Summary
Diphthamide is a post-translational derivative of histidine in protein synthesis elongation factor-2 (eEF-2) that is present in all eukaryotes with no known normal physiological role. Five proteins Dph1-Dph5 are required for the biosynthesis of diphthamide. Chinese hamster ovary (CHO) cells mutated in the biosynthetic genes lack diphthamide and are resistant to bacterial toxins such as diphtheria toxin. We found that diphthamide-deficient cultured cells were 3-fold more sensitive than their parental cells towards ricin, a ribosome-inactivating protein (RIP). RIPs bind to ribosomes at the same site as eEF-2 and cleave the large ribosomal RNA, inhibiting translation and causing cell death. We hypothesized that one role of diphthamide may be to protect ribosomes, and therefore all eukaryotic life forms, from RIPs, which are widely distributed in nature. A protective role of diphthamide against ricin was further demonstrated by complementation where dph mutant CHO cells transfected with the corresponding DPH gene acquired increased resistance to ricin in comparison with the control transfected cells, and resembled the parental CHO cells in their response to the toxin. These data show that the presence of diphthamide in eEF-2 provides protection against ricin and suggest the hypothesis that diphthamide may have evolved to provide protection against RIPs.
Introduction
Diphthamide, a unique amino acid, is a post-translational derivative of histidine that is present only in protein synthesis elongation factor 2 of eukaryotes (eEF-2) (Bodley et al., 1984; Moehring et al., 1980; Van Ness et al., 1980). Diphthamide is the target of the bacterial toxins diphtheria toxin (DT) and Pseudomonas exotoxin A (ETA) (Liu and Leppla, 2003; Oppenheimer and Bodley, 1981) and of fusion proteins derived from them that are being developed as anticancer agents (Liu et al., 2003; Pastan et al., 2007). These toxins transfer ADP-ribose from NAD to diphthamide (Fig. 1). The ADP-ribosylated eEF-2 can no longer perform its normal function in the translational process and intoxicated cells that are unable to synthesize proteins eventually die. DT and ETA are single polypeptides which bind the receptors heparin-binding epidermal growth factor-like growth factor precursor and low-density lipoprotein receptor-related protein-1, respectively (FitzGerald et al., 1995; Naglich et al., 1992).
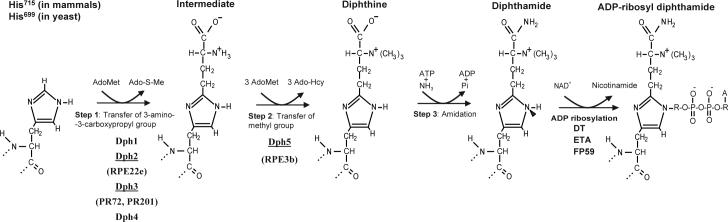
Fig. 1. Diphthamide biosynthetic pathway and ADP-ribosylation.
Dph1 to Dph5 are required for the biosynthesis of diphthamide. Dph1 to Dph4 are involved in the first step, the transfer of 3-amino-3-carboxypropyl to the His715 (His699 in yeast) of eEF-2. Dph5 acts as methyl transferase in the next step, yielding diphthine. Amidation of diphthine is the last step of diphthamide biosynthesis. Arrowhead (N-1 of the histidine imidazole ring of diphthamide) indicates the site of ADP-ribosylation by ADP-ribosylating toxins. AdoMet, S-adenosyl methionine; Ado-S-Me, methylthioadenosine; Ado-Hcy, S-adenosylhomocysteine. In ADP-ribosyl diphthamide, A, adenine moiety; R, ribosyl moiety. The CHO cell mutants lacking corresponding functional Dph proteins used in this study are shown in parentheses.
Our laboratory has made extensive use of a fusion protein designated FP59, which contains the ADP-ribosylating catalytic domain of ETA fused to a portion of anthrax toxin. Anthrax toxin consists of the single cell-binding moiety protective antigen (PA) and two alternate catalytic subunits known as lethal factor (LF) and edema factor (EF) (Leppla, 2006). PA is the central component which is essential for translocation of LF and EF into the cytosol. PA uses tumor endothelium marker 8 (TEM8) and capillary morphogenesis protein 2 (CMG2) as receptors to gain entry into the cells (Leppla, 2006). In the fusion protein FP59, LF amino acids 1−254 are fused to the catalytic domain of ETA such that administration with PA causes delivery of the ADP-ribosylating activity to the cytosol (Arora et al., 1992). Because PA receptors are widely distributed, the combination of PA and FP59 is highly toxic to nearly all cell types. The three ADP-ribosylating toxins (DT, ETA, PA+FP59) have diphthamide as their sole target, as shown by the facile selection of somatic mutant cells cross-resistant to all three toxins, and the demonstration that these have eEF-2 lacking diphthamide (Liu et al., 2006; Moehring et al., 1980; Moehring and Moehring, 1979).
Diphthamide biosynthesis requires five proteins, Dph1 to Dph5, which work cooperatively and sequentially to assemble the side chain on the precursor His715 (His699 in yeast) in eEF-2, leading to diphthamide (Fig. 1) (Chen and Bodley, 1988; Liu et al., 2004; Moehring et al., 1984). The first step is transfer of a 3-amino-3-carboxypropyl group to the imidazole C-2 of the precursor histidine using S-adenosyl methionine (AdoMet) as donor. This step requires a coordinated action by Dph-1 to −4 (Liu et al., 2004; Liu and Leppla, 2003). The second step, catalyzed by Dph5, is trimethylation of the 3-amino-3-carboxypropyl group to produce diphthine (Mattheakis et al., 1992). In the final step, amidation of carboxyl group, by a yet unknown amidase, yields diphthamide. Mutations have been found in yeast and somatic cells that fail to perform many of these steps.
Because diphthamide-deficient somatic cell and yeast mutants are viable (Chen et al., 1985; Liu et al., 2004), it is evident that eEF-2 lacking diphthamide is still able to perform its basic role in protein synthesis. The question then arises as to why this complex, apparently dispensable post-translational modification has evolved and been maintained in all eukaryotes, and also in the archaea (Pappenheimer, Jr. et al., 1983). Many studies have been done that attempt to decipher the role of diphthamide in eEF-2, but they are not conclusive. Site directed mutagenesis studies of the precursor histidine residue are consistent in showing that eEF-2 lacking diphthamide at His699 (in yeast) or His715 (in mammals) retains activity in protein synthesis while failing to be a substrate for DT (Ivankovic et al., 2006; Kimata and Kohno, 1994; Phan et al., 1993). The first direct evidence of a beneficial action was the recent demonstration that diphthamide increases translational accuracy, because yeast strains having diphthamide-deficient eEF-2 showed increased −1 frameshifting (Ortiz et al., 2006).
The region on the ribosome at which eEF-2 acts overlaps with the site targeted by “ribosome inactivating proteins” (RIPs). RIPs comprise a family of toxic proteins with representatives in fungi (alpha-sarcin) and pathogenic bacteria (Shiga toxin), and a large number of both single and two-chain proteins in plants, where they may act to discourage consumption by animals (Barbieri et al., 1993; Perentesis et al., 1992). RIPs act as N-glycosidases that inactivate 60 S ribosomal subunits by hydrolyzing the N-glycosidic bond of an adenosine residue (A4324) in the “ricin-sarcin loop” of 28 S rRNA. Ricin, a toxic protein from the seeds of Ricinus communis, is representative of two-chain RIPs which have a receptor-binding chain that facilitates cell entry and high potency (Olsnes and Kozlov, 2001). In contrast, a larger and diverse set of RIPs are single-chain proteins (e.g., saporin) which lack a receptor-binding domain and rely on non-specific mechanisms to gain entry to cells (Perentesis et al., 1992).
Previous studies in vitro have shown that ricin-induced inactivation of ribosomes can be overcome by adding higher concentrations of eEF-2 (Brigotti et al., 1989; Fernandez-Puentes et al., 1976; Holmberg and Nygard, 1994). Binding of eEF-2 in the presence of non-hydrolyzable GTP analogue GuoPP[CH2]P completely protected ribosomes from ricin (Holmberg and Nygard, 1994). However, there appear to be no studies in which the binding of eEF-2 lacking diphthamide to ribosomes was measured. In the present study, we analyzed the comparative sensitivity to ricin of the various CHO cells having wild type or diphthamide-deficient eEF-2 and found that diphthamide protects against ricin. Based on this result, we speculate that diphthamide may have evolved to protect against ribosome inactivating proteins.
Results
CHO cells having DPH3 inactivated show increased sensitivity towards ricin
To identify mammalian genes required for anthrax toxin action, we previously performed retroviral insertional mutagenesis in CHO cells (Liu and Leppla, 2003). The mutagenized CHO cell population was selected with PA plus FP59 and many toxin resistant mutants were obtained. These mutant cells were further screened for their sensitivity against various toxins including DT, ETA and ricin. Most of the mutant clones were resistant only to PA plus FP59 and were determined to be PA receptor-deficient mutants (data not shown), whereas two of the mutants, PR72 and PR201, showed a different phenotype. PR72 and PR201 were resistant not only to PA plus FP59 (Fig. 2a), but also were completely resistant to DT and ETA (data not shown). These cells were shown to be mutated in a gene now designated DPH3 (Liu et al., 2006) that is required for diphthamide biosynthesis (Fig. 1). Using PR72 as representative of the two dph3 mutant cell lines, we confirmed the inability of eEF-2 to be ADP-ribosylated by ADP ribosylating toxin, FP59 (Fig. 2a, inset). Surprisingly, PR72 and PR201 showed increased sensitivity to ricin; the EC50 values for PR72 and PR201 cells were reduced by 3-fold as compared to their parental cells, WTP4 (Fig. 2b, and Table 1). Since both PR72 and PR201 cells lack diphthamide on eEF-2, we considered whether the absence of this unique post-translational modification might be responsible for increased sensitivity to ricin.
Fig. 2. Toxin sensitivity of diphthamide-deficient CHO mutant cells compared to their parental cell lines.
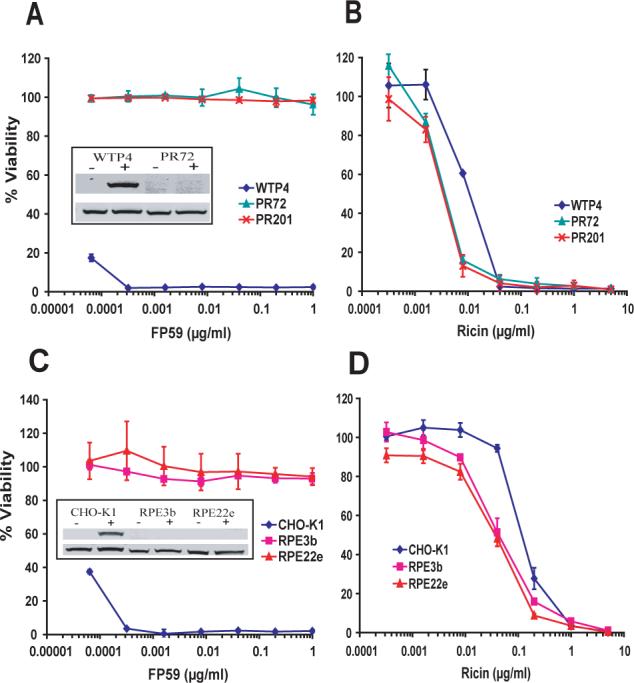
Cells were incubated with the indicated concentrations of FP59 + 500 ng/ml PA (a, c) or with ricin (b, d) for 48 h and MTT assay was done to measure cell viability. The A540 values obtained for cells that received no toxin were considered as 100 % and used to calculate the percent viability of other data points. Insets show western blots for in vitro ADP-ribosylation assay with the cell lysates. In the inset, top panels show ADP-ribosylated eEF-2 while the bottom panel shows the blot using antibody against the carboxy terminus of eEF-2 of human origin. − and + denote the absence or addition of FP59 respectively.
Table 1.
Ricin sensitivity of diphthamide-deficient CHO cells and their parental cells.
| CHO Cell Line | Gene mutated | Parental cells | #EC50 (ng/ml) | *Ratio | Reference |
|---|---|---|---|---|---|
| WTP4 | - | - | 115 | - | Liu and Leppla, 2003 |
| PR72 | DPH3 | WTP4 | 38 | 3 | Liu and Leppla, 2003 |
| PR201 | DPH3 | WTP4 | 39 | 3 | Liu and Leppla, 2003 |
| CHO-K1 | - | - | 120 | - | Moehring et al., 1984 |
| RPE3b | DPH5 | CHO-K1 | 40 | 3 | Moehring et al., 1980 |
| RPE22e | DPH2 | CHO-K1 | 40 | 3 | Moehring et al., 1980 |
All the cell lines used are derivatives of CHO cells and were obtained by chemical or retroviral mutagenesis as reported earlier (Liu and Leppla, 2003).
EC50 is the effective concentration of toxin required to kill 50% of cells. Data is taken from Fig. 2.
Ratio was calculated by dividing EC50 value of parental cells by the EC50 of their corresponding diphthamide-deficient cells.
CHO cells with other DPH genes inactivated are also hypersensitive towards ricin
Recently, the DPH3 gene has been shown to be required for modifying certain tRNAs (Huang et al., 2005), and therefore the Dph3 defect in PR72 and PR201 cells could theoretically have altered sensitivity to ricin in other ways. Therefore we analyzed additional CHO mutant cells lacking diphthamide because of defects in other DPH genes. RPE22e and RPE3b cells were generated from their parental CHO-K1 cells by chemical mutagenesis and selection for resistance to DT (Moehring et al., 1980; Moehring et al., 1984). RPE3b cells do not have functional Dph5 while RPE22e cells lack functional Dph2 protein (Liu et al., 2004; Moehring et al., 1984). RPE3b and RPE22e cells showed complete resistance towards the ADP-ribosylating toxin FP59 in combination with PA, while CHO-K1 cells were highly sensitive (Fig. 2c), confirming that CHO-K1 cells have wild type eEF-2, while RPE3b and RPE22e cells have eEF-2 lacking diphthamide. In vitro ADP-ribosylation assays further confirmed the presence of ADP-ribosylatable eEF-2 specific to CHO-K1 cells (Fig. 2c, inset). In cytotoxicity experiments, RPE3b and RPE22e cells exhibited significant hypersensitivity towards ricin as compared to their parental cell line, CHO-K1 (Fig. 2d). The EC50 was 3-fold lower for both RPE22e and RPE3b as compared to CHO-K1 cells (Table 1).
Complementation of diphthamide biosynthesis in diphthamide-deficient cells restores their resistance to ricin
To further verify that the hypersensitivity of the mutant cells towards ricin is due to the absence of the diphthamide modification on eEF-2, the cells were transfected with the corresponding DPH genes to complement diphthamide biosynthesis. The empty expression plasmid or a plasmid harboring an unrelated gene was also transfected into each cell type to be used for comparative analysis. Many independent stable cell lines were generated by expansion of individual clones from each transfection. RPE3b cells were transfected with either pIRES-DPH5 or empty vector (pIRES-hyg-2). DPH5 encodes a 300-residue methyl transferase (Mattheakis et al., 1992). Expression of Dph5 in RPE3b cells made them sensitive to PA plus FP59 while control transfected cells were still resistant, as expected (Fig. 3a). Complementation was further confirmed by an in vitro ADP-ribosylation assay (Fig. 3a, inset). For toxicity assays with ricin, a number of DPH5 and control vector transfected cell lines were assayed to assure that any changes observed were not restricted to single cell line. All the DPH5 transfectants were more resistant than any of the control transfectants (Fig. 3b). The average EC50 for DPH5 transfected RPE3b cells was 4.7-fold higher than for vector transfected RPE3b cells (Table 2).
Fig. 3. Toxin sensitivity of diphthamide-deficient CHO mutant cells complemented by transfection of the corresponding DPH genes.
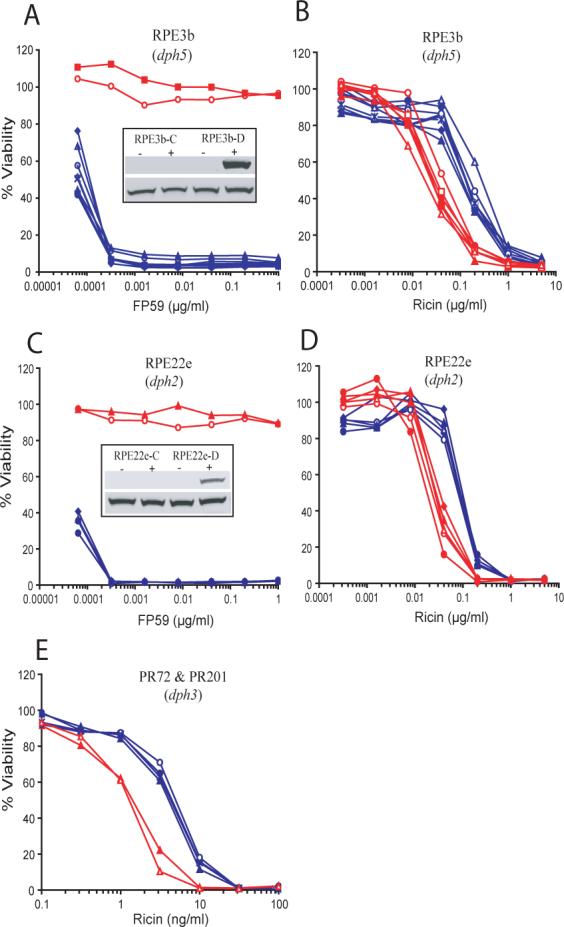
Cells transfected with corresponding DPH gene have been designated by adding “D” after the name of cell lines and are shown in blue, while control transfectants for same cell types have been shown by adding “C” and are shown in red. Each line is derived from an independent stable transformant cell line. Toxin sensitivities of representative individual clones were measured as in Fig. 2 for “PA + FP59” (a, c) and “ricin” (b, d, e). In panels b, d, and e the blue lines (“D” clones) are clustered to the right, indicating increased resistance to ricin. Insets show the western blot for in vitro ADP-ribosylation assays done with the cell lysates, as in Fig. 2. In the experiment of panel e, the “-C” clones are those transfected with an unrelated gene, VPS11, rather than empty vector. Also, in the experiment of panel e, both mutant and transfected cells show increased sensitivity towards ricin compared to other experiments (e.g., Fig. 2, 3b and 3d) due to use of a more potent batch of ricin, but the effect of complementation was again to increase resistance.
Table 2.
Ricin sensitivity of diphthamide-deficient cells transfected with the corresponding DPH gene or control vector.
| CHO Cell Line | Transfected with | #EC50 of individual transfectants [ng/ml] (average EC50) | *Ratio | p Value |
|---|---|---|---|---|
| RPE3b-D | DPH5 gene | 130,100,120,100,300,120,150,120 (143) | 4.7 | 0.0014 |
| RPE3b-C | Empty vector | 25,25,32,20,30,50 (30) | ||
| RPE22e-D | DPH2 gene | 100,90,80,90, 90 (90) | 3.3 | <.0001 |
| RPE22e-C | Empty vector | 22,28,20,35,30 (27) | ||
| PR72-D, PR201-D& (2 clones each) | DPH3 gene | 4.5, 4, 5, 4.5 (4.5) | 3.2 | 0.0001 |
| PR72-C, PR201-C& | Unrelated gene, VPS11 | 1.5, 1.2 (1.4) |
Cell lines were generated by transfecting the diphthamide-deficient CHO cells either with the corresponding DPH gene (cell line names having the extension “-D”) or with the control (empty vector or unrelated VPS11 gene; cell line names having the extension “-C”).
EC50 is the effective concentration of toxin required to kill 50 % of cells. Data is taken from Fig. 3.
Fold difference was calculated by dividing average EC50 of DPH transfectants by the average of control transfectants.
Assays for these cell lines used a more potent batch of ricin than that used for the other toxicity assays.
RPE22e cells, which lack functional Dph2, were complemented with the wild type gene. DPH2 encodes a 534-residue protein having an unknown role in the first step of diphthamide biosynthesis (Mattheakis et al., 1993). As expected, cells transfecting with DPH2 became sensitive to PA plus FP59 in combination with PA while control transfected cells remained resistant (Fig. 3c). Complementation also restored the activity of eEF-2 to ADP-ribosylation by FP59 (Fig. 3c, inset). Assays for sensitivity to ricin yielded results paralleling those for Dph5, in that restoration of diphthamide biosynthesis led to increased resistance to ricin (Fig. 3d). The average EC50 for DPH2 transfected cells was 3.3-fold higher than for the control transfected cells (Table 2).
Finally, PR72 and PR201 cells, described above as lacking functional Dph3, were transfected with pIRES-DPH3 or the same vector carrying an unrelated gene, VPS11. Cells stably transfected with DPH3 had the expected sensitivity to PA plus FP59 confirming the expression of Dph3 (data not shown). Cytotoxicity assays with ricin showed that PR72 and PR201 cells transfected with DPH3 had increased resistance as compared to the control transfected cells (Fig. 3e). The average EC50 of DPH3 transfected cells increased 3.2-fold in comparison with control transfected cells (Table 2). These results for Dph3 are consistent with those for Dph2 and Dph5 in showing that the presence of diphthamide in eEF-2 increases the resistance of CHO cells to ricin by about 3-fold.
Discussion
Diphthamide, the post-translational side chain modification to His715 in mammalian eEF-2 (His699 in yeast) is unique in several respects. This modification occurs only on eEF-2, it is conserved in all eukaryotes and apparently in archaea, and it serves as the single target for several ADP-ribosylating toxins that are key virulence factors for the bacteria that secrete them. The retention by eukaryotes of a unique target that makes them susceptible to attack by pathogenic bacteria clearly implies that diphthamide serves some important function in normal cellular physiology. However, the role of diphthamide in cellular physiology is not defined yet. The viability of somatic cell (CHO) mutants like those used here shows that eEF-2 lacking diphthamide can function in protein synthesis at a rate and with a degree of accuracy that are sufficient to maintain life. It follows that any important functions of eEF-2 that depend on its containing diphthamide must be relatively subtle, and are likely to become evident only under special circumstances (e.g., stress) or in the context of a multicellular organism.
Disruption of DPH1 and DPH3 in mice showed that diphthamide or its biosynthetic genes play important roles during early stages of development. Thus, DPH1 knockout mice died at an early age (Chen and Behringer, 2004) while DPH3 knockout mice showed embryonic lethality (Liu et al., 2006). Later work showed that Dph3 is also required for tRNA modification (Huang et al., 2005), which could explain the more severe phenotype of this knockout mouse. However, the early death of the DPH1-deficient mice, where the deficit appears restricted to diphthamide biosynthesis, suggests that even modest changes to the accuracy or efficiency of the protein synthetic process can have profound effects in the whole animal at certain stage. However, the evidence presented here suggests an alternative hypothesis regarding the role of diphthamide. Several CHO cell lines have been isolated which lack one of the functional Dph proteins. We noted that two independent CHO cells mutants, PR72 and PR201, lacking functional Dph3 and therefore diphthamide (Liu and Leppla, 2003), showed increased sensitivity to ricin. Because several previous studies showed that binding of eEF-2 to ribosomes prevents access by ricin to the susceptible loop in the rRNA (Brigotti et al., 1989; Fernandez-Puentes et al., 1976) we formed the simple hypothesis that diphthamide-containing eEF-2 might block ricin access to ribosomes and thereby protect intact cells against ricin. To test the hypothesis, other diphthamide-deficient CHO mutant cells were analyzed. We selected RPE3b and RPE22 cells which have been well characterized for their DT resistance phenotype and lack functional Dph5 and Dph2, respectively (Liu et al., 2004; Moehring et al., 1980; Moehring et al., 1984). These cells were also found to be hypersensitive to ricin and showed 3-fold lower EC50 as compared to their parental cells CHO-K1. To further confirm that observed hypersensitivity is a result from the diphthamide deficiency, not some other unrecognized mutations in diphthamide-deficient cells, these cells were complemented for diphthamide biosynthesis.
Cells were transfected with the plasmid pIRES-hyg2 harboring the specific DPH gene or control vector. RPE3b and RPE22e cells transfected with DPH5 and DPH2, respectively, regained the parental degree of resistance to ricin. In another experiment, expression of DPH3 in PR72 and PR201 cells restored the parental cell resistance towards ricin. In each case, the differences in sensitivity were about 3-fold, regardless of the parental line. All these results clearly indicate that diphthamide-deficient cells show increased sensitivity to ricin as compared to the corresponding diphthamide-containing cells.
Studies cited above showed that ricin and eEF-2 have overlapped binding sites on the ribosome. We speculate that eEF-2 containing the diphthamide modification may have greater affinity for the ribosome, and therefore may more effectively occlude the site targeted by ricin and other RIPs. Given the wide distribution of RIPs in nature, and especially of the single-chain RIPs in plants, it is reasonable to assume that they impose a modest but continuous selective pressure on other organisms, and especially on those species like eukaryotes that eat plants (Barbieri et al., 1993; Perentesis et al., 1992). Although the single-chain RIPs do not enter eukaryotic cells efficiently, their high concentration in some plants suggests that they are toxic under some circumstances (Peumans et al., 2001). The cytotoxicity of the single-chain pokeweed antiviral protein toward virally-infected cells demonstrates the potential of single-chain RIPs to damage cells (Parikh and Tumer, 2004). This ability would seem to be sufficient to select for heritable changes to the protein synthetic machinery that protect it from inactivation. The questions arises whether creation of the diphthamide residue provided such a selective advantage at some stage during evolution, and also whether the 3-fold level of resistance observed here is sufficient to drive and maintain this post-translational change. The organisms competing by modifying these sites may not have resembled those now in existence, so reconstruction of the early evolutionary events must remain speculative. It does appear reasonable to argue that a 3-fold selective advantage is sufficient to assure gene retention when multiplied over many generations in which the selective pressure from RIPs is constant. Some evidence for (or against) the hypothesis that diphthamide evolved in response to pressure from RIPs may eventually be obtained by careful analyses of the extensive genomic DNA sequence information now becoming available, but this is beyond the scope of this study.
Experimental Procedures
CHO cell lines and cell culture
All the cell lines used in this study are derivatives of CHO cells. CHO-K1, RPE3b and RPE22e were derived by Thomas and Joan Moehring and were provided by Gary Ward (University of Vermont), as described previously (Liu et al., 2004). All cell lines were grown in α-minimal essential medium (MEM) supplemented with 8% fetal bovine serum, 2 mM glutamine, 50 μg/ml gentamicin, and 25 mM HEPES.
Transfection
All the DPH constructs used for the transfection were described earlier (Liu et al., 2004; Liu and Leppla, 2003). All DPH genes were from mouse origin. The expression plasmids were transfected into CHO cells using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions and selected by growing them in medium with hygromycin- B (500 μg/ml) for 2 weeks. Individual clones were picked and expanded.
Cytotoxicity assays
PA and FP59 were produced as described previously (Arora et al., 1992; Liu et al., 2007). Ricin was purchased from Boehringer Mannheim. Cytotoxicity assays were done as described previously (Liu and Leppla, 2003). In brief, the cells were sub-cultured in 96-well plate one day prior to experiment. For cytotoxicity assays, cells were incubated with various concentrations of FP59 in combination with a fixed amount (500 ng/ml) of PA for 48 h. At the end of incubation, 0.5 mg/ml MTT (3-[4,5-dimethylthiazol-2,5-diphenyltetrazolium bromide) dissolved in MEM was used to measure cell viability. A540 values obtained for cells which were not treated with toxin were considered as 100 % viability. Ricin toxicity assays were conducted in the same way.
ADP-ribosylation assay
The assay for ADP-ribosylation of eEF-2 in CHO cell extracts was performed as described previously with some modification (Liu et al., 2004). Biotin-NAD was used as source of ADP-ribose for ADP-ribosylation and transfer of biotin-ADP-ribose to eEF-2 in the presence of toxin was detected by Western blotting using streptavidin-conjugates. In brief, cells were lysed in RIPA buffer containing protease inhibitors and cell lysate (5 μl containing 50 μg protein) was mixed with 500 ng of FP59 in ADP-ribosylation buffer (20 mM Tris-HCl, pH 7.4; 1 mM EDTA; 50 mM DTT) with 5 μM 6-Biotin-17-NAD (Trevigen) followed by incubation at 25°C for 30 min. Samples were then mixed with SDS sample buffer, boiled for 5 min and run on 4−25 % SDS-PAGE gels (Invitrogen). The proteins were transferred to nitrocellulose membranes using the iBlot system (Invitrogen) and western blotting was performed using streptavidin-IR conjugate (Rockland Immunochemicals, Gilbertsville, PA) and scanned on an Odyssey Infrared Imager (LICOR Biosciences, Lincoln, NE).
Acknowledgements
We thank Rasem Fattah for assistance with protein purification and Jiamo Lu for technical assistance to maintain cell lines. This research was supported by the Intramural Research Program of the National Institutes of Health (NIH), National Institute of Allergy and Infectious Diseases (NIAID).
References
- Arora N, Klimpel KR, Singh Y, Leppla SH. Fusions of anthrax toxin lethal factor to the ADP-ribosylation domain of Pseudomonas exotoxin A are potent cytotoxins which are translocated to the cytosol of mammalian cells. J Biol Chem. 1992;267:15542–15548. [PubMed] [Google Scholar]
- Barbieri L, Battelli MG, Stirpe F. Ribosome-inactivating proteins from plants. Biochim Biophys Acta. 1993;1154:237–282. doi: 10.1016/0304-4157(93)90002-6. [DOI] [PubMed] [Google Scholar]
- Bodley JW, Dunlop PC, VanNess BG. Diphthamide in elongation factor 2: ADP-ribosylation, purification, and properties. Methods Enzymol. 1984;106:378–387. doi: 10.1016/0076-6879(84)06040-7. [DOI] [PubMed] [Google Scholar]
- Brigotti M, Rambelli F, Zamboni M, Montanaro L, Sperti S. Effect of alpha-sarcin and ribosome-inactivating proteins on the interaction of elongation factors with ribosomes. Biochem J. 1989;257:723–727. doi: 10.1042/bj2570723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CM, Behringer RR. Ovca1 regulates cell proliferation, embryonic development, and tumorigenesis. Genes Dev. 2004;18:320–332. doi: 10.1101/gad.1162204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JY, Bodley JW. Biosynthesis of diphthamide in Saccharomyces cerevisiae. Partial purification and characterization of a specific S-adenosylmethionine:elongation factor 2 methyltransferase. J Biol Chem. 1988;263:11692–11696. [PubMed] [Google Scholar]
- Chen JY, Bodley JW, Livingston DM. Diphtheria toxin-resistant mutants of Saccharomyces cerevisiae. Mol Cell Biol. 1985;5:3357–3360. doi: 10.1128/mcb.5.12.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Puentes C, Benson S, Olsnes S, Pihl A. Protective effect of elongation factor 2 on the inactivation of ribosomes by the toxic lectins abrin and ricin. Eur J Biochem. 1976;64:437–443. doi: 10.1111/j.1432-1033.1976.tb10320.x. [DOI] [PubMed] [Google Scholar]
- FitzGerald DJ, Fryling CM, Zdanovsky A, Saelinger CB, Kounnas M, Winkles JA, et al. Pseudomonas exotoxin-mediated selection yields cells with altered expression of low-density lipoprotein receptor-related protein. J Cell Biol. 1995;129:1533–1541. doi: 10.1083/jcb.129.6.1533. [published erratum appears in J Cell Biol 1995 Aug;130(4):1015]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg L, Nygard O. Interaction sites of ribosome-bound eukaryotic elongation factor 2 in 18S and 28S rRNA. Biochemistry. 1994;33:15159–15167. doi: 10.1021/bi00254a027. [DOI] [PubMed] [Google Scholar]
- Huang B, Johansson MJ, Bystrom AS. An early step in wobble uridine tRNA modification requires the Elongator complex. RNA. 2005;11:424–436. doi: 10.1261/rna.7247705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivankovic M, Rubelj I, Matulic M, Reich E, Brdar B. Site-specific mutagenesis of the histidine precursor of diphthamide in the human elongation factor-2 gene confers resistance to diphtheria toxin. Mutat Res. 2006;609:34–42. doi: 10.1016/j.mrgentox.2006.06.027. [DOI] [PubMed] [Google Scholar]
- Kimata Y, Kohno K. Elongation factor 2 mutants deficient in diphthamide formation show temperature-sensitive cell growth. J Biol Chem. 1994;269:13497–13501. [PubMed] [Google Scholar]
- Leppla SH. Bacillus anthracis toxins. In: Alouf JE, Popoff MR, editors. The Comprehensive Sourcebook of Bacterial Protein Toxins. Academic Press; Burlington, MA: 2006. pp. 323–347. [Google Scholar]
- Liu S, Leppla SH. Retroviral insertional mutagenesis identifies a small protein required for synthesis of diphthamide, the target of bacterial ADP-ribosylating toxins. Mol Cell. 2003;12:603–613. doi: 10.1016/j.molcel.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Liu S, Leung HJ, Leppla SH. Characterization of the interaction between anthrax toxin and its cellular receptors. Cell Microbiol. 2007;9:977–987. doi: 10.1111/j.1462-5822.2006.00845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Milne GT, Kuremsky JG, Fink GR, Leppla SH. Identification of the proteins required for biosynthesis of diphthamide, the target of bacterial ADP-ribosylating toxins on translation elongation factor 2. Mol Cell Biol. 2004;24:9487–9497. doi: 10.1128/MCB.24.21.9487-9497.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Schubert RL, Bugge TH, Leppla SH. Anthrax toxin: structures, functions and tumour targeting. Expert Opin Biol Ther. 2003;3:843–853. doi: 10.1517/14712598.3.5.843. [DOI] [PubMed] [Google Scholar]
- Liu S, Wiggins JF, Sreenath T, Kulkarni AB, Ward JM, Leppla SH. Dph3, a small protein required for diphthamide biosynthesis, is essential in mouse development. Mol Cell Biol. 2006;26:3835–3841. doi: 10.1128/MCB.26.10.3835-3841.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattheakis LC, Shen WH, Collier RJ. DPH5, a methyltransferase gene required for diphthamide biosynthesis in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:4026–4037. doi: 10.1128/mcb.12.9.4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattheakis LC, Sor F, Collier RJ. Diphthamide synthesis in Saccharomyces cerevisiae: structure of the DPH2 gene. Gene. 1993;132:149–154. doi: 10.1016/0378-1119(93)90528-b. [DOI] [PubMed] [Google Scholar]
- Moehring JM, Moehring TJ. Characterization of the diphtheria toxin-resistance system in Chinese hamster ovary cells. Somatic Cell Genet. 1979;5:453–468. doi: 10.1007/BF01538880. [DOI] [PubMed] [Google Scholar]
- Moehring JM, Moehring TJ, Danley DE. Posttranslational modification of elongation factor 2 in diphtheria-toxin-resistant mutants of CHO-K1 cells. Proc Natl Acad Sci U S A. 1980;77:1010–1014. doi: 10.1073/pnas.77.2.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moehring TJ, Danley DE, Moehring JM. In vitro biosynthesis of diphthamide, studied with mutant Chinese hamster ovary cells resistant to diphtheria toxin. Mol Cell Biol. 1984;4:642–650. doi: 10.1128/mcb.4.4.642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naglich JG, Metherall JE, Russell DW, Eidels L. Expression cloning of a diphtheria toxin receptor: identity with a heparin-binding EGF-like growth factor precursor. Cell. 1992;69:1051–1061. doi: 10.1016/0092-8674(92)90623-k. [DOI] [PubMed] [Google Scholar]
- Olsnes S, Kozlov JV. Ricin. Toxicon. 2001;39:1723–1728. doi: 10.1016/s0041-0101(01)00158-1. [DOI] [PubMed] [Google Scholar]
- Oppenheimer NJ, Bodley JW. Diphtheria toxin. Site and configuration of ADP-ribosylation of diphthamide in elongation factor 2. J Biol Chem. 1981;256:8579–8581. [PubMed] [Google Scholar]
- Ortiz PA, Ulloque R, Kihara GK, Zheng H, Kinzy TG. Translation elongation factor 2 anticodon mimicry domain mutants affect fidelity and diphtheria toxin resistance. J Biol Chem. 2006;281:32639–32648. doi: 10.1074/jbc.M607076200. [DOI] [PubMed] [Google Scholar]
- Pappenheimer AM, Jr., Dunlop PC, Adolph KW, Bodley JW. Occurrence of diphthamide in archaebacteria. J Bacteriol. 1983;153:1342–1347. doi: 10.1128/jb.153.3.1342-1347.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh BA, Tumer NE. Antiviral activity of ribosome inactivating proteins in medicine. Mini Rev Med Chem. 2004;4:523–543. doi: 10.2174/1389557043403800. [DOI] [PubMed] [Google Scholar]
- Pastan I, Hassan R, FitzGerald DJ, Kreitman RJ. Immunotoxin treatment of cancer. Annu Rev Med. 2007;58:221–237. doi: 10.1146/annurev.med.58.070605.115320. [DOI] [PubMed] [Google Scholar]
- Perentesis JP, Miller SP, Bodley JW. Protein toxin inhibitors of protein synthesis. Biofactors. 1992;3:173–184. [PubMed] [Google Scholar]
- Peumans WJ, Hao Q, Van Damme EJ. Ribosome-inactivating proteins from plants: more than RNA N-glycosidases? FASEB J. 2001;15:1493–1506. doi: 10.1096/fj.00-0751rev. [DOI] [PubMed] [Google Scholar]
- Phan LD, Perentesis JP, Bodley JW. Saccharomyces cerevisiae elongation factor 2. Mutagenesis of the histidine precursor of diphthamide yields a functional protein that is resistant to diphtheria toxin. J Biol Chem. 1993;268:8665–8668. [PubMed] [Google Scholar]
- Van Ness BG, Howard JB, Bodley JW. ADP-ribosylation of elongation factor 2 by diphtheria toxin. Isolation and properties of the novel ribosyl-amino acid and its hydrolysis products. J Biol Chem. 1980;255:10717–10720. [PubMed] [Google Scholar]