Abstract
Previous reports showed that urokinase plasminogen activator (uPA) converts plasminogen to plasmin which then activates matrix metalloproteinases (MMPs). Here, we report that uPA directly cleaved pro-MMP-9 in a time-dependent manner at both C- and N-terminus and generated two gelatinolytic bands. uPA-activated-MMP-9 efficiently degraded fibronectin and blocked by uPA inhibitor B428 and recombinant tissue inhibitor of metalloproteinase-1 (TIMP-1). B428 inhibited basal and PMA-induced active MMP-9 in glioblastomas (GBM) U1242 cell media as well as cell invasion in vitro. A combination of MMP-9 and uPA antibodies more significantly inhibited U1242 cell invasion than uPA or MMP-9 antibody alone. Both uPA and MMP-9 were highly expressed in U1242 cell and GBM patient specimens. Furthermore, two active MMP-9 fragments with identical molecular weights to the uPA-activated MMP-9 products were detected in GBM patient specimens. These results suggest that uPA-mediated direct activation of MMP-9 may promote GBM cell invasion.
Keywords: MMP-9, uPA, Activation, Cell invasion, Glioblastoma, Mass spectrum
Extracellular proteolysis is critical for tumor invasion, metastasis and angiogenesis. The two best-characterized groups of extracellular proteolytic enzymes are urokinase plasminogen activator (uPA) and matrix metalloproteinases (MMPs) [1]. MMPs, a family of zinc-dependent enzymes that proteolytically degrade various components of the ECM, play a critical role in a variety of malignant tumor invasive processes [2]. The expression and activation of gelatinase B (MMP-9) are involved in tumor progression [2]. Similarly, increased expression of uPA and MMP-9 has been found in human malignant brain tumors in vivo [3,4] and these proteases play an important role in human glioblastomas (GBM) invasion and tumorigenesis [5,6]. MMP-9 is secreted as an inactive precursor and requires activation by other proteases or autocatalysis [7] which is critical for its activity and biological function.
Previous study reported that uPA interacts with MMP-9 indirectly through uPA/plasminogen/plasmin system, in which uPA activates plasminogen to plasmin. The latter subsequently acts as a potential activator of pro-MMP-9 in various human cancer cell lines [6,8–10]. Other reports demonstrated that plasmin is not a direct [10] or an efficient [11] activator of pro-MMP-9. These findings led us to investigate whether uPA can directly activate MMP-9.
In this study, we provide evidence that uPA directly activates pro-MMP-9 at both N-terminus and C-terminus in vitro. Neutralization of uPA and MMP-9 activities with specific antibodies attenuated GBM U1242 cell invasion. Our results suggest that uPA-evoked MMP-9 activation and increased invasion may be partly mediated through the plasminogen/plasmin-independent pathway.
Materials and methods
Biochemical cleavage assay of MMP-9 by uPA
Purified recombinant uPA (0.02 μM, a gift from Drs. Jack Henkin and Andrew Mazar of Abbott Laboratories, Abbott Park, IL) was incubated with purified pro-MMP-9 (pMMP-9, 0.2 μM), purified MMP-9/TIMP-1 (0.2 μM), and purified MMP-9–lipocalin complex (0.2 μM), MMP-9 monomer (mMMP-9, 0.2 μM) in the presence or absence of purified recombinant N-terminal domain of TIMP-1 (N-TIMP-1, 0.2 μM) [12] in glycine buffer (0.1 M glycine, pH 8.0) at 37 °C for 24 h. To compare this novel activation with other known MMP-9 activators mediated activation, recombinant MMP-3 catalytic domain (0.02 μM), and MMP-26 (0.02 μM), and uPA-activated plasmin (0.02 μM) were incubated with latent MMP-9 (0.2 μM) in the incubation buffer [15] at 37 °C for 24 h. The purified MMPs (except MMP-26 [13]) were purchased from Calbiochem, San Diego, CA. The selective uPA inhibitor 4-iodobenzo[b]thiophene-2-carboxamidine (B428, 7.5 μM) was added to inhibit the cleavage. To verify whether latent forms of MMP-9, MMP-9/TIMP-1 complex and MMP-9/NGAL complex can be activated, 4-aminophenylmercuric acetate (APMA, 1 mM), a well-known pro-MMP activator in vitro, was added and incubated for 4 h. For the time-dependent assay of MMP-9 cleavage by uPA, MMP-9 (0.2 μM) was incubated with uPA (0.02 μM) for the indicated time periods.
Substrate cleavage assay, TIMP-1 inhibition and silver staining
To further determine the uPA cleavage of MMP-9 is activation, substrate cleavage assays were performed. Fibronectin is one of the major ECM components, which are elevated in the brain of the human glioblastoma patients [14]. For fibronectin cleavage assays, pro-MMP-9 was pre-incubated with uPA, MMP-26, MMP-3, plasmin and APMA and for 24 h to generate active MMP-9 solution. Fibronectin (1 mg/ml, Sigma) was incubated with 2 μl of the active MMP-9 solution in incubation buffer [13] at 37 °C for another 24 h. Fibronectin incubated with uPA (0.02 μM), pro-MMP-9 (0.2 μM), MMP-26 (0.02 μM), or MMP-3 (0.02 μM) alone under the same experimental condition were served as control. To test the effects of TIMP-1 on activity of uPA-activated MMP-9, N-TIMP-1 (0.03 μM) was pre-incubated MMP-9 monomer (0.02 μM) at 37 °C for 6 h, followed by adding uPA (0.002 μM) to the pre-incubated solution and incubated at 37 °C for another 24 h. The silver staining was performed according to our previous report [13].
Edman protein N-terminal sequencing and phenyl isocyanate (PIC) N-terminal labeling and mass spectrum (MS) analysis
Edman N-terminal sequencing was performed as previously reported [13] at the Biomolecular Research Facility, University of Virginia. The reaction solution of uPA and MMP-9, and MMP-9 alone incubated with 2.5 mM PIC (Sigma) in 10 mM Hepes (pH 7.5) for 10 min at room temperature, the reaction was stopped by addition of 1 μl of 100 mM ammonium bicarbonate buffer. Under these conditions PIC has been shown to label only the N-terminal amines [15]. The mixture was loaded onto 9% SDS–PAGE for separation and the protein bands were revealed with silver staining [13]. The interested band was excised from the gel. The detailed method of MS was provided in Supplementary Method 1.
Gelatin and fibrinogen zymography
MMP-9 activity was detected by gelatin zymography as described previously [13]. uPA activity was detected using fibrinogen zymography. Briefly, cell media or protein extracts were resolved under non-reducing conditions on 10% SDS–PAGE gels containing 1 mg/ml fibrinogen and 20 μg/ml plasminogen (Sigma). The gels were rinsed, incubated, stained and destained as described previously [13] except the incubation buffer was 0.1 M glycine (pH 8.0).
Cell invasion assay
The cell invasion assay was performed as previously reported [13]. Briefly, cell suspension (1 × 105 cells) was added to each insert, which were pre-coated with 0.25 mg/ml fibronectin in the presence or absence of specific uPA inhibitor, B428 (7.5 μM), phorbol 12-myristate 13-acetate (PMA, 50 nM), uPA antibody (25 μg/ml, American Diagnostica Inc.) and MMP-9 antibody (25 μg/ml, CalBiochem). DMSO or normal IgG was used as controls. The invaded cells were stained and counted as previous report [13].
Results
uPA directly cleaves latent MMP-9
Our biochemical assays revealed that purified 92 kDa pro-MMP-9 was cleaved directly by uPA, generating two new bands (86 and 80 kDa) showed gelatinolytic activity (Fig. 1). A specific uPA inhibitor B428 (7.5 μM) completely blocked the cleavage of MMP-9 by uPA (Fig. 1). Furthermore, uPA-mediated cleavage of pro-MMP-9 was time-dependent. These results suggest that pro-MMP-9 can be cleaved by uPA directly and generated two new bands with gelatinolytic activity in vitro.
Fig. 1.
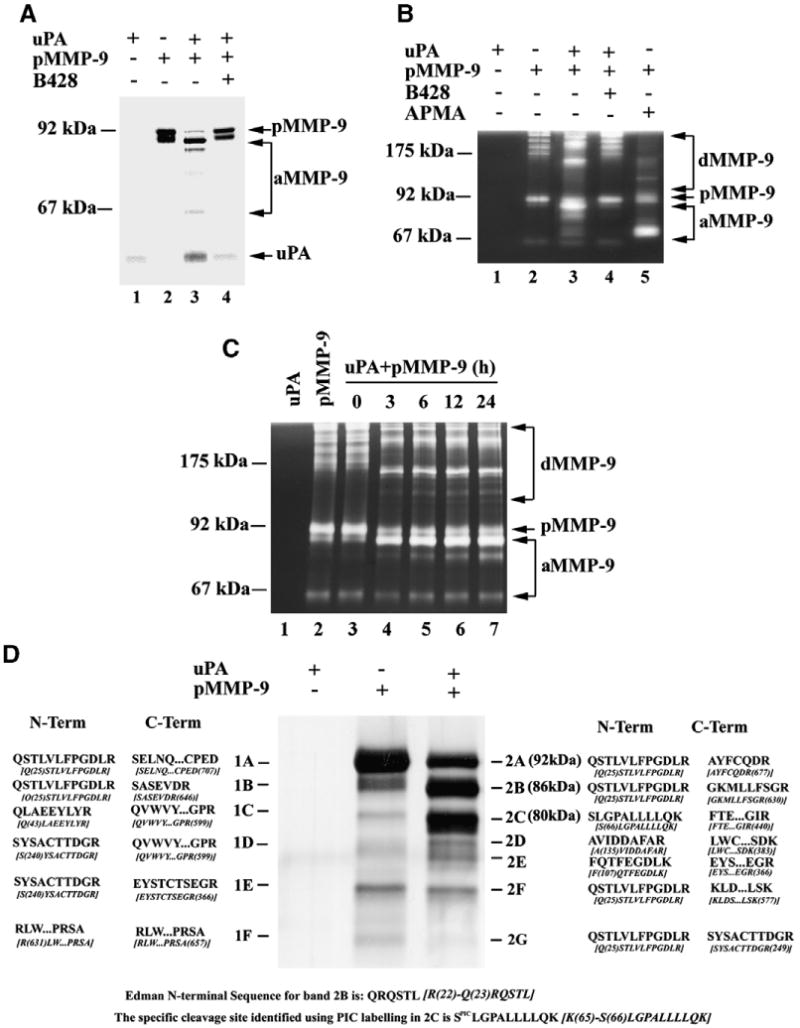
Direct cleavage of pro-MMP-9 by uPA. (A) Purified recombinant uPA directly cleaved pro-MMP-9 in vitro as detected by silver staining. The molar concentration of pro-MMP-9 and uPA are 0.2 μM and 0.02 μM, respectively. pMMP-9, pro-MMP-9; aMMP-9, active MMP-9. (B) Gelatin zymography of uPA-activated MMP-9 under non-reducing conditions. uPA inhibitor B428 (7.5 μM) blocked uPA-activated MMP-9. 0.25 ng MMP-9 protein/lane was loaded. dMMP-9, dimeric MMP-9. (C) Gelatin zymography of time-dependent analysis of MMP-9 cleavage by uPA. (D) Edman N-terminal sequence and mass spectrum (MS) analysis of uPA-activated MMP-9 products. The number in the brackets indicates the position of amino residue. N-Term/C-Term, N-terminal/C-terminal peptide sequence. MMP-9 sequence was listed under each MS sequence. The N-terminal sequence (Edman) of band 2B and PIC N-terminal labeling sequence of band 2C were listed at the bottom.
To identify the cleavage sites of MMP-9 by uPA, Edman N-terminal sequencing and MS analyses using N-terminal PIC-labeling were performed. N- and C-terminal amino acids of 86 kDa band (band 2B) and 80 kDa (band 2C) were shown in Fig. 1D. MS analyses showed that the N-terminal amino acids of band 2B was very similar to the data generated from Edman N-terminal sequencing, which matched the known sequence of MMP-9. Our results explain why bands 2B and C showed gelatinolytic activity because they still had the zinc-binding motif in the catalytic domain of MMP-9. N-terminal and C-terminal fragments of band 1A indicate that band 1A is a full length MMP-9. These results suggest that latent MMP-9 can be cleaved by uPA at both N-terminus and C-terminus. The N- and C-terminal amino acids of other bands were shown as in Fig. 1D.
Comparison of uPA and other known MMP-9 activators on gelatin and fibronectin cleavage
Since a number of previous studies show that uPA converts inactive plasminogen to active plasmin, the latter further cleaves extracellular matrix components and activates some MMPs [6,8–10], we compared the cleavage of uPA-activated MMP-9 and uPA–plasmin-activated MMP-9. The results revealed that uPA–plasmin-activated MMP-9 fragment was 84 kDa, indicating the cleavage site is different from uPA-activated MMP-9 (Fig. 2). Next, we designed experiment to identify enzymatic activity of uPA-activated MMP-9 against its substrate and compare with MMP-26-, MMP-3- and APMA-activated MMP-9 substrate cleavage. We chose fibronectin as substrate because it is a common extracellular matrix component and elevated in human brain glioblastoma [14]. Biochemical substrate digestion assay showed that uPA-activated MMP-9 cleaved fibronectin and generated at least nine new products compared with control (Fig. 2). MMP-26-activated MMP-9 generated four fragments, MMP-3-activated MMP-9 generated three fragments and APMA-activated-MMP-9 even did not show any cleavage; while uPA, pro-MMP-9, MMP-26 and MMP-3 alone exhibited no catalytic activity to fibronectin (Fig. 2C). The silver staining of uPA-, MMP-26-, MMP-3- and APMA-activated MMP-9 and uPA, pro-MMP-9, MMP-26 and MMP-3 alone exhibited no additional bands under the same experimental condition (molar concentration ratio enzyme:fibronectin = 1:50) (data not shown). These results indicate that uPA-activated MMP-9 cleaved fibronectin more efficiently than MMP-26-, -3- and APMA-activated MMP-9 or uPA, pro-MMP-9, MMP-26, and -3 alone in in vitro.
Fig. 2.
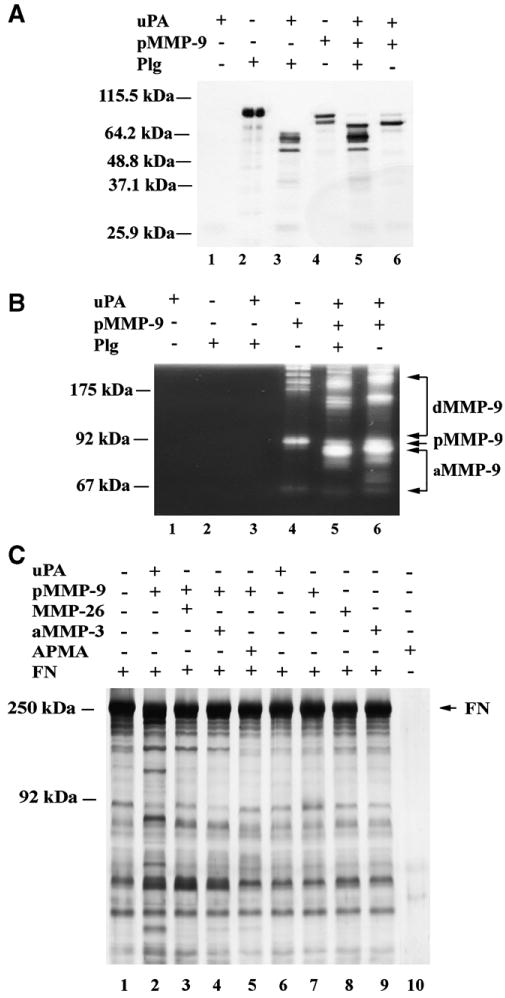
Comparison of uPA- and other activators-activated-MMP-9. (A) Silver staining gel of uPA-activated MMP-9 and uPA–plasmin-activated-MMP-9. The molar concentration of pro-MMP-9, uPA and plasminogen (Plg) are 0.2 μM, 0.02 μM and 0.02 μM, respectively. pMMP-9, pro-MMP-9. (B) Gelatin zymography analysis of the samples in (A). (C) Comparison of fibronectin (FN, 5 μM) cleavage assay amonst uPA-, MMP-26-, MMP-3- and APMA-activated-MMP-9 or uPA, pro-MMP-9, MMP-26 and aMMP-3 alone. The molar concentration of pro-MMP-9, uPA, MMP-26-, MMP-3- and APMA are 0.2 μM, 0.02 μM, 0.02 μM, 0.02 μM and 1 mM, respectively. aMMP-3, purified MMP-3 catalytic domain.
uPA-mediated MMP-9 activation is regulated by TIMP-1 and NGAL
Since MMP-9 preferentially forms complexes with its regulators (TIMP-1 and NGAL), we determined the effects of these regulators on uPA-mediated MMP-9 activation, using purified pro-MMP-9/TIMP-1 and pro-MMP-9/NGAL complexes. Both MMP-9/TIMP-1 and MMP-9/NGAL complexes were not cleaved by uPA compared with control (Fig. 3A), while APMA activated the pro-MMP-9/NGAL complex and reduced the 175 kDa pro-MMP-9/TIMP-1 complex (data not shown). Purified MMP-9 monomer, and the purified recombinant N-TIMP-1 were used to further confirm the regulation of uPA-mediated-pro-MMP-9 cleavage by TIMP-1. The results showed that pre-incubation with both N-TIMP-1 and uPA specific inhibitor B428 for 2 h abolished MMP-9 monomer activation by uPA when compared with control (Fig. 3B). These results suggest that uPA-mediated MMP-9 activation is inhibited by TIMP-1.
Fig. 3.
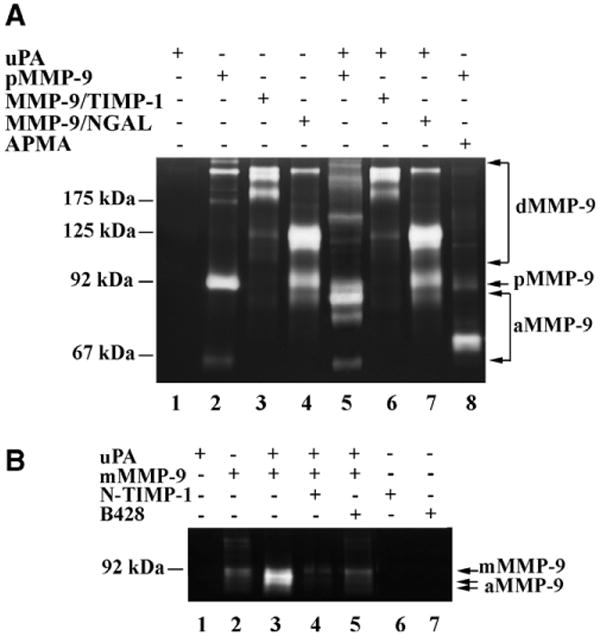
uPA-mediated MMP-9 cleavage is regulated by its regulators. (A) Latent MMP-9 cleavage by uPA is inhibited by forming complex with tissue inhibitor of metalloproteinase-1 (TIMP-1) and neutrophil gelatinase-associated lipocalin (NGAL). Latent MMP-9 (0.2 μM) cleaved by uPA (0.02 μM, lane 5) and APMA (1 mM, lane 8), in contrast, MMP-9/TIMP-1 and MMP-9/NGAL complexes were not cleaved by uPA (lanes 6 and 7) compared with controls (lanes 3 and 4) at the same molar concentration. (B) Latent MMP-9 monomer (0.2 μM) was cleaved by uPA (0.02 μM) (lane 3). Both recombinant N-terminal of TIMP-1 (N-TIMP-1, 0.4 μM, lane 4) and B428 (7.5 μM, lane 5) blocked the cleavage. dMMP-9, dimeric MMP-9; mMMP-9, MMP-9 monomer; aMMP-9, activated MMP-9.
uPA-activated MMP-9 promotes U1242 GBM cell invasion
We then explored whether uPA-mediated-MMP-9 activation is involved in GBM cell invasion. First we identified that both uPA and MMP-9 were highly expressed in U1242 GBM cell media (Fig. 4A and B). Treatment of U-1242 GBM cells with uPA specific inhibitor (B428, 7.5 μM) for 24 h inhibited formation of active MMP-9 in U1242 cell media (Fig. 4A). Following treatment of U1242 cells with PMA (50 nM), a stimulator of uPA and MMPs via activation of protein kinase C pathway, secreted uPA, pro-MMP-9 and active MMP-9 were highly increased compared with DMSO (Fig. 4B and C). B428 blocked PMA-induced pro-MMP-9 and active MMP-9 when compared with control. However, both PMA and B428 had no effect on activity of MMP-2 (Fig. 4C). To further explore the role of uPA-activated MMP-9 in in vitro invasion of U1242 cells, fibronectin invasion assay was used, because uPA-activated MMP-9 degraded fibronectin more efficiently (Fig. 2C). Fibronectin is one of the ECM proteins found in GBM patient specimens [14]. The results showed that B428 significantly inhibited basal invasion (20.1%, p < 0.05) and PMA-stimulated invasion (55.6%, p < 0.001) in U1242 cells as compared with DMSO (Fig. 4D). In addition, a combination of functional neutralizing antibodies of MMP-9 and uPA synergically inhibited U1242 cell invasion by 63.74% (P < 0.001) compared with uPA or MMP-9 antibody alone. Treatment of U-1242 cells with uPA or MMP-9 antibody significantly reduced invasive potential of the cells by 40% (P < 0.001) or 47.63% (P < 0.001), respectively, when compared with normal IgG (Fig. 4D). These results show that uPA-mediated MMP-9 activation promotes U1242 GBM cell invasion.
Fig. 4.
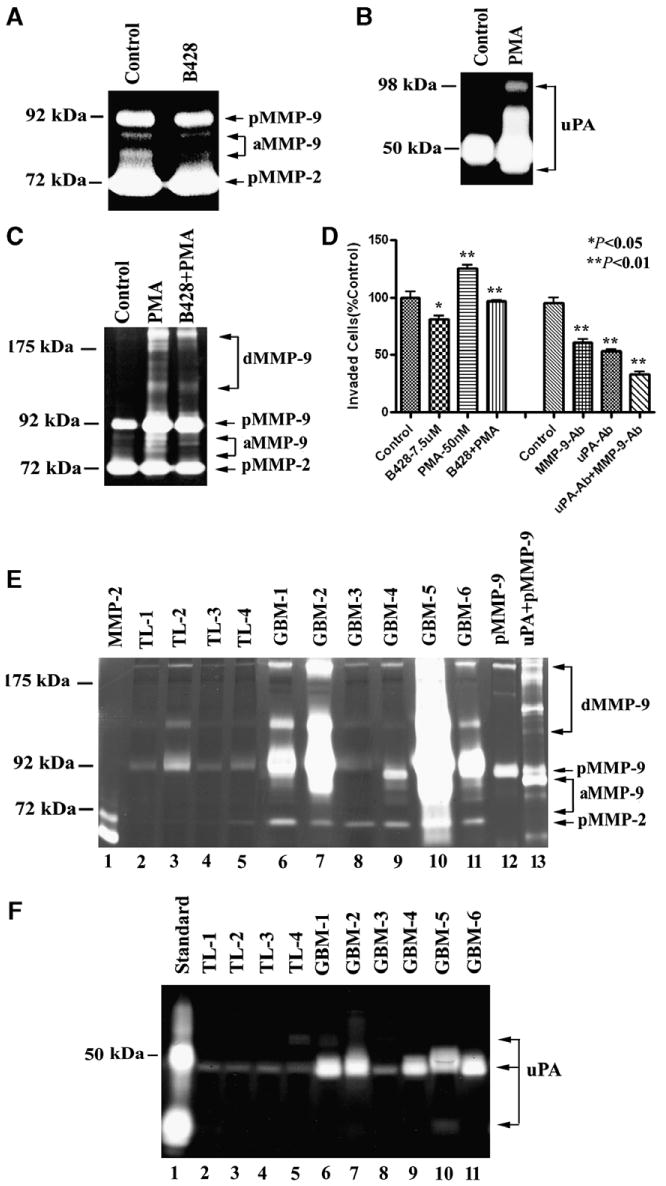
uPA-activated MMP-9 increased U1242 cell invasion and expression of uPA and MMP-9 in GBM patients' specimens and non-neoplastic temporal lobes (TL). (A) B428 (7.5 μM) reduced MMP-9 activity in U1242 cell media. pMMP-9, pro-MMP-9, aMMP-9, active MMP-9; pMMP-2. pro-MMP-2 (B) PMA (50 nM) induced uPA activity in U1242 cell media. (C) B428 (7.5 μM) blocked PMA-induced MMP-9 activity in U1242 cell media. dMMP-9, dimeric MMP-9 (D) Inhibition of uPA and MMP-9 activities reduced U1242 cell invasion. B428 (7.5 μM) blocked both U1242 and PMA-induced U1242 cell invasion. Combination of functional neutralizing MMP-9 and uPA antibodies more significantly inhibited U1242 cell invasion through fibronectin than uPA or MMP-9 antibody alone. The concentration of antibody was 25 μg/ml. Normal IgG was used as control. The numbers of control were used as the 100% invasiveness. Data shown are means ± SEM values from three separate experiments for each group. *p < 0.05; **p < 0.01. (E) Gelatin zymography analysis of MMP-2 and MMP-9 activities in TLs and GBMs. The recombinant MMP-2, latent MMP-9 and uPA-activated MMP-9 were used as controls. (F) Fibrinogen zymography analysis of uPA activities in TLs and GBMs. Recombinant uPA was loaded as control. Equal amount of protein (25 μg) was loaded each lane.
Both uPA and MMP-9 are highly expressed in patient glioblastoma specimens
To explore whether uPA-mediated MMP-9 cleavage exists in vivo, we determined the activities of MMP-9 and uPA in glioblastoma patient specimens as well as non-neoplastic temporal lobes. The results revealed 83% of GBM patient samples showed higher MMP-9 and uPA activities when compared with non-neoplastic temporal lobes (Fig. 4). These results were also confirmed by Western blot analysis (data not shown). In addition, the molecular weights of active MMP-9 fragments in GBM patient samples are similar to the uPA-activated MMP-9 (Fig. 4). This indicates that the uPA-mediated MMP-9 activation may be involved in GBM invasion in vivo via plasminogen/plasmin-independent pathway.
Discussion
Our current results demonstrate that uPA directly cleaved latent MMP-9 to generate two new bands (86 kDa and 80 kDa) that showed gelatinolytic activity (Fig. 1B). The 86 kDa band was the product of MMP-9 cleaved by uPA at the C-terminus, while the 80 kDa band was cleaved at both N-terminus (at Lys65-Ser66 site) and C-terminus. Although we do not know exactly the cleavage sites of 86 kDa and 80 kDa bands at C-terminus from the mass spectrum analysis, the detected C-terminus fragments of 86 kDa and 80 kDa bands contain the zinc-binding motif in catalytic domain, which is essential for metalloproteinase activity. This might explain why these two bands showed gelatinolytic activity in gelatin zymography (Fig. 1B). The time-dependent cleavage assay demonstrates that uPA-cleaved MMP-9 products were very stable for at least 24 h (Fig. 1C). These results indicate that uPA-cleaved MMP-9 is activation rather than random cleavage process.
Our data show that uPA-activated MMP-9 fragments contain cysteine-switch residue and efficiently cleaved gelatin and fibronectin (Figs. 1B and 2C). We also reported previously that MMP-26 cleaved MMP-9 at the Ala (93)-Met (94) site, which is before the cysteine [Cys (99)]-switch residue, but showed a great activity [13]. Both pro-MMP-1 and pro-MMP-3 can be activated via cleavage at the N-terminal region by plasmin, which does not eliminate the Cys-switch residue [8]. These data suggest that it is possible that the cysteine-switch residue remains in its position after an initial cleavage and the enzyme still show activity. These findings appear to be in conflict with the original Cys-switch hypothesis [16], which states that the cysteine residue in the conserved cysteine-switch motif PRCGVPD of the prodomain has to be removed for MMP activation. uPA-activated MMP-9 cleavage sites are different from the cleavage with MMP-1, -7 [17], -2(-14/-2/-3) [18], MMP-3 [19], MMP-26 [13], APMA [20] (Fig. 1B)] and plasmin [7] (Fig. 2B). uPA-activated MMP-9 cleaved fibronectin more efficiently than MMP-26-, -3- and APMP-activated MMP-9, or pro-MMP-9, uPA, MMP-26, and -3 alone. uPA-mediated MMP-9 activation is uPA specific because the uPA specific inhibitor (B428) completely blocked the activation in vitro (Fig. 1). Our results indicate that uPA is a direct and efficient activator of MMP-9 in vitro and uPA-activated MMP-9 efficiently degraded fibronectin in vitro.
It has been proposed that uPA-activated plasminogen/plasmin is the natural or physiological activator(s) of pro-MMP-9 in human fibrosarcoma HT1080 cells [9]. uPA/plasmin-mediated MMP-9 activation promotes invasion of MDA-MB-231 cells produced by the addition of exogenous plasminogen [10]. Several reports have implicated plasmin as a direct activator of pro-MMP-9 [9], while others have reported that plasmin is not a direct [10] or efficient [8] activator of pro-MMP-9. Interestingly, our current results showed that uPA and MMP-9 were highly expressed in U1242 cell line and GBM patient specimens. In addition, GBM patient samples displayed active MMP-9 fragments with identical molecular weights to the uPA-activated MMP-9 products. We further proved that uPA selective inhibitor B428 blocked secreted MMP-9 activity in U1242 cells and cell invasion through fibronectin, as well as inhibited PMA-induced secreted MMP-9 activity in U1242 and cell invasion (Fig. 4). However, both PMA and B428 had no effect on secreted MMP-2 activity. Interestingly, a combination of functional neutralizing MMP-9 and uPA antibodies significantly inhibited U1242 cell invasion more than uPA or MMP-9 antibody alone. In addition, plasminogen/plasmin was not detected in U1242 cell media, lysis and GBM patient specimens (data not shown). These results suggest that uPA-mediated MMP-9 activation promotes U1242 GBM cell in vitro invasion. uPA is an efficient activator of MMP-9 in U1242 GBM cells and may contribute to GBM in vivo invasion via plasminogen/plasmin-independent activation pathway.
pro-MMP-9 forms a preferential complex with TIMP-1 via its C-terminal domain [11]. Our findings showed that the MMP-9/TIMP-1 complex was not cleaved by uPA and the purified N-terminal domain of TIMP-1 completely blocked MMP-9 monomer cleavage by uPA. These results indicate that TIMP-1 may inhibit the uPA-mediated MMP-9 cleavage at both N-terminus and C-terminus by forming MMP-9/TIMP-1 complex. Therefore, the C-terminal hemopexin-like domain of pro-MMP-9 may interact with TIMP-1 to prevent C-terminal cleavage of MMP-9 by uPA. This hypothesis is based on: (1) Our current results that showed uPA cleavage of pro-MMP-9 at both N-terminus and C-terminus, while pro-MMP-9/TIMP-1 complex prevent MMP-9 activation by uPA. (2) TIMP-1 is involved in pro-MMP-3 activation through interaction with the C-terminal hemopexin-like domain of pro-MMP-3 [21]. (3) The prodomain interacts with the C-terminal hemopexin-like domain, which directly affects the activation of pro-MMP-1 [22].
In summary, uPA directly cleaved the latent form of MMP-9 at both N- the C-terminus and this novel activation pathway promotes U1242 GBM cell invasion. The expression and activities of uPA and MMP-9, but not plasminogen/plasmin, were elevated in U1242 cell and patient GBM specimens. These results indicate that uPA-mediated MMP-9 cleavage maybe a patho-physiological event during local invasion of brain tumor cells and these two proteases could be potential targets for GBM treatment.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.bbrc.2008.03.038.
Acknowledgments
We gratefully acknowledge Dr. Galina Kuznetsov at Eisai Research Institute for providing the uPA inhibitor (B428). This work was supported by the Farrow Fellowship and the UVA Cancer Center Support Grant, P30 CA44570 (to Y.G.Z.), NIH Grants NS35122 and CA90851 (to I.M.H.) and AR40994 (to K.B.).
References
- 1.Duffy MJ, Duggan C. The urokinase plasminogen activator system: a rich source of tumour markers for the individualised management of patients with cancer. Clin Biochem. 2004;37:541–548. doi: 10.1016/j.clinbiochem.2004.05.013. [DOI] [PubMed] [Google Scholar]
- 2.Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer. 2002;2:161–174. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- 3.Rao JS, Yammamoto M, Mohanam S, Gokaslan ZL, Stetler-Stevenson WG, Rao VH, Fuller GN, Liotta LA, Nicholson GL, Sawaya RE. Expression and localization of 92 kDa type IV collagenase/gelatinase B (MMP-9) in human gliomas. Clin Exp Metastasis. 1996;14:12–18. doi: 10.1007/BF00157681. [DOI] [PubMed] [Google Scholar]
- 4.Hsu DW, Efird JT, Hedley-Whyte ET. Prognostic role of urokinase-type plasminogen activator in human gliomas. Am J Pathol. 1995;147:114–123. [PMC free article] [PubMed] [Google Scholar]
- 5.Rao JS. Molecular mechanisms of glioma invasiveness: the role of proteases. Nat Rev Cancer. 2003;3:489–501. doi: 10.1038/nrc1121. [DOI] [PubMed] [Google Scholar]
- 6.Murphy G, Atkinson S, Ward R, Gavrilovic J, Reynolds J. The role of plasminogen activators in the regulation of connective tissue metalloproteinases. Ann NY Acad Sci. 1992;667:1–12. doi: 10.1111/j.1749-6632.1992.tb51590.x. [DOI] [PubMed] [Google Scholar]
- 7.Fridman R, Toth M, Chvyrkova I, Meroueh SO, Mobashery S. Cell surface association of matrix metalloproteinase-9 (gelatinase B) Cancer Metastasis Rev. 2003;22:153–166. doi: 10.1023/a:1023091214123. [DOI] [PubMed] [Google Scholar]
- 8.DeClerck YA, Laug WE. Cooperation between matrix metalloproteinases and the plasminogen activator–plasmin system in tumor progression. Enzyme Protein. 1996;49:72–84. doi: 10.1159/000468617. [DOI] [PubMed] [Google Scholar]
- 9.Baramova EN, Bajou K, Remacle A, L'Hoir C, Krell HW, Weidle UH, Noel A, Foidart JM. Involvement of PA/plasmin system in the processing of pro-MMP-9 and in the second step of pro-MMP-2 activation. FEBS Lett. 1997;405:157–162. doi: 10.1016/s0014-5793(97)00175-0. [DOI] [PubMed] [Google Scholar]
- 10.Ramos-DeSimone N, Hahn-Dantona E, Sipley J, Nagase H, French DL, Quigley JP. Activation of matrix metalloproteinase-9 (MMP-9) via a converging plasmin/stromelysin-1 cascade enhances tumor cell invasion. J Biol Chem. 1999;274:13066–13076. doi: 10.1074/jbc.274.19.13066. [DOI] [PubMed] [Google Scholar]
- 11.Goldberg GI, Strongin A, Collier IE, Genrich LT, Marmer BL. Interaction of 92-kDa type IV collagenase with the tissue inhibitor of metalloproteinases prevents dimerization, complex formation with interstitial collagenase, and activation of the proenzyme with stromelysin. J Biol Chem. 1992;267:4583–4591. [PubMed] [Google Scholar]
- 12.Wei S, Chen Y, Chung L, Nagase H, Brew K. Protein engineering of the tissue inhibitor of metalloproteinase 1 (TIMP-1) inhibitory domain. In search of selective matrix metalloproteinase inhibitors. J Biol Chem. 2003;278:9831–9834. doi: 10.1074/jbc.M211793200. [DOI] [PubMed] [Google Scholar]
- 13.Zhao YG, Xiao AZ, Newcomer RG, Park HI, Kang T, Chung LW, Swanson MG, Zhau HE, Kurhanewicz J, Sang QX. Activation of pro-gelatinase B by endometase/matrilysin-2 promotes invasion of human prostate cancer cells. J Biol Chem. 2003;278:15056–15064. doi: 10.1074/jbc.M210975200. [DOI] [PubMed] [Google Scholar]
- 14.Chintala SK, Sawaya R, Gokaslan ZL, Fuller G, Rao JS. Immunohistochemical localization of extracellular matrix proteins in human glioma, both in vivo and in vitro. Cancer Lett. 1996;101:107–114. doi: 10.1016/0304-3835(96)04124-9. [DOI] [PubMed] [Google Scholar]
- 15.Mason DE, Liebler DC. Quantitative analysis of modified proteins by LC–MS/MS of peptides labeled with phenyl isocyanate. J Proteome Res. 2003;2:265–272. doi: 10.1021/pr0255856. [DOI] [PubMed] [Google Scholar]
- 16.Van Wart HE, Birkedal-Hansen H. The cysteine switch: a principle of regulation of metalloproteinase activity with potential applicability to the entire matrix metalloproteinase gene family. Proc Natl Acad Sci USA. 1990;87:5578–5582. doi: 10.1073/pnas.87.14.5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sang QX, Birkedal-Hansen H, Van Wart HE. Proteolytic and non-proteolytic activation of human neutrophil progelatinase B. Biochim Biophys Acta. 1995;1251:99–108. doi: 10.1016/0167-4838(95)00086-a. [DOI] [PubMed] [Google Scholar]
- 18.Toth M, Chvyrkova I, Bernardo MM, Hernandez-Barrantes S, Fridman R. Pro-MMP-9 activation by the MT1-MMP/MMP-2 axis and MMP-3: role of TIMP-2 and plasma membranes. Biochem Biophys Res Commun. 2003;308:386–395. doi: 10.1016/s0006-291x(03)01405-0. [DOI] [PubMed] [Google Scholar]
- 19.Ogata Y, Enghild JJ, Nagase H. Matrix metalloproteinase 3 (stromelysin) activates the precursor for the human matrix metalloproteinase 9. J Biol Chem. 1992;267:3581–3584. [PubMed] [Google Scholar]
- 20.Stack MS, Itoh Y, Young TN, Nagase H. Fluorescence quenching studies of matrix metalloproteinases (MMPs): evidence for structural rearrangement of the proMMP-2/TIMP-2 complex upon mercurial activation. Arch Biochem Biophys. 1996;333:163–169. doi: 10.1006/abbi.1996.0377. [DOI] [PubMed] [Google Scholar]
- 21.Suzuki K, Kan CC, Huang W, Gehring MR, Brew K, Nagase H. Expression of human pro-matrix metalloproteinase 3 that lacks the N-terminal 34 residues in Escherichia coli: autoactivation and interaction with tissue inhibitor of metalloproteinase 1. Biol Chem. 1998;379:185–191. doi: 10.1515/bchm.1998.379.2.185. [DOI] [PubMed] [Google Scholar]
- 22.Jozic D, Bourenkov G, Lim NH, Visse R, Nagase H, Bode W, Maskos K. X-ray structure of human proMMP-1: new insights into procollagenase activation and collagen binding. J Biol Chem. 2005;280:9578–9585. doi: 10.1074/jbc.M411084200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.bbrc.2008.03.038.


