such as vessel attenuation and alterations in the RPE were noted. Fundus photographs were taken with a Kowa Genesis small animal fundus camera (Kowas, Tokyo, Japan) (Hawes, Smith et al., 1999).
Histology and morphometrics
Plastic or paraffin sections of eyes from mice ranging in age from one week to three months were studied histologically. For plastic embedding, eyes were immersed in cold fixative (1% paraformaldehyde, 2% glutaraldehyde, and 0.1 M cacodylate buffer) while maintaining orientation. Eyes were left in fixative for 24 hours, after which time they were transferred to cold 0.1 M cacodylate buffer solution for an additional 24 hours. Samples were embedded in methacrylate historesin in order to obtain superior to inferior cross-sections of the eyecup. Tissue was cut at 0.05 μm on an ultramicrotome (Reichert Ultracut, IL, USA) using a histo-diamond knife and heat fixed to glass slides. For paraffin embedding, eyes were fixed by injecting approximately 1 μl of 10% buffered formalin (Stephens Scientific, Riverdale, NJ), followed by immersion in the same fixative for 30 min at 4°C, then stored in PBS for later processing. Eyes were dehydrated in consecutive washes of ethanol and xylene, washed in melted paraffin (TissuePrep 2, Fisher Scientific, Fair Lawn, NJ) and embedded into blocks. Eyes were sectioned at 5 μm on an American Optical 820 rotary microtome (Sathbridge, MA). Plastic and paraffin sections were stained with hematoxylin and eosin (H&E) or 1% toluidine blue.
Sections were analyzed using light microscopy (Leica DMRB, Bannockburn, IL). Areas approximately 500 microns were acquired as Adobe Photoshop images at 20X magnification. Four areas spanning the superior-inferior retinal axis were images such that two areas were above the optic nerve and two below. Three measurements of photoreceptor nuclei counts and retinal thickness (the inner nuclear layer, the ganglion cell layer, photoreceptor inner segments, and photoreceptor outer segments) were made on each image and averaged as the representative value for that area. A total of five sections were averaged for each eye using Image Pro Plus 5.0 (Media Cybernetics, Silver Springs, MD). To ensure that the choice of fixation protocol did not alter the number of nuclei counted per field, P18 rd10 retinal ONL nuclei were counted from six sections (four fields per section) from plastic embedded eyes and from six sections from paraffin embedded eyes. Mean counts per field ± SEM were 428±44 and 501±50 for paraffin- versus plastic-embedded samples, respectively. Two-tailed t-test analysis indicated that these means were not statistically different (p = 0.274).
Electroretinography
After overnight dark-adaptation, mice were anesthetized with an intraperitoneal injection of normal saline solution containing ketamine (80 mg/kg) and xylazine (16 mg/kg). Electroretinograms (ERGs) were recorded from the corneal surface of one eye after pupil dilation (1% atropine sulfate) using a gold or platinum loop electrode referenced to a gold wire in the mouth or a needle electrode in the cheek. A needle electrode placed in the tail served as ground. A drop of methylcellulose (2.5%) was placed on the corneal surface to ensure electrical contact and to maintain corneal integrity. Body temperature was maintained at a constant temperature of 38°C using a heated water pad. All stimuli were presented in a Ganzfeld dome (LKC Technologies, Gaithersburg, MD). Rod-dominated responses to white flashes of light over a 4.0 to 5.0 log unit range of intensities were recorded. Cone-dominated responses were obtained with white flashes over a 2.0 log unit range of intensities at 2.1 Hz on a rod-saturating background (1.46 log cd/m2) after 10 minutes of exposure to the background light to allow for complete light adaptation.
Responses were amplified and filtered (0.03 to 10,000 Hz) and digitized using an I/O board (model PCI-1200; National Instruments, Austin, TX) in a personal computer. Signals were sampled every 0.5 msec over a response window of 240 msec. For each stimulus condition, responses were computer-averaged with up to 20 records averaged for the weakest signals. Responses were recorded from 33 rd10 mice ranging in age from P14 to P63. Comparisons with wild type mice were performed on 14 C57Bl/6J mice from P18 to P40.
β-PDE Immunoreactivity
Protein isolated from retinas of C57BL/6, C57BL/6Jrd10/rd10, and C3H/HeJrd1/rd1 mice at postnatal days 9 through 11 (P9–11), P12–14, P15–17, P18–20, and P60 and from P60 C57BL/6 whole brain were subjected to western immunoblot analysis using a WesternBreeze™ Chemiluminescent Kit (Invitrogen, Carlsbad, CA) following manufacturer’s instructions. The primary antibody was an affinity-purified polyclonal antibody generated against a mixed set of synthetic peptides to accommodate a single mismatch between the human and mouse amino acid sequences of the genes encoding the β subunit of cGMP-PDE, H Q Y F G (K/R) K L S P E N V A G A C (Affinity Bioreagents, Golden, CO). Three to four mice were sacrificed at each stage, eyes enucleated, and retinas removed. Each pair of retinas was homogenized in manufacturer’s lysis buffer using a handheld, battery-powered pestle (Pellet Pestle Motor; Kimble/Kontes, Vineland, New Jersey) in microcentrifuge tubes, then centrifuged for 10 min at 10,000 g. A portion of a macerated whole brain from a P60 wild type mouse was similarly homogenized. Supernatants were transferred to new tubes and assayed for protein concentration by the BCA method (Pierce, Rockford, IL). Aliquots containing 30 μg of protein were subjected to SDS polyacrylamide gel electrophoresis using 4–12% gradient Bis-Tris ZOOM™ gel (Invitrogen). Proteins were transferred onto nitrocellulose membranes, blocked per manufacturer’s instructions, then incubated with PDE6b polyclonal antibody (1 μg/ml) for 90 min. Washing, incubation with secondary antibody, chemiluminescent substrate, and enhancer, were as per manufacturer instructions. Kodak BioMax X-ray films (Fisher Scientific, Pittsburgh, PA) were then exposed to blots for various times. Transmission scans of exposures within the linear range of the films were done using an Epson Perfection 4870 Photo scanner connected to an Apple Macintosh G5 computer (Apple Computers, Cupertino, CA) running Adobe Photoshop CS v.8 (Adobe Systems, Inc., San Jose, CA). Density of signal was calculated by determining the net intensity of a band area on an Adobe Photoshop image.
Immunohistochemistry was conducted on mouse retina sections using the PDE6b polyclonal antibody. Crysections (7 μm) were made of eyes from P22 C57BL/6, rd10, and rd1 mice. Sections were fixed in 4% paraformaldehyde in 0.1 M phosphate buffer with 0.03 M sucrose on ice for 20 min then permeabilized in 0.1% Triton X-100 in PBS at room temperature for 5 min. Sections were blocked with normal goat serum for 15 min, incubated with primary antibody (1 μg/ml) for 1 hr, then incubated with FITC-conjugated goat anti-rabbit IgG (1:1000 dilution; Calbiochem). Following a wash with PBS-BSA and two washes with SlowFade Light buffer (Molecular Probes, Eugene, OR), a drop of SlowFade Light antifade medium was put on the section and a cover slip placed on the slide. Sections were then observed and photographed by confocal microscopy.
Apoptosis Assays
Apoptosis was assessed in paraffin sections by TUNEL (TdT-mediated dUTP Nick-End Labeling) assay and by anti-active caspase-3 immunoreactivity (DeadEnd™ Fluorometric TUNEL System and Anti-ACTIVE® Caspase-3 antibody, respectively; Promega, Madison, WI), with confocal microscopy per manufacturer’s instructions. For TUNEL, sections were deparaffinized, rehydrated, treated with Proteinase K, reacted with TdT/nucleotide mix (containing fluorescein-12-dUTP), and counter-stained with propidium iodide. For active caspase-3 detection, sections were permeabilized in 0.1% triton x-100/PBS then reacted with Anti-ACTIVE® Caspase-3 antibody followed by fluorescein conjugated donkey anti-rabbit antibody (Jackson Immunoresearch, West Grove, PA). One drop of Anti-Fade solution (Molecular Probes, Eugene, OR) was added to the area containing tissue and cover slips were mounted and edges sealed with nail polish. Sections were viewed by fluorescent confocal microscopy using a Nikon EFD-3 microscope (Nikon, Melville, NY) with Biorad MRC 1024 confocal unit controlled by Biorad LaserSharp 2000 v5.2 software (Bio-Rad Laboratories, Inc., Hercules, CA) using 520 nm emission and 495 nm excitation for fluorescein detection and 617 nm emission and 535 nm excitation for propidium iodide (counter stain) detection.
Gene Mapping
To determine the chromosomal location of the rd10 gene we mated C57BL/6Jrd10/rd10 mice to CAST/Ei mice. The F1 mice, which exhibited no retinal abnormalities, were backcrossed to C57BL/6Jrd10/rd10 mice. Tail DNA was isolated as previously reported(Johnson, Gagnon et al., 2003). For PCR (polymerase chain reaction) amplification, 25 ng DNA was used in a 10-μl volume containing 50 mM KCl, 10 mM 200 μM dNTP, and 0.02 U Tris–Cl, pH 8.3, 2.5 mM MgCl2, 0.2 mM oligonucleotides, AmpliTaq DNA polymerase. The reactions, which were initially denatured for 3 minutes at 94°C, were subjected to 40 cycles of 15 seconds at 94°C, 1-minute at 51°C, 1-minute at 72°C, and then a final 7-minute extension at 72°C. PCR products were separated by electrophoresis on 3% MetaPhor (FMC, Rockland, ME) agarose gels and visualized under UV light after staining with ethidium bromide. Initially a genome scan of microsatellite (Mit) DNA markers was carried out on pooled DNA samples (Taylor, Navin et al. 1994). After detection of linkage on Chromosome 5, the microsatellite markers D5Mit157, D5Mit91, D5Mit25 were scored on individual DNA samples.
Gene Identification/Genomic sequencing
To test the Pde6b gene as a candidate, we designed four pairs of PCR primers based on mRNA sequence from GenBank accession numbers X60133, X57656, and X87952 (Table 1). For direct sequencing, the PCR reaction was scaled up to 30 μl; amplification was done during 36 cycles with a 15 sec denaturing step at 94°C, a 2 min. annealing step at 60°C, and a 2 min. extension step at 72°C. PCR products were purified from agarose gels using a Qiagen kit (Qiagen Co.). Sequencing reactions were carried out with automated fluorescence tag sequencing. Genomic DNA was prepared from spleens of 3-week-old mice according to standard procedures. Total RNA was isolated from retinas of newborn mice by TRIZOL LS Reagent (GIBCO BRL) and SuperScript™ preamplification system (GIBCO BRL) was used to make first strand cDNA. For CfoI digestion, we designed another pair of primers to amplify genomic DNA to confirm the mutation: Pde6b13F 5′-CTTTCTATTCTCTGTCAGCAAAGC-3′ and Pde6b13R 5′-CATGAGTAGGGTAAACATGGTCTG-3′. Amplification was done during 36 cycles with a 15 sec denaturing step at 94°C, a 1 min. annealing step at 51°C, and a 1 min. extension step at 72°C. CfoI digestion was carried out directly in a 10-μl volume by adding 8 μl of PCR products, 1 μl of 10x buffer (SE Buffer 5), and 2–5 units of CfoI (SibEnzyme).
Table 1.
PCR Primers Used for Murine Pde6b cDNA Amplification and Sequencing
| Designation | Sequence (5′-3′) | Fragment size | Exons |
|---|---|---|---|
| Pdeb1f | TGTGAAGATGGTTGGCTGGC | 760 | 1–4 |
| Pdeb1r | CATCAAAGAACTCCTTCTCCTTGG | ||
| Pdeb2f | CTGGTCAGCCAATAAGGTGTTTG | 748 | 4–11 |
| Pdeb2r | CAAGATTTTCCCCAGCTCATCC | ||
| Pdeb3f | AAAGAGCCTGCTGACTGTGAGG | 901 | 11–19 |
| Pdeb3r | TGAAACCAACTTGCAGCTTAGGG | ||
| Pdeb4f | AGGCACCACCTGGAATTTGG | 700 | 15–22 |
| Pdeb4r | ACACGGCTTATAGGATACAGCAGG |
Complementation tests between the rd1 strain and the rd10 and nmf137 strains were performed. Affected rd10 or nmf137 homozygotes were mated to C3H/HeJ mice that were homozygous for the Pde6b rd1 mutation. Three experienced observers (B.C., N.L.H., and J.H.) visually inspected the posterior eyes of F1 offspring by funduscopy for signs of retinal degeneration.
RESULTS
Clinical Phenotype
rd10 mice showed an early onset retinal degeneration with sclerotic retinal vessels at four weeks of age. Nmf137 mice showed a mottled retina and white retinal vessels at 24 days of age. The retinal degeneration in the two strains was easily distinguished from normal and rd1 retinal appearance at two months of age by fundoscopy (Fig. 1).
Histological Phenotype
Histological examination revealed progressive retinal outer nuclear layer (ONL) degeneration in rd10 mice that started in the central retina at 16 days and spread to peripheral retina by 20 days of age (data not shown). By 60 days of age, no ONL was left (data not shown). Nuclei counts of the ONL mirrored this decline (Fig. 2). However, inner nuclear layer and ganglion cell layer thicknesses were not affected (Fig. 3). The nmf137 mutants showed extensive degeneration of the outer nuclear layer of the retina starting at 16 days of age and no ONL was left in the retina by 30 days. nmf137 and rd1 appeared to be more severe than the degeneration observed in rd10 mutants (Fig. 3). Retinas from dark-reared rd10 mice showed no degeneration through P24 (Fig. 3). Loss of nuclei in the ONL became apparent by P30 and variably progressed at later times (data not shown). Dark-rearing had no effect on rd1 or nmf137 retinal degeneration.
Figure 2.
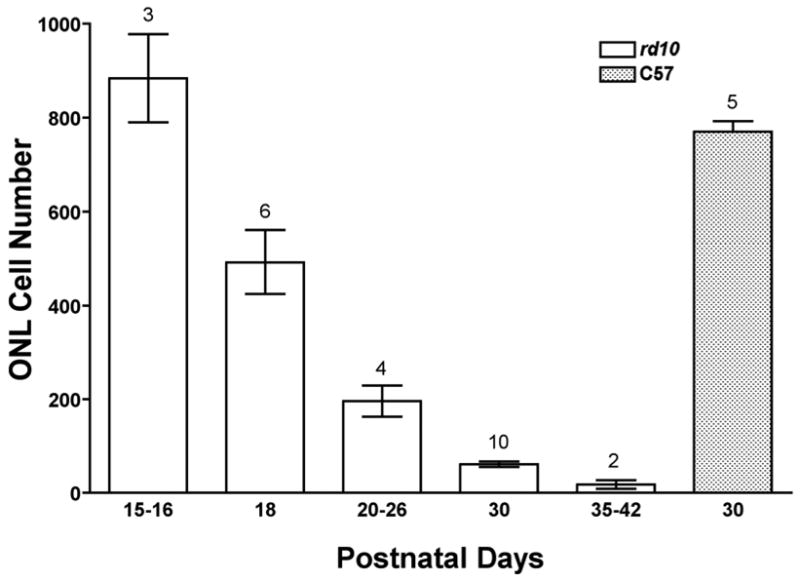
Nuclei counts in the outer nuclear layer (ONL) of rd10 mouse retinas as a function of time. Plastic or paraffin sections of eyes from mice ranging in age from one week to three months were studied using light microscopy. Counts from rd10 sections are significantly lower than those from C57/Bl6 wild type (“C57”) at all stages expect for postnatal day15–16 samples (p<0.001). Data are means±SEM; numbers above bars are sampling sizes. Statistical analysis was by ANOVA with post-hoc Student-Newman-Keuls multiple comparisons testing.
Figure 3.
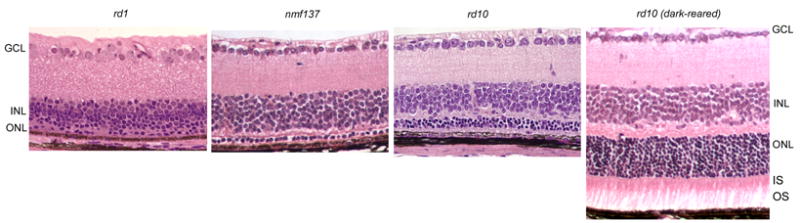
Histology of rd10, rd1, and nmf137 mouse retina at 24 days of age showed comparative degrees of retinal degeneration. By this stage, rd10 outer nuclear layer (ONL) of cyclic light reared mice was about four nuclei thick, whereas nmf137 was only one nuclei thick and rd1 had no photoreceptor nuclei. Conversely, ONL of dark-reared rd10 mice showed no thinning at 24 days of age and had substantial inner segments (IS) and outer segments (OS). Retinal ganglion cell layer (GCL) and inner nuclear layer (INL) did not vary with strain or lighting regimen.
ERG Phenotype
ERGs recorded from homozygous rd10 mice showed reduced rod and cone responses compared to wild-type mice. Figure 4 shows a series of ERG waveforms recorded in response to increasing flash intensities under dark- and light-adapted conditions. Panel A, first column, shows the rod dominated responses from a C57BL/6J wild-type animal. Note the increase in amplitude and decrease in implicit time of the positive b-wave and the negative a-wave with increasing flash intensities. The same flash intensities resulted in much smaller responses in rd10 mice at P18 and P30 (second and third columns of Panel A). At P18, the a-wave was not visible until 0.6 log cd sec/m2 flash intensity. This loss of sensitivity in the dark-adapted response with degeneration was further illustrated by the appearance of a small a-wave only at the brightest flash intensity by P30. Panel B illustrates the cone-isolated responses from C57Bl/6J and rd10 mice. The rd10 response was smaller at all flash intensities at P18 compared to wild type responses and there was continued loss of cone function by P30. However, note that the loss of cone function was not as fast with the rd10 light-adapted b-wave compared to the dark-adapted b-wave. At P18, the rd10 dark-adapted response was 30% of the C57Bl/6 response while the light-adapted response was 50% of the wild type response. Similarly, at P30, the rd10 dark-adapted b-wave was only 10% while the light-adapted response was 30% compared to wild type. Figure 5 shows the reduction in a- and b-wave amplitude with age for dark- and light-adapted recordings. The dark-adapted a-wave declined rapidly from P14 to P30 and was non-recordable by P50 (Fig. 5A, circles). The dark-adapted b-wave steadily declined in amplitude from P14 to less than 15 μV at P63 (Fig. 5A, triangles). The loss of cone-isolated responses was slower. By P63 the cone-isolated a-wave was nonrecordable and the b-wave was less than 6 μV in amplitude. There were no detectable rod and cone ERG responses in nmf137 mice at any ages.
Figure 4.
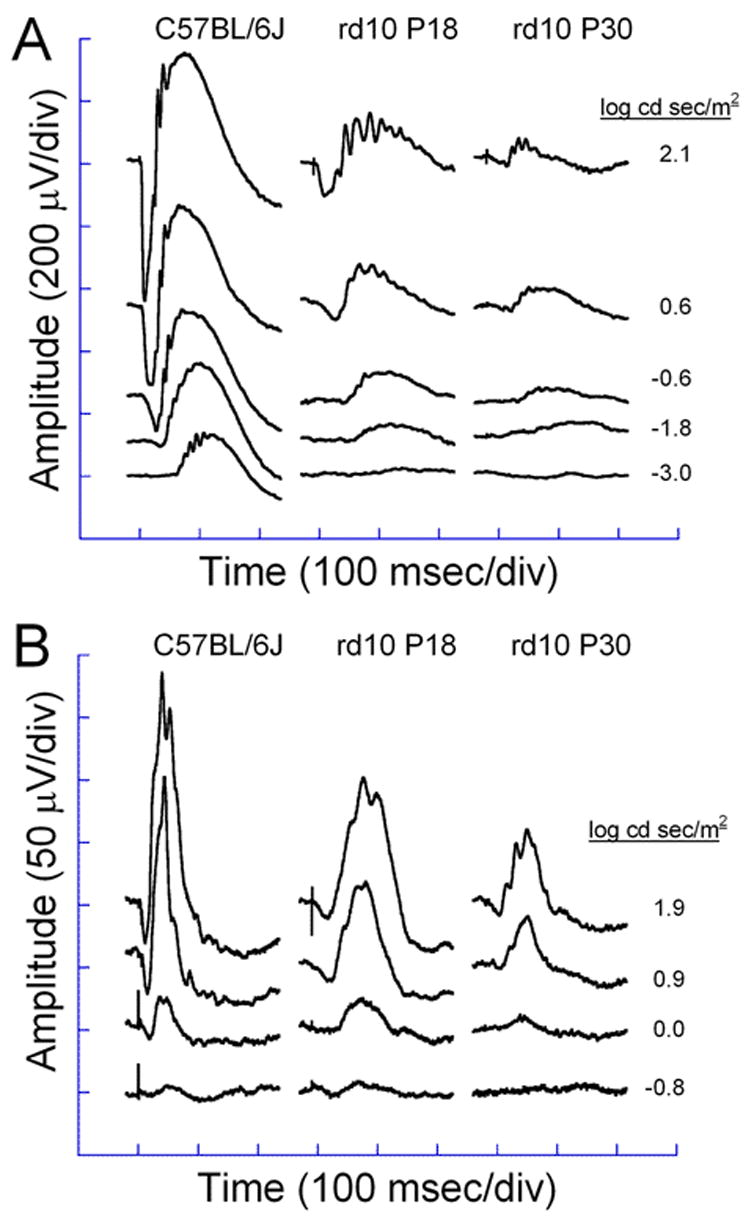
Electroretinograms of C57BL/6J and rd10 mice at P18 and P30 dark-adapted (A) and light-adapted (B) conditions. Dark-adapted responses were recorded after overnight dark-adaptation, while light-adapted responses were recorded with a background light of 1.46 log cd/m2 following a 10 minutes exposure to the same intensity.
Figure 5.
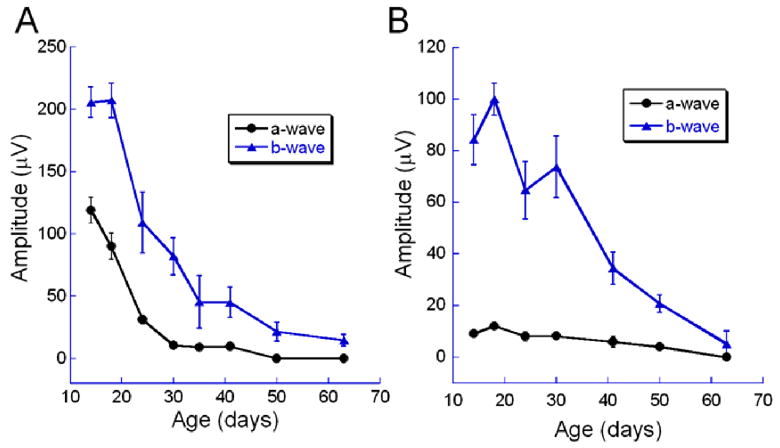
ERG a- and b-wave amplitude of rd10 mice declined with age. A) Rod dominated dark-adapted responses. B) Cone-isolated light-adapted responses. Error bars represent the standard error of the mean.
TUNEL analysis
Retina sections from P18 rd10 mice were positive for TUNEL compared to wild type retinas (Fig. 6). Signal was specific to the ONL, with very little to no signal in the INL and GCL (Fig. 6). Anti-active caspase-3 immunoreactivity was found in P18 rd10 inner segments but not wild type. Little to no anti-active caspase-3 activity was found in other rd10 retina layers (Fig. 6).
Figure 6.
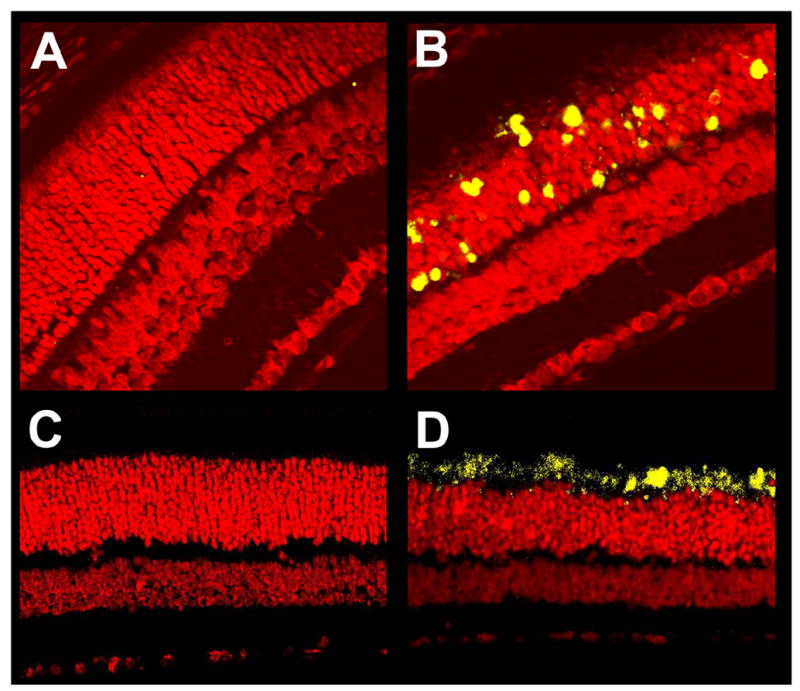
Apoptosis in P18 mouse retinas. Paraffin sections of P18 C57 wild type (A, C) and rd10 (B, D) mouse eyes were subjected to fluorescent TUNEL (A, B) or fluorescent immunochemistry using an antibody specific to activated caspase-3 (C, D). Sections were counterstained with propidium iodide. TUNEL-positive and activated caspase-3-positive cells fluoresced yellow-green. Wild type sections showed almost no TUNEL signal (A), whereas rd10 sections had abundant TUNEL signal in the outer nuclear layer and occasionally inner segments of photoreceptor cells, but not in other layers of the retina (B). Wild type sections showed almost no activated caspase-3 immunofluorescence (C), whereas rd10 sections had abundant signal in the inner segments of photoreceptor cells, but not in other layers of the retina (D). Control stainings lacking primary antibody had no signal (data not shown).
β-PDE immunoreactivity
β-PDE immunoreactivity corresponding to a protein size of about 86 kDa was visualized in extracts from wild type and rd10 retinas, but not rd1retinas or wild type brain (Fig. 7, top). Immunoreactivity to rd10 and wild type retina extract was detectable in P9–11 extracts. Though equal amounts of protein were loaded in all lanes, the signal with rd10 extract was less than that of wild type at all stages (Fig. 7, top and middle). No immunoreactivity was visualized in extracts from rd1 retina at any stage or P60 wild type whole brain (Fig. 7). The Pde6b antibody labeled only inner and outer segments of photoreceptors in cryosections of P22 wild type and rd10 retina, but did not label anywhere in rd1 retina cryosections (Fig. 7, bottom). Omitting primary antibody in immunostains of wild type retina cryosections resulted in no signal (Fig. 7, bottom).
Figure 7.
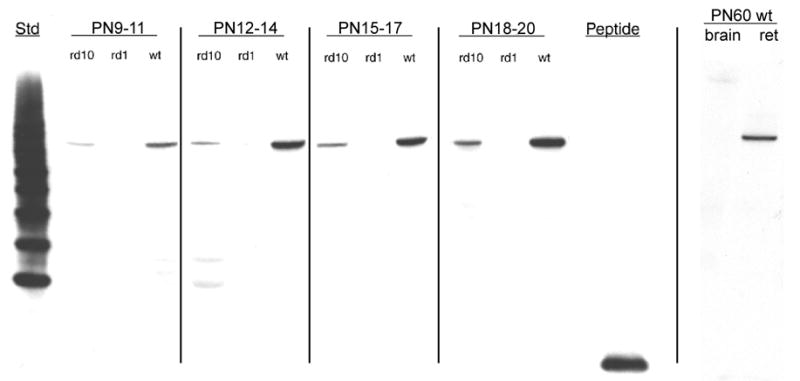
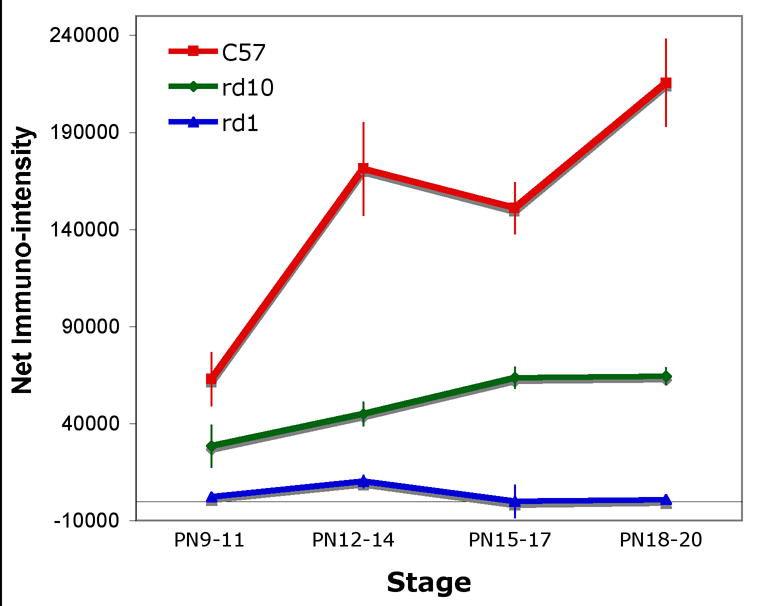
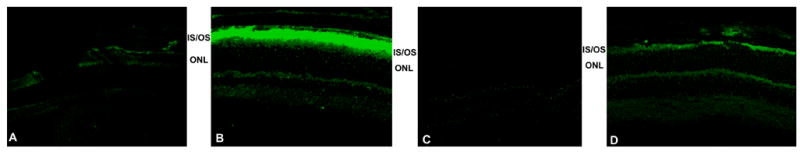
β-PDE immunoreactivity across development. Protein extracts from retinas removed from C57BL/6 (wild type; “wt”), rd1, and rd10 mice at postnatal days (P) 9 through 20 were analyzed by western immunoblotting using an antibody specific for the β subunit of rod cGMP PDE. Additionally, protein extract from retinas and whole brain of a P60 C57BL/6 were probed. Top Panel: Representative autoradiographs of western immunoblots. A band of approximately 86 kDa was present in the wt and rd10 retina protein extracts, but not in rd1 retina or wt brain extracts. Source for protein extracts is indicated above the lanes. “Std” is a “Magic Mark™” molecular weight standard (Invitrogen). “Peptide” is the synthetic peptide mix against which the primary antibody was raised (see text for details of this). As indicated by vertical lines, blots were taken from separate experiments and are representative. Middle Panel: Quantification of P9–20 data from western blots. Autoradiographs were scanned. Net intensities of bands were determined by subtracting background pixel densities from pixel densities of bands. Resulting net intensities were averaged for 3–4 mice per stage per strain. Bottom Panel: Immunohistochemistry shows specificity of β-PDE primary antibody. Cryosections of eyes from P22 rd1 (A), wt (B), and rd10 (D) mice were probed with the β-PDE primary antibody and FITC-conjugated secondary antibody. β-PDE antibody labeled the inner and outer segments of wt (B) and rd10 (D) photoreceptors, but no signal was apparent in rd1 sections (A) or in wt sections incubated with secondary antibody but not primary antibody (C).
Genetic analysis
Genetic analysis shows that the two mutant phenotypes were each inherited as single autosomal recessive genes. The mutation in the rd10 strain was mapped to mouse chromosome (Chr) 5 between D5Mit7 and D5Mit291 (Fig. 8) and the gene symbol Pde6brd10/rd10 (again, abbreviated rd10 here) was assigned in the series of mouse retinal degenerations. In complementation testing in which an affected rd10 mouse was mated to a C3H/HeJ mouse that was homozygous for the rd1 mutation, all F1 offspring were observed to be affected at an early age, with some showing a few white retinal vessels at 21 days, all showing white vessels and mottled retinas at 28 days, and all identical in retinal appearance to rd1 homozygotes at 66 days. Similarly, a complementation test mating nmf137 with rd1 mice resulted in all F1 offspring being affected at 26 days old, suggesting that the retinal degeneration phenotype observed in nmf137 represents a possible new allele of Pde6b that is different from rd1.
Figure 8.
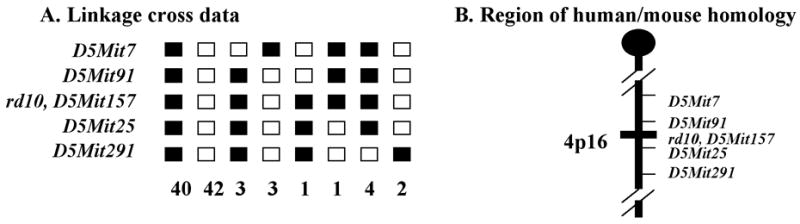
A. 96 mice from a backcross between rd10/rd10 and CAST/Ei were phenotyped and genotyped. Linkage to several markers on mouse Chr 5 was observed. The columns of squares represent haplotypes (filled boxes, rd10/rd10 allele; open boxes, CAST/Ei allele). The number of chromosomes with each haplotype is indicated below each column. B. Genetic map of Chr 5 in the rd10 region showing the closest markers and the region of human homology. Recombination estimates (± standard error) and order for the closest markers were D5Mit91 − 1.04±1.04 − rd10, D5Mit157 − 1.04±1.04 −D5Mit25.
Gene identification and genomic sequencing
Since the rd10 retinal degeneration mapped to mouse Chr 5 near the rd1 mutant gene and testing for allelism was positive, the Pde6b gene was very likely a candidate. Sequence analysis shows that the rd10 retinal degeneration is caused by a missense mutation in exon 13 (a novel mutation changes codon 560 CGC to TGC; changing an arginine to a cysteine, Arg560Cys) in the Pde6b gene (Fig. 9A). The nmf137 retinal degeneration is also caused by a missense mutation in exon 16 (a novel mutation changes codon 659 CTC to CCC; changing an leucine to a proline, Leu659Pro) in the Pde6b gene (Fig. 9B). The rd10 mutation results in the loss of a CfoI site that enabled us to genotype the rd10 mutation by PCR/RFLP (Polymerase Chain Reaction/Restriction Fragment Length Polymorphism) (Fig. 9). ). To confirm the presence of the missense codon in the rd10 Pde6b gene, we re-examined 96 DNAs (49 affected and 47 unaffected mice) from our linkage analysis for the CfoI RFLP. We amplified a Pde6b13F/Pde6b13R 97-bp genomic fragment that contains one CfoI site in the normal allele and none in the rd10 allele. Digestion of the PCR amplified products with CfoI from wildtype, homozygous, and heterozygous rd10 DNA revealed the predicted RFLP pattern (wild-type: two bands of 54 and 43 bp; homozygous rd10: one band of 97 bp; heterozygous rd10: three bands of 97, 54, and 43 bp) (Fig. 9C). This analysis showed that there was absolute concordance between the rd10/rd10 phenotype and the missense mutation. The RFLP pattern thus provides a tool for verification of the presence or absence of the rd10 allele in genetic analysis.
Figure 9.
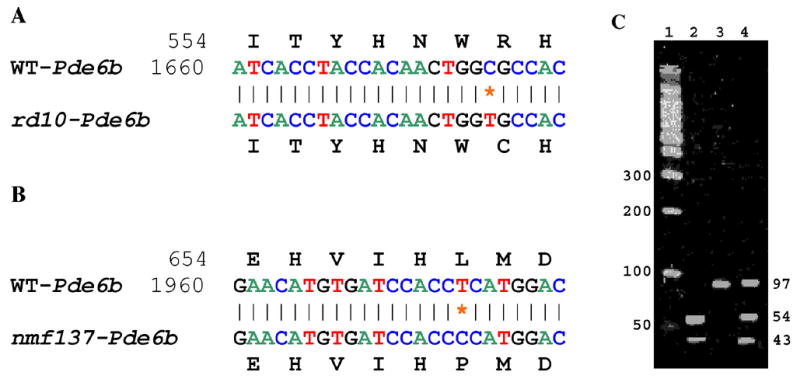
The nucleotide sequences around the single base substitution at position 1678 (C to T) in exon 13 are shown for the wild type (WT) allele and the rd10 allele of Pde6b gene (panel A) and at position 1976 (T to C) in exon 16 are shown for the wild type allele and the nmf137 allele of Pde6b gene (panel B). The novel mutations changed codon 560 CGC to TGC (amino acid change: Arg560Cys) in the Pde6b gene in rd10 mice (panel A) and codon 659 CTC to CCC (amino acid change: Leu659Pro) in the Pde6b gene in nmf137 mice (panel B). Digestion of the PCR amplified products with CfoI from DNA of wild type C57BL/6J (lane 2), homozygous rd10/rd10 (lane 3) and heterozygous CXB-1xB6-rd10/+ F1 (lane 4) revealed the predicted RFLP pattern (panel C). Lane 1 is a 100 base pair (bp) DNA size marker. Numbers that flank the gel image are predicted fragment sizes in base pairs (bp).
DISCUSSION
Mice with mutations in the gene encoding the β subunit of rod cGMP phosphodiesterase, the Pde6b gene, comprise some of the oldest and most-studied animal models of retinal degeneration. Mice homozygous for the rd1 mutation harbor an intronic retrotransposon insertion and a nonsense mutation in exon 7 of the Pde6b gene. This pair of mutations is widely distributed through inbred and wild-derived mouse strains from different parts of the world (Bowes et al. 1993; Pittler & Baehr 1991). In affected animals, the retinal rod photoreceptor cells begin degenerating at about postnatal day 8, and by 4 weeks no photoreceptors are left.
We identified two additional strains of mice carrying mutations in the Pde6b gene, Pde6brd10 (rd10) and Pde6bnmf137 (nmf137), that lead to retinal degenerations. The retinal degeneration caused by a missense mutation in exon 13 of the Pde6b gene in rd10 mice is the first known occurrence of a spontaneously arising mutant allele in the mouse Pde6b gene that is not rd1. Mice homozygous for the rd10 mutation display autosomal recessive hereditary retinal degeneration caused by a missense mutation in exon 13 of the Pde6b gene. Rod photoreceptor cells start degenerating at 16 days of age in the central retina, at 20 days of age in the peripheral retina, and by 60 days of age no photoreceptors are left.
As opposed to the rd10 mutation, the nmf137 mutation arose in an ENU-induced scan. The T to C change in the nmf137 mutation is consistent with mutagenization by ENU, which typically alkylates purines. In this case, the ENU treatment likely modified the A opposite the T of the wild type sequence shown in Figure 9, resulting in the A depurinating followed by an error-prone enzymatic repair replacing the A with a G, and subsequently a complementing C in the indicated mutant gene sequence shown in Figure 9. The nmf137 mutants show extensive degeneration of the ONL starting at 16 days of age and no ONL is left in the retina by 30 days. The hereditary pattern in nmf137 mice is autosomal recessive. This appears to be a more severe degeneration than observed in rd10 mutation and about the same as the rd1 mutant.
The rd10 strain has retinal β-PDE immunoreactivity from about ten days of age, but signal is substantially below that seen in age-matched wild type retina. The lack of immunoreactivity to proteins from rd1 retina at any stage, wild type brain at P60, and in retina cell types other than photoreceptors indicates the specificity of the antibody. Though the immunoreactivity was lower than wild type, it was substantially above that of background or that of rd1 mouse retina at any stage, suggesting that, as opposed to the rd1 mouse, a significant amount of β-PDE is produced in the rd10 retina.
Electroretinograms (ERGs) showed detectable rod and cone ERG responses in rd10 mutants, but there are no detectable rod and cone ERG responses in nmf137 and rd1 mutants (data not shown). The persistence of rod visual cycle function measured by the ERG along with the persistence of β-PDE protein expression in the rd10 mutant well past degeneration onset suggests that the rd10 retinal degeneration is not caused by absence of β-PDE protein but may be caused by insufficient expression and/or low unit enzymatic activity. This could lead to intracellular cGMP accumulation, slower than but similar to that shown in rd1 retina, and eventual cell death. That is, these data indicate that the Pde6b exon 13 missense mutation in rd10 mutants may still have some function while the Pde6b exon 7 nonsense mutation in rd1 mice and Pde6b exon 16 missense mutation in nmf137 mice have lost function. Future experiments are planned to compare rod cGMP PDE enzymatic activity and cGMP accumulation in these strains and to explore the effects of dark-rearing on these parameters.
The rd10 mouse may provide a better model for studying the pathogenesis of autosomal recessive retinitis pigmentosa (arRP) and for experimental therapy than rd1 and nmf137 because of its later onset and milder retinal degeneration. Indeed, the rd10 mouse is being used to study the effects of neurotrophin expression following virally-mediated gene delivery (Rex et al., 2004) and stem cell transplantation (Otani et al., 2004) on retinal degeneration. Further utility may derive from being able to delay degeneration onset by rearing rd10 mice in darkness.
Acknowledgments
Supported by Foundation Fighting Blindness, Research to Prevent Blindness, Knights Templar Education Foundation, a Merit Review Award from the Veteran’s Administration Health Services Research and Development Service, and NIH grants R01 EY007758, P30 EY06360, T32 EY007092, R01 EY014026, R01 EY12514, and R01 EY016470. Parts of this work were presented in abstract form at annual meetings of The Association for Research in Vision and Ophthalmology, Ft. Lauderdale, FL.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Beavo JA, Conti M, Heaslip RJ. Multiple cyclic nucleotide phosphodiesterases. Mol Pharmacol. 1994 Sep;46:399–405. [PubMed] [Google Scholar]
- Bowes C, Li T, Frankel WN, Danciger M, Coffin JM, Applebury ML, Farber DB. Localization of a retroviral element within the rd gene coding for the beta subunit of cGMP phosphodiesterase. Proc Natl Acad Sci U S A. 1993;90:2955–9. doi: 10.1073/pnas.90.7.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang B, Hawes NL, Hurd RE, Davisson MT, Nusinowitz S, Heckenlively JR. Retinal degeneration mutants in the mouse. Vision Res. 42:517–25. doi: 10.1016/s0042-6989(01)00146-8. [DOI] [PubMed] [Google Scholar]
- Clapcote SJ, Lazar NL, Bechard AR, Wood GA, Roder JC. NIH Swiss and Black Swiss mice have retinal degeneration and performance deficits in cognitive tests. Comp Med. 2005 Aug;55:310–6. [PubMed] [Google Scholar]
- Errijgers V, Van Dam D, Gantois I, Van Ginneken CJ, Grossman AW, D'Hooge R, De Deyn PP, Kooy RF. FVB.129P2-Pde6b(+) Tyr(c-ch)/Ant, a sighted variant of the FVB/N mouse strain suitable for behavioral analysis. Genes Brain Behav. 2006 Nov 3 doi: 10.1111/j.1601-183X.2006.00282.x. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Farber DB, Lolley RN. Cyclic guanosine monophosphate: elevation in degenerating photoreceptor cells of the C3H mouse retina. Science. 1974;186:449–51. doi: 10.1126/science.186.4162.449. [DOI] [PubMed] [Google Scholar]
- Farber DB, Lolley RN. Enzymic basis for cyclic GMP accumulation in degenerative photoreceptor cells of mouse retina. J Cyclic Nucleotide Res. 1976;2:139–48. [PubMed] [Google Scholar]
- Hart AW, McKie L, Morgan JE, Gautier P, West K, Jackson IJ, Cross SH. Genotype- phenotype correlation of mouse pde6b mutations. Invest Ophthalmol Vis Sci. 2005 Sep;46:3443–50. doi: 10.1167/iovs.05-0254. [DOI] [PubMed] [Google Scholar]
- Hawes NL, Smith RS, et al. Mouse fundus photography and angiography: a catalogue of normal and mutant phenotypes. Mol Vis. 1999;5:22. [PubMed] [Google Scholar]
- Johnson KR, Gagnon LH, Webb LS, Peters LL, Hawes NL, Chang B, Zheng QY. Mouse models of USH1C and DFNB18: phenotypic and molecular analyses of two new spontaneous mutations of the Ush1c gene. Hum Mol Genet. 2003;12:3075–3086. doi: 10.1093/hmg/ddg332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeler C. Retinal degeneration in the mouse is rodless retina. J Hered. 1966;57:47–50. doi: 10.1093/oxfordjournals.jhered.a107462. [DOI] [PubMed] [Google Scholar]
- Keeler CE. The Inheritance of a Retinal Abnormality in White Mice. PNAS. 1924;10:329–333. doi: 10.1073/pnas.10.7.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaVail MM, Sidman RL. C57BL-6J mice with inherited retinal degeneration. Arch Ophthalmol. 1974;91:394–400. doi: 10.1001/archopht.1974.03900060406015. [DOI] [PubMed] [Google Scholar]
- McLaughlin ME, Ehrhart TL, Berson EL, Dryja TP. Mutation spectrum of the gene encoding the beta subunit of rod phosphodiesterase among patients with autosomal recessive retinitis pigmentosa. Proc Natl Acad Sci U S A. 92:3249–53. doi: 10.1073/pnas.92.8.3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin ME, Sandberg MA, Berson EL, Dryja TP. Recessive mutations in the gene encoding the beta-subunit of rod phosphodiesterase in patients with retinitis pigmentosa. Nat Genet. 1993;4:130–4. doi: 10.1038/ng0693-130. [DOI] [PubMed] [Google Scholar]
- Pittler SJ, Baehr W. Identification of a nonsense mutation in the rod photoreceptor cGMP phosphodiesterase beta-subunit gene of the rd mouse. Proc Natl Acad Sci U S A. 1991;88:8322–6. doi: 10.1073/pnas.88.19.8322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittler SJ, Keeler CE, Sidman RL, Baehr W. PCR analysis of DNA from 70-year-old sections of rodless retina demonstrates identity with the mouse rd defect. Proc Natl Acad Sci U S A. 1993;90:9616–9. doi: 10.1073/pnas.90.20.9616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BA, Navin A, Phillips SJ. PCR-Amplification of Simple Sequence Repeat Variants from Pooled DNA Samples for Rapidly Mapping New Mutations of the Mouse. Genomics. 21:626. doi: 10.1006/geno.1994.1323. [DOI] [PubMed] [Google Scholar]
- Yau KW, Baylor DA. Cyclic GMP-activated conductance of retinal photoreceptor cells. Annu Rev Neurosci. 1989;12:289–327. doi: 10.1146/annurev.ne.12.030189.001445. [DOI] [PubMed] [Google Scholar]


