Abstract
Background
The rodent malaria parasite Plasmodium chabaudi has proven of great value in the analysis of fundamental aspects of host-parasite-vector interactions implicated in disease pathology and parasite evolutionary ecology. However, the lack of gene modification technologies for this model has precluded more direct functional studies.
Methods
The development of in vitro culture methods to yield P. chabaudi schizonts for transfection and conditions for genetic modification of this rodent malaria model are reported.
Results
Independent P. chabaudi gene-integrant lines that constitutively express high levels of green fluorescent protein throughout their life cycle have been generated.
Conclusion
Genetic modification of P. chabaudi is now possible. The production of genetically distinct reference lines offers substantial advances to our understanding of malaria parasite biology, especially interactions with the immune system during chronic infection.
Background
The development of transfection technologies in Plasmodium have led to substantial advances in malaria research [1-3]. The first successful transient transfection to be reported was performed in Plasmodium gallinaceum, followed by Plasmodium falciparum [4,5]. However, stable gene disruption and replacement technologies were developed first for the rodent malaria parasite, Plasmodium berghei [3,6-9], and have subsequently proved particularly useful in analysis of proteins mediating sexual development and transmission of the parasite through the mosquito vector [10-13]. Stable transformation has also been achieved for Plasmodium yoelii [14] but P. berghei has been the focus for the development of technical advances in transfection [2].
However, P. berghei causes a rapid and virulent infection leading to widespread tissue pathology and early death without effective host immune control making the investigation of host adaptive immune responses and chronic malaria infections challenging [15,16]. The lack of genetically distinct P. berghei clones limits its use as a model for investigations of the evolution and ecology of host-parasite interactions [17,18]. Furthermore, transmission of P. berghei through the mosquito vector occurs at lower temperatures (18–21°C) and is longer (21 days) than transmission of human or other rodent malaria parasites (> 24°C for 10–14 days) and P. falciparum. This may influence studies of parasite biology and development in the vector as parasite-vector interactions are sensitive to temperature (e.g. [19]).
In contrast, the rodent malaria parasite P. chabaudi could provide a more relevant model for investigating anti-malarial host immune responses because infections are usually controlled by host immunity (reviewed in [16,20,21]). Plasmodium chabaudi also shares a number of life history features with the most severe human malaria parasite, P. falciparum, including mature erythrocyte preference, synchronous schizogony, sequestration, rosetting, antigenic variation, and acute parasitaemia which becomes chronic [22-26]. Also, as in P. falciparum infections, the inability to control the consequences of the first peak of parasitaemia is a major determinant of severe disease and death. In P. chabaudi and P. falciparum most investment into sexual stages occurs after the initial peaks of parasitaemia and commitment to gametocytes appears to follow changes in anaemia and red cell dynamics [27-30]. In addition, the bank of genetically distinct and phenotypically well-characterized clones available for P. chabaudi is substantially larger than for all other rodent malaria models [17].
The development of gene transformation technologies for P. chabaudi will, therefore, provide opportunities for experimental and analytical advances in fields as diverse as immunology and evolutionary ecology. This paper presents the first report of the generation of fluorescent P. chabaudi lines that constitutively express high levels of Green Fluorescent Protein (GFP) throughout their life-cycle.
Methods
Preparation of parasites for transfection
To obtain parasites for transfection, male MF1 mice (10 weeks) were infected i.p. with 1 × 107 P. chabaudi parasites from clone AJ4916. 500 μl of blood containing ring and early trophozoite-stage parasites were collected by cardiac puncture at 3 days post infection (5–10% parasitaemia). Parasites were cultured for 17–18 hours at a 1.5% dilution in complete culture medium (RPMI medium (with NaHCO3, Hepes and L-glutamine, Invitrogen) containing 25% heat inactivated foetal calf serum (Gibco), at pH 7.25) in the presence of 10% O2, 5% CO2, 85% N2, at 32°C in upright 200 ml flasks (Ikawa) in a horizontal shaking incubator at 30 rpm. Parasitized blood forming a layer at the bottom of the flask was gently removed and centrifuged at 1500 rpm for 30 sec. 5 μl of pelleted cells (1 × 106 schizonts) were used per transfection.
Transfection and selection of transformed parasites
Gfp was introduced into the genome of P. chabaudi parasites, using the PbGFPCON plasmid previously described [7]. 5 μg of plasmid DNA, linearized at the Apa1 site, in 5 μl of dH2O was added to 100 μl of Amaxa nucleofectortm test solution 88A6 (Basic Parasite Nucleofectortm solution 2) in the manufacturers' cuvette (Amaxa Biosystems). 5 μl of schizont mix was added and electroporation was carried out in an Amaxa nucleofectortm using program U33. After electroporation, 50 μl of pre-warmed complete culture media was added and the transfection mix was immediately injected i.v. into an MF1 mouse. Recipient mice received 35 μg/ml pyrimethamine in their drinking water (pH 3.5–5) for seven days post infection; the minimum pyrimethamine dose required to clear P. chabaudi infections of clone AJ in 24 hours (J. Thompson, personal observation). Giemsa-stained smears were scanned every one to two days and parasites that produced patent infections (days 14–20) were immediately passaged to further mice for the production of stabilate stocks.
Analysis of transformed parasites
For genetic analysis of the integration locus, DNA was isolated as previously described [2] and Southern blot and PCR analyses were performed. PCR amplifications were carried out using Qiagen Taq DNA polymerase in the presence of 'Q solution' under the following conditions; 35 cycles; anneal, 54°C, 1 min; extend, 68°C, 3 min, denature, 94°C, 10 sec. To demonstrate integration into a ssu-rrna locus at the 5' region, amplification was carried out with primer Pc5'F (TTGTAAGAACGTGCTTGGTG) that is specific for P. chabaudi ssu-rrna sequence on P. chabaudi genome contig827, in the target region, and primer Pl5'R (TTCCCAGTCACGACGTTG) that anneals to P. berghei d-ssu rrna sequence in the PbGFPCON plasmid. To demonstrate integration into the contig827 ssu-rrna locus at the 3' region, amplification was carried out with primer Pc3'R (AGAGCCCAGCGATGAC) that is specific for P. chabaudi contig827 ssu-rrna sequence in the integration site, and primer Pl3'F (CAATGATTCATAAATAGTTGGAC) that anneals to P. berghei d-ssu rrna sequence in the PbGFPCON plasmid. To demonstrate the presence of tg-dhfr sequence, amplification was carried out with primers L190 (CGGGATCCATGCATAAACCGGTGTGTC) + L191; CGGGATCCAAGCTTCTGTATTTCCG. To amplify circular PbGFPCON plasmid, amplification was carried out with primers PlF2 (AATCATGACTTCTGTCACTGC) and Pl5'R. Primers Pc5'R and Pc3'R anneal specifically to sequences within P. chabaudi contig827 ssu-rrna and not to sequences within the PbCON vector. Probe template for the detection of tgdhfr by Southern blots was amplified using primers L190/L191. DNA was digested with HindIII and NheI, transferred to Hybond N+ membrane (Amersham) and hybridized according to the manufacturers methods. Wet preparations of live parasites expressing GFP were visualized using Openlab digital imaging (Improvision). The development and progression of parasitaemia in transformed parasites was compared to the wild type ancestor by following six mice infected with 106 parasitized red blood cells for each line. Infections were monitored daily to collect red blood cell density and parasitaemia data until day 14 post infection, when the acute phase parasites had been cleared. The infection dynamics of the two lines were analysed using linear mixed-effects models, which account for repeated measures across infections. One mouse from each line was euthanized (day 10 and 11) so these infections did not contribute data for the whole time course.
Results and discussion
The optimal stage for DNA uptake by Plasmodium is thought to be the free merozoites, released at schizogony, that are not surrounded by red blood cell cytoplasm and membranes.P. berghei may, therefore, be relatively amenable to genetic transformation because schizonts developing in reticulocytes do not rupture in in vitro culture conditions, so high numbers can be purified. These rupture, releasing merozoites, during electroporation. In contrast, P. chabaudi schizonts, developing in mature red blood cells, do not arrest in culture and cannot be purified in such large numbers. For this reason, the transformation efficiency of P. chabaudi is likely to be considerably lower that that of P. berghei. The major improvements in rodent malaria parasite transformation efficiency obtained with the Amaxa Nucleofectortm technology [9], however, together with the development of culture conditions that generate mature P. chabaudi schizonts suggested to us that transformation of P. chabaudi may now be possible.
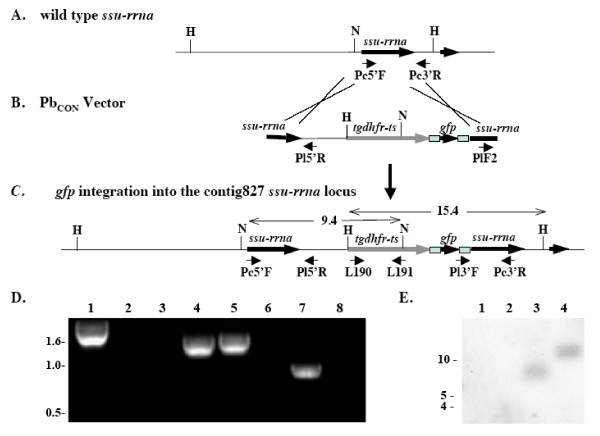
To obtain appropriate numbers of P. chabaudi schizonts, the in vitro culture protocol described by Mackinnon et al [24] was adapted and scaled-up. Under these conditions, synchronous P. chabaudi ring-stage parasites/young trophozoites develop over 17–18 hours into a population of parasites that contain > 10% mature schizonts (Figure 1A). Schizonts produced by in vitro culture of P. chabaudi AJ blood stage parasites were transfected with the PbGFPCON plasmid [7] that contains an incomplete copy of the P. berghei d-ssu-rrna as a target region for integration, and the pyrimethamine-resistant tgdhfr-ts selectable cassette (pyrR2) for selection of transgenic parasites (Figure 2). Blast analysis of the P. chabaudi genome http://www.sanger.ac.uk/cgi-bin/blast/submitblast/p_chabaudi showed that there is highest identity (94%) between the P. berghei d-ssu-rrna target region and P. chabaudi ssu-rrna sequences on genome contig827, indicating that these are orthologous loci. Linearized PbGFPCON DNA was introduced into the parasite genome by electroporation and recombinant parasites that express tghfr were selected by treatment with pyrimethamine.
Figure 1.
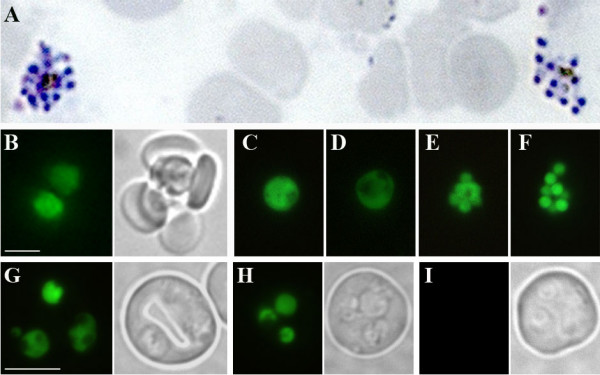
(A) Cultured P. chabaudi schizonts used for transfection (giemsa stain) (B-F) Fluorescent and bright field images of P. chabaudi PcGFPcon blood-stage parasites. (B) two trophozoites in a rosetting red blood cell; (C) young trophozoite; (D) gametocyte; (E) schizont; (F) rupturing schizont; multiple infection of young and mature trophozoites (G); multiple infection of rings and young trophozoite (H); wild-type AJ P. chabaudi (I). Scale bar = 5 μm.
Figure 2.
Integration of PbCON into P. chabaudi ssu-rrna. Schematic representation of P. chabaudi ssu-rrna locus on contig827 (A). Schematic representation of linearized plasmid PbCON containing the Tgdhfr-ts cassette, conferring pyrimethamine resistance and gfp, flanked by P. berghei d-ssu-rrna target sequences for integration (B). Schematic representation of the contig827 ssu-rrna locus in PcGFPCON clone 2.3, following integration of the PbCON plasmid (C). D: tgdhfr is present in PcGFPCON (lane 1) but not wt (lane 2) DNA; amplification with L190/L191. Circular PbGFPCON plasmid is absent in PcGFPCON (lane 3); circular PbGFPCON (lane 4); amplification with PlF2/Pl5'R. Verification of the 5' integration site; Pc5'F/Pl5'R amplify a product in PcGFPCON (lane 5) but not in wt (lane 6) DNA. Verification of the 3' integration site; Pl3'F/Pc3'R amplify a product in PcGFPCON (lane 7) but not wt (lane 8) DNA. E: Tgdhfr is present in the genome of PcGFPCON but not wt parasites. wt (lanes 1 and 2) and PcGFPCON (lanes 3 and 4) genomic DNA digested with NheI (lanes 1 and 3) or HindIII (lanes 2 and 4). Integration of PcGFP into the P. chabaudi ssu-rrna locus on contig827 results in restriction enzyme digestion products of 9.4 and 15.4 kb following NheI and HindIII digestion respectively. N, NheI; H, HindIII.
Integration of the PbGFPCON cassette into the P. chabaudi genome was confirmed by PCR analysis in four independent lines and line '2.3' was selected for further genetic and phenotypic analysis. In P. chabaudi line 2.3, PbGFPCON integrated into the genome of contig 827 rRNA subunit, orthologous to the P. berghei c or d-ssu-rrna that have previously been shown to be non-essential genes in rodent malaria parasites [7]. Recombinant parasites develop to blood-stages with no apparent defects (Figure 1B–E) and form gametocytes that exflagellate and can infect mosquitoes.
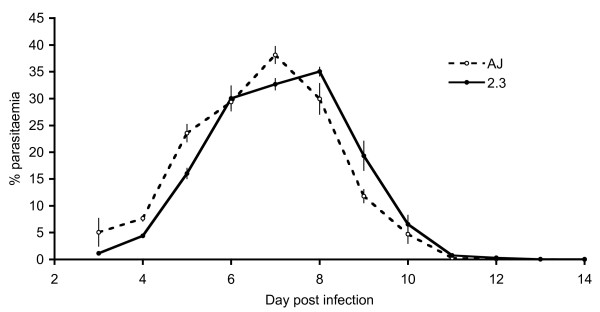
The in vivo asexual dynamics of line 2.3 was not significantly different to wild type P. chabaudi AJ4916 ancestral parasites that had undergone comparable numbers of passages (Figure 3). Infections initiated with line 2.3 and AJ4916 parasites both reached patency by microscopy on day 3 post infection, peaked on day 7–8 and the acute phase lasted for 14 days. More detailed analysis of parasiteamia revealed that the lines did not differ significantly in their average parasitaemia (F(1,10)= 0.02; P = 0.901). The patterns observed throughout infections were similar, though parasitaemia of line AJ4916 was significantly lower on days 8 and 9 post infection (F(10,91) = 4.96; P < 0.0001). The lines did not differ in the patterns or the average levels of anaemia they caused (F(1,10) = 0.001; P = 0.975). Although reversion to the wild-type genotype was observed at a low rate after multiple blood-stage passages, all Pc-GFPCON (line 2.3) parasites observed at day 8 of infection were GFP-positive.
Figure 3.
Blood stage dynamics of wild type and PcGFPCON parasites. Average (± standard error) parasitaemia of six infections with PcGFPCON (2.3) and wild type AJ (AJ4916) parasites. Infections were initiated with 106 parasitized red blood cells and followed by microscopy throughout the acute phase.
Conclusion
Plasmodium chabaudi is reproducibly accessible for genetic transformation at an efficiency that is sufficient for genomic integration of introduced genes. The development of technologies that allow disruption or modification of gene expression in P. chabaudi, thus, opens the way for direct functional analysis of parasite proteins throughout both acute and chronic stages of an in vivo malaria infection, including those that have been implicated in modulation of the host immune response [31]. The generation of stable fluorescent P. chabaudi parasite lines also offers the opportunity for imaging of direct interactions between the parasite and host cells within a variety of host tissues.
Abbreviations
GFP: green fluorescent protein; Tgdhfr: Toxoplasma gondii Dihydrofolate reductase.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
Both authors conceived and designed the project and prepared the manuscript. SR prepared and characterized parasites and JT undertook the transfection and molecular analyses. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
We thank Blandine Franke-Fayard, Chris Janse, Margaret Mackinnon and Andy Waters for constructs and helpful discussion, and Sittiporn Pattaradilokrat, Ronnie Mooney, Sally Moore and Will Chadwick for assistance. Sequence data were produced by the P. chabaudi Sequencing Group at the Sanger Institute and can be obtained from http://www.sanger.ac.uk/cgi-bin/blast/submitblast/p_chabaudi. This work was funded by the MRC, BBSRC and Wellcome Trust.
Contributor Information
Sarah E Reece, Email: sarah.reece@ed.ac.uk.
Joanne Thompson, Email: Joanne.Thompson@ed.ac.uk.
References
- Carvalho TG, Menard R. Manipulating the Plasmodium genome. Curr Issues Mol Biol. 2005;7:39–55. [PubMed] [Google Scholar]
- de Koning-Ward TF, Janse CJ, Waters AP. The development of genetic tools for dissecting the biology of malaria parasites. Annu Rev Microbiol. 2000;54:157–185. doi: 10.1146/annurev.micro.54.1.157. [DOI] [PubMed] [Google Scholar]
- Vandijk MR, Waters AP, Janse CJ. Stable transfection of malaria parasite blood stages. Science. 1995;268:1358–1362. doi: 10.1126/science.7761856. [DOI] [PubMed] [Google Scholar]
- Goonewardene R, Daily J, Kaslow D, Sullivan TJ, Duffy P, Carter R, Mendis K, Wirth D. Transfection of the malaria parasite and expression of firefly luciferase. Proc Natl Acad Sci USA. 1993;90:5234–5236. doi: 10.1073/pnas.90.11.5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YM, Sifri CD, Lei HH, Su XZ, Wellems TE. Transfection of Plasmodium falciparum within human red blood cells. Proc Natl Acad Sci USA. 1995;92:973–977. doi: 10.1073/pnas.92.4.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning-Ward TF, Thomas AW, Waters AP, Janse CJ. Stable expression of green fluorescent protein in blood and mosquito stages of Plasmodium berghei. Mol Biochem Parasitol. 1998;97:247–252. doi: 10.1016/s0166-6851(98)00134-0. [DOI] [PubMed] [Google Scholar]
- Franke-Fayard B, Trueman H, Ramesar J, Mendoza J, Keur M van der, Linden R van der, Sinden RE, Waters AP, Janse CJ. A Plasmodium berghei reference line that constitutively expresses GFP at a high level throughout the complete life cycle. Mol Biochem Parasitol. 2004;137:23–33. doi: 10.1016/j.molbiopara.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Janse CJ, Franke-Fayard B, Mair GR, Ramesar J, Thiel C, Engelmann S, Matuschewski K, van Gemert GJ, Sauerwein RW, Waters AP. High efficiency transfection of Plasmodium berghei facilitates novel selection procedures. Mol Biochem Parasitol. 2006;145:60–70. doi: 10.1016/j.molbiopara.2005.09.007. [DOI] [PubMed] [Google Scholar]
- Janse CJ, Franke-Fayard B, Waters AP. Selection by flow-sorting of genetically transformed, GFP-expressing blood stages of the rodent malaria parasite, Plasmodium berghei. Nat Protoc. 2006;1:614–623. doi: 10.1038/nprot.2006.88. [DOI] [PubMed] [Google Scholar]
- Menard R, Sultan AA, Cortes C, Altszuler R, vanDijk MR, Janse CJ, Waters AP, Nussenzweig RS, Nussenzweig V. Circumsporozoite protein is required for development of malaria sporozoites in mosquitoes. Nature. 1997;385:336–340. doi: 10.1038/385336a0. [DOI] [PubMed] [Google Scholar]
- Sultan AA, Thathy V, de Koning-Ward TF, Nussenzweig V. Complementation of Plasmodium berghei TRAP knockout parasites using human dihydrofolate reductase gene as a selectable marker. Mol Biochem Parasitol. 2001;113:151–156. doi: 10.1016/s0166-6851(01)00209-2. [DOI] [PubMed] [Google Scholar]
- van Dijk MR, Douradinha B, Franke-Fayard B, Heussler V, van Dooren MW, van Schaijk B, van Gemert GJ, Sauerwein RW, Mota MM, Waters AP, Janse CJ. Genetically attenuated, P36p-deficient malarial sporozoites induce protective immunity and apoptosis of infected liver cells. Proc Natl Acad Sci USA. 2005;102:12194–12199. doi: 10.1073/pnas.0500925102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk MR, Janse CJ, Thompson J, Waters AP, Braks JAM, Dodemont HJ, Stunnenberg HG, van Gemert GJ, Sauerwein RW, Eling W. A central role for P48/45 in malaria parasite male gamete fertility. Cell. 2001;104:153–164. doi: 10.1016/s0092-8674(01)00199-4. [DOI] [PubMed] [Google Scholar]
- Mota MM, Thathy V, Nussenzweig RS, Nussenzweig V. Gene targeting in the rodent malaria parasite Plasmodium yoelii. Mol Biochem Parasitol. 2001;113:271–278. doi: 10.1016/s0166-6851(01)00228-6. [DOI] [PubMed] [Google Scholar]
- Augustijn KD, Kleemann R, Thompson J, Kooistra T, Crawford CE, Reece SE, Pain A, Siebum AHG, Janse CJ, Waters AP. Functional characterization of the Plasmodium falciparum and P. berghei homologues of macrophage migration inhibitory factor. Infect Immun. 2007;75:1116–1128. doi: 10.1128/IAI.00902-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langhorne J, Quin SJ, Sanni LA. Chem Immunol. 2. Vol. 80. 2002. Mouse models of blood-stage malaria infections: Immune responses and cytokines involved in protection and pathology; pp. 204–228. [DOI] [PubMed] [Google Scholar]
- Mackinnon MJ, Read AF. Genetic relationships between parasite virulence and transmission in the rodent malaria Plasmodium chabaudi. Evolution. 1999;53:689–703. doi: 10.1111/j.1558-5646.1999.tb05364.x. http://www.jstor.org/stable/2640710 [DOI] [PubMed] [Google Scholar]
- Reece SE, Drew DR, Gardner A. Sex ratio adjustment and kin discrimination in malaria parasites. Nature. 2008;453:609–614. doi: 10.1038/nature06954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noden B, Kent M, Beier J. The impact of variations in temperature on early Plasmodium falciparum development in Anopheles stephensi. Parasitology. 1995;111:539–545. doi: 10.1017/s0031182000077003. [DOI] [PubMed] [Google Scholar]
- Langhorne J, Albano FR, Hensmann M, Sanni L, Cadman E, Voisine C, Sponaas AM. Dendritic cells, pro-inflammatory responses, and antigen presentation in a rodent malaria infection. Immunol Rev. 2004;201:35–47. doi: 10.1111/j.0105-2896.2004.00182.x. [DOI] [PubMed] [Google Scholar]
- Stevenson MM, Urban BC. Antigen presentation and dendritic cell biology in malaria. Parasite Immunol. 2006;28:5–14. doi: 10.1111/j.1365-3024.2006.00772.x. [DOI] [PubMed] [Google Scholar]
- Fischer K, Chavchich M, Huestis R, Wilson DW, Kemp DJ, Saul A. Ten families of variant genes encoded in subtelomeric regions of multiple chromosomes of Plasmodium chabaudi, a malaria species that undergoes antigenic variation in the laboratory mouse. Mol Microbiol. 2003;48:1209–1223. doi: 10.1046/j.1365-2958.2003.03491.x. [DOI] [PubMed] [Google Scholar]
- Gilks CF, Walliker D, Newbold CI. Relationships between sequestration, antigenic variation and chronic parasitism in Plasmodium chabaudi chabaudi – a rodent malaria model. Parasite Immunol. 1990;12:45–64. doi: 10.1111/j.1365-3024.1990.tb00935.x. [DOI] [PubMed] [Google Scholar]
- Mackinnon MJ, Walker PR, Rowe JA. Plasmodium chabaudi: rosetting in a rodent malaria model. Exp Parasitol. 2002;101:121–128. doi: 10.1016/s0014-4894(02)00103-0. [DOI] [PubMed] [Google Scholar]
- McLean SA, Pearson CD, Phillips RS. Plasmodium chabaudi – antigenic variation during recrudescent parasitemias in mice. Exp Parasitol. 1982;54:296–302. doi: 10.1016/0014-4894(82)90038-8. [DOI] [PubMed] [Google Scholar]
- Mota MM, Jarra W, Hirst E, Patnaik PK, Holder AA. Plasmodium chabaudi-infected erythrocytes adhere to CD36 and bind to microvascular endothelial cells in an organ-specific way. Infect Immun. 2000;68:4135–4144. doi: 10.1128/iai.68.7.4135-4144.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece SE, Duncan AB, West SA, Read AF. Host cell preference and variable transmission strategies in malaria parasites. Proc Biol Sci. 2005;272:511–517. doi: 10.1098/rspb.2004.2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautret P, Miltgen F, Chabaud AG, Landau I. The production of gametocytes by rodent Plasmodium species in mice during phenylhydrazine induced reticulocytosis. Acta Parasitologica. 1997;42:65–7. http://psjc.icm.edu.pl/psjc/cgi-bin/getdoc.cgi?B97-6-92 [Google Scholar]
- Gautret P, Miltgen F, Gantier JC, Chabaud AG, Landau I. Enhanced gametocyte formation by Plasmodium chabaudi in immature erythrocytes: Pattern of production, sequestration, and infectivity to mosquitoes. J Parasitol. 1996;82:900–906. [PubMed] [Google Scholar]
- Paul REL, Ariey F, Robert V. The evolutionary ecology of Plasmodium. Ecology Letters. 2003;6:866–880. doi: 10.1046/j.1461-0248.2003.00509.x. [DOI] [Google Scholar]
- Thompson J, Millington OR, Garside P, Brewer JM. What can transgenic parasites tell us about the development of Plasmodium-specific immune responses? Parasite Immunol. 2008;30:223–233. doi: 10.1111/j.1365-3024.2007.01011.x. [DOI] [PubMed] [Google Scholar]