Abstract
Background
This study was designed to investigate, for the first time, the short-term molecular evolution of the HIV-2 C2, V3 and C3 envelope regions and its association with the immune response. Clonal sequences of the env C2V3C3 region were obtained from a cohort of eighteen HIV-2 chronically infected patients followed prospectively during 2–4 years. Genetic diversity, divergence, positive selection and glycosylation in the C2V3C3 region were analysed as a function of the number of CD4+ T cells and the anti-C2V3C3 IgG and IgA antibody reactivity
Results
The mean intra-host nucleotide diversity was 2.1% (SD, 1.1%), increasing along the course of infection in most patients. Diversity at the amino acid level was significantly lower for the V3 region and higher for the C2 region. The average divergence rate was 0.014 substitutions/site/year, which is similar to that reported in chronic HIV-1 infection. The number and position of positively selected sites was highly variable, except for codons 267 and 270 in C2 that were under strong and persistent positive selection in most patients. N-glycosylation sites located in C2 and V3 were conserved in all patients along the course of infection. Intra-host variation of C2V3C3-specific IgG response over time was inversely associated with the variation in nucleotide and amino acid diversity of the C2V3C3 region. Variation of the C2V3C3-specific IgA response was inversely associated with variation in the number of N-glycosylation sites.
Conclusion
The evolutionary dynamics of HIV-2 envelope during chronic aviremic infection is similar to HIV-1 implying that the virus should be actively replicating in cellular compartments. Convergent evolution of N-glycosylation in C2 and V3, and the limited diversification of V3, indicates that there are important functional constraints to the potential diversity of the HIV-2 envelope. C2V3C3-specific IgG antibodies are effective at reducing viral population size limiting the number of virus escape mutants. The C3 region seems to be a target for IgA antibodies and increasing N-linked glycosylation may prevent HIV-2 envelope recognition by these antibodies. Our results provide new insights into the biology of HIV-2 and its relation with the human host and may have important implications for vaccine design.
Background
The etiologic agents of AIDS, HIV-1 and HIV-2, are two distinct human lentiviruses with similar structural and genomic organization but sharing only 50% of genetic similarity [1]. Compared to HIV-1, the infection by HIV-2 is associated with better prognosis, slower disease progression and transmission, longer latency period and reduced mortality rate [2-6]. Moreover, most HIV-2 patients have normal CD4+ T cell counts and low or undetectable plasmatic viral levels [7,8]. Two possible explanations for these differences may be the slower replication capacity of HIV-2 and a more efficient immune control of HIV-2 [9-13].
The env gene codes for the viral envelope glycoproteins, which are responsible for HIV entry into cells [14]. Rapid evolutionary changes and high genetic variability are two major characteristics of the HIV env gene [15]. In HIV-1 infection, conflicting associations have been reported between disease status and within-patient env gene evolution. Hence, some studies have shown that genetic diversity and divergence from the infecting strain increase during HIV-1 infection but become stable or even decrease in the advanced stage of disease, with the lower CD4+ T cell counts and progression to AIDS [16-18]. Other authors have shown that higher genetic diversity and divergence are found in patients with rapid progression to disease than in slow- or non-progressors [19,20]. There is also a positive correlation between viral replication and intrahost HIV-1 evolution in elite controllers and long-term nonprogressors [21].
The number of studies investigating within-patient HIV-2 molecular evolution and their association with clinical and immunological evolution is limited. In one transversal study, we have shown that the genetic diversity of the HIV-2 env may be directly related to the period of infection [22]. Longitudinal studies performed in Senegal have shown that higher variability in the env V3 region is generally found in patients with faster disease progression to AIDS [23] and that in elite controllers (patients infected for ≈ 10 years with normal CD4+ T cell counts without antiretroviral therapy and with low or undetectable viral load) the rate of env gene diversification may be positively associated with the rate of CD4+ T cell number decrease [24].
Higher rate of molecular evolution, with predominance of nonsynonymous amino acid substitutions, tends to occur in regions of the HIV-1 env gene submitted to strong selective pressure from the immune system [15,25-28]. A structure of particular importance in this process is the V3 loop of the surface glycoprotein which is essential for HIV coreceptor usage [29-32] and for inducing the production of neutralizing and nonneutralizing antibodies in HIV infected individuals [33]. Neutralizing antibody responses, both autologous [34-36] and heterologous [36,37] may be more common in HIV-2 than in HIV-1 infection. Still, little is known about the role of humoral immunity in the evolution of the HIV-2 env gene. In the present study we analyze, for the first time, the molecular evolution of the env C2V3C3 regions in chronically HIV-2 infected patients over a two to four year period in the context of their antibody response (IgG and IgA) against the same envelope region.
Methods
Patients
Eighteen HIV-2 patients attending different hospitals in Lisbon, Portugal, were followed prospectively during 2–4 years (Table 1). Fourteen patients were taking reverse transcriptase and/or protease inhibitors. During the follow-up period three patients (PTHCC20, PTHSM9 and PTHSM10) had detectable plasma viral load. Eight patients had < 200 CD4+ T cells/μl (AIDS defining condition).
Table 1.
Virological and immunological characterization of the patients
| Patient | Year of diagnosis | Sample | CD4+ T cells/μl | RNA copies/ml | Antiretroviral therapy | Antibody reactivity against C2V3C3 (OD/cut-off) | |
| IgG | IgA | ||||||
| PTHCC1 | 2001 | 2003 | 308 | <200 | + | 14.4 | 1.41 |
| 2005 | 319 | na | 13.3 | 1.49 | |||
| PTHCC2 | 2003 | 2003 | 358 | <200 | + | 24.3 | 1.69 |
| PTHCC4 | 2000 | 2003 | 240 | <200 | + | 9.6 | 3.26 |
| PTHCC5 | 1993 | 2003 | 480 | <200 | + | 20.1 | 2.40 |
| 2004 | na | <200 | 22.4 | 2.14 | |||
| PTHCC7 | 2002 | 2003 | 144 | <200 | + | 22.9 | 1.93 |
| 2005 | 43 | <200 | 22.1 | 2.42 | |||
| PTHCC8 | 2000 | 2003 | 141 | <200 | + | 19.4 | 3.98 |
| 2005 | 350 | na | 17.1 | 3.69 | |||
| PTHCC12 | 1995 | 2003 | 66 | <200 | - | 28.0 | 5.79 |
| 2004 | 84 | na | 25.5 | 2.67 | |||
| PTHCC13 | 2004 | 2005 | 954 | <200 | - | na | na |
| PTHCC14 | 1998 | 2003 | 184 | <200 | + | 23.6 | 3.82 |
| PTHCC17 | 1998 | 2003 | 367 | <200 | + | 22.5 | 2.74 |
| 2004 | 270 | <200 | 18.5 | 2.66 | |||
| PTHCC19 | 2003 | 2003 | 175 | na | + | 26.5 | 1.82 |
| 2004 | 400 | <200 | 22.3 | 2.22 | |||
| 2005 | 60 | na | 18.7 | 2.01 | |||
| PTHCC20 | 1998 | 2003 | 78 | na | + | 24.5 | 1.57 |
| 2004 | 73 | 5246 | 20.0 | 1.66 | |||
| 2005 | 85 | <200 | 19.9 | 1.37 | |||
| PTHSM2 | 2002 | 2003 | 275 | <200 | + | 5.9 | 1.55 |
| 2004 | 65 | <200 | 6.2 | 1.57 | |||
| 2005 | 122 | <200 | 10.9 | 1.93 | |||
| 2006 | 172 | <200 | 4.7 | 2.09 | |||
| PTHSM3 | 1993 | 2005 | 1452 | <200 | - | 7.7 | 3.47 |
| PTHSM6 | 2001 | 2005 | 471 | <200 | + | 13.9 | 5.24 |
| PTHSM7 | 1996 | 2003 | 587 | na | - | 11.4 | 2.39 |
| PTHSM9 | 1996 | 2003 | 15 | <200 | + | neg | 0.82 |
| 2004 | na | 484 | neg | 0.79 | |||
| PTHSM10 | 2001 | 2003 | 342 | 5804 | + | neg | 3.56 |
| 2004 | 265 | 4792 | neg | 3.54 | |||
| 2005 | 212 | na | neg | 3.79 | |||
na, not available; neg, no reactivity; +, yes; -, no.
Quantification of HIV-2 plasma viremia
HIV-2 viremia in the plasma was quantified with a quantitative-competitive RT-PCR assay as described elsewhere [38].
DNA extraction, PCR amplification, cloning and sequencing
PBMCs from all patients were co-cultivated with normal PBMCs to try to isolate virus [39]. At the end of the culture period, which is when the culture was positive (mean, 15 days), cells were harvested and DNA was extracted with the Wizard® Genomic DNA Purification kit (Promega) for subsequent analysis. A fragment of the C2V3C3 region (378 bp) of the HIV-2 env gene was amplified in a nested Polymerase Chain Reaction (PCR) as described previously [22]. PCR fragments were cloned into pCR®4-TOPO® vector (Invitrogen) and transformed into One Shot® Match1™-T1R competent cells (Invitrogen). Cloned plasmids were extracted [40], purified and sequenced using BigDye Terminator Cycle sequencing kit (Applied Biosystems), with M13 Forward and Reverse primers, and an automated sequencer (ABI Prism 3100, Applied Biosystems). For each patient an average of 13 clones (range 7–21) was sequenced per sampling year.
Sequence analysis and phylogenetic studies
The nucleotide sequences were aligned using Clustal X [41] and manual adjustments were made using Genedoc [42]. Genetic distances between sequences were calculated using the maximum composite likelihood method implemented in the MEGA version 4 [43]. Inter- and intra-sample synonymous (dS) and nonsynonymous (dN) distances were estimated using the modified Nei-Gojobory method with the Jukes-Cantor correction, also implemented in the MEGA software package.
Maximum likelihood analyses [44] were performed using the best-fit model of molecular evolution estimated by Modeltest under the Akaike information criterion [45]. The chosen model was TVM+G+I. Tree searches were conducted in PAUP version 4.0 using the nearest-neighbor interchange (NNI) and tree bisection and reconnection (TBR) heuristic search strategies [46], and bootstrap resampling [47]. The nucleotide divergence rate was estimated using an adaptation of the methodology previously described by Salazar-Gonzalez et al. [48]. Firstly, maximum likelihood trees were constructed for each patient using all clonal sequences from each time point and rooted with the consensus sequences from other patients. Then, assuming a molecular clock, the branch lengths between the leafs and the root of the tree were calculated by using Branchlength Calculator [49] and plotted against time in years.
Natural selection of specific amino acids was examined using Codeml, models M0 and M3, with the HYPHY package [50]. Potential N-glycosylation sites were identified using N-Glycosite [51]. The entropy at each position in protein alignment was measured with Shannon Entropy [52].
Humoral antibody response against the env C2V3C3 regions
IgG and IgA antibody response against the env C2V3C3 region was quantified with the ELISA-HIV2 test developed in our laboratory, as described elsewhere with some modifications [53]. Briefly, microtiter plates (96-well) were coated with rgp36 and rpC2-C3 by overnight incubation at 4°C and blocked with 1% gelatine in Tris-buffered saline (TBS). HIV-2-positive plasma samples were added to the antigen coated wells at a 1:100 dilution. Bound antibodies were detected by using alkaline phosphatase (AP)-conjugated goat anti-human IgG (diluted 1:2000 in TBS) or horseradish peroxidase (HPR)-conjugated rabbit anti-human IgA (diluted 1:2000 in phosphate-buffer saline) (Sigma-Aldrich). The colour was developed using p-nitrophenilphosphate (p-NPP Tablets, Sigma-Aldrich) as chromogenic substrate to AP and o-phenylenediamine dihydrochloride (OPD) to HPR. Optical density (OD) was measured with an automated microplate reader LP 400 (Bio-Rad) at 405 and 492 nm against a reference wavelength of 620 nm. The clinical cut-off value of the assay, calculated as the mean OD value of HIV-seronegative samples plus three times the standard deviation [SD], was determined using samples from healthy HIV-seronegative subjects. The results of the assay are expressed quantitatively as ODclinical sample(S)/ODcut-off(CO) ratios. For ratio values >1 the sample is considered as seroreactive.
Statistical analysis
Statistical analysis was performed in GraphPad Prism version 4.00 for Windows (GraphPad Software), with a level of significance of 5%. For the inter-patient statistical analysis across time, only information obtained from one time point (one sample) per patient was considered in order to guarantee the independence of the data analyzed. Thus, to maximize the number of observations in the analysis, we chose the first sample (first time point) available for each patient. Nonparametric tests were used to compare means and medians between variables: paired data was analyzed with Wilcoxon-matched pairs test and Friedman test; unpaired variables were tested with Mann Whitney U test and Kruskal-Wallis test. To study how two variables varied together linear regression was performed and Spearman correlation coefficients were computed. Finally, Deming linear regression was used to study the overall variation (slopes) of intra-patient data with time (longitudinal analysis).
GenBank accession numbers
Sequences have been assigned the following GenBank accession numbers: EU358115–EU358499, EU358501, EU358504, EU358507, EU358509, EU358513, EU358517, EU358519–EU358521, EU358524, EU358525, EU358527–EU358531, EU358533, EU358536–EU358538, EU358541, EU358543, EU358546–EU358549, EU358551–EU358567, EU360797–EU360799.
Results
Phylogenetic relationships, genetic diversity and divergence
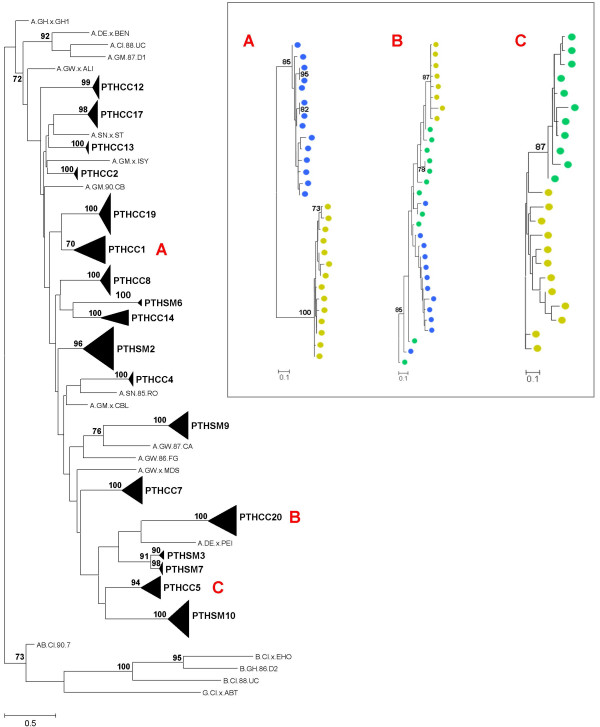
To investigate the molecular evolution of the HIV-2 env gene we have amplified, cloned and sequenced the env gene fragment coding for the C2, V3 and C3 regions using yearly samples collected from 18 patients followed prospectively for 2–4 years. A total of 431 clonal sequences were obtained from 18 patients (average of 13 sequences per patient per sampling year). Phylogenetic analysis showed that all sequences clustered together within HIV-2 group A and that each patient sequences formed monophyletic sub-clusters with high bootstrap supporting values (Figure 1). Phylogenetic analysis also showed that with the exceptions of patients PTHCC1, PTHCC5 and PTHCC20, sequences from most patients were not segregated according to sampling years, a clear indication that there were no major shifts in virus population structure from one year to the other.
Figure 1.
Maximum-likelihood phylogenetic analysis. The phylogenetic tree was constructed with reference sequences from HIV-2 groups A, B and G, under the TVM+G+I evolutionary model, using the NNI heuristic search strategy and 1000 bootstrap replications. The triangles represent the compressed subtrees containing clonal sequences obtained from all samples collected for each patient. The length of the triangle represents the intra-patient nucleotide diversity and its thickness is proportional to the number of sequences. The bootstrap values supporting the internal branches are shown. The scale bar represents evolutionary distances in substitutions per site. The inset contains the subtrees of patient PTHCC1 (A), PTHCC20 (B) and PTHCC5 (C) (Yellow circle – 2003; green circle – 2004; blue circle – 2005).
The mean evolutionary distance between different nucleotide sequences from each sample/year (nucleotide diversity) was 2.1% (standard deviation = 1.1) (additional file 1). Nucleotide diversity was neither associated with clinical status (2.1% mean median genetic distance in AIDS patients vs 1.4% in the other patients; p = 0.203) nor with plasma viremia (2.3% in viremic patients vs 1.8% in aviremic patients; p = 0.386) (n = 18).
Considering the first and the last samples of each patient, nucleotide diversity increased along the course of infection in all patients, except for patient PTHCC5 (additional file 1). Shannon's entropy was used to measure the relative amino acid variability in our set of sequences [52]. The sum of entropy values of the amino acid alignments varied between regions (p < 0.001), being significantly lower for the V3 region (p < 0.001) and higher for the C2 region (p < 0.005) (additional file 1).
Within-patient nucleotide divergence rate was on average 0.014 substitutions per site per year for the C2V3C3 region, but it varied widely between patients (SD = 0.011). There was no association between the divergence rate and the variation in the number of CD4+ T cells over time (Deming regression analysis, F = 0.058, p = 0.816). Likewise, the divergence rate of the C2V3C3 regions was not related with the level of IgG antibodies produced against the homologous peptide over time (F = 0.192, p = 0.675).
Selection analysis and adaptation rate of the C2, V3, and C3 regions
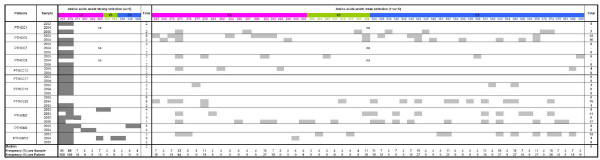
Intra-patient analysis showed that the overall C2V3C3 region was under purifying selection (dN/dS ratio < 1) along the course of infection in all patients (additional file 1). Analysis of the number and location of positively selected codons is useful to identify particular amino acids that may be under the selective pressure of the immune system, regions that can define potential neutralizing epitopes or that are functionally important for the protein [15,25-28]. In the present study, higher number of sites under positive selection tended to be found in patients with detectable viremia compared to patients with undetectable viremia (median, 15 sites vs 2; p = 0.061) (n = 18) (additional file 1). Otherwise, the number of positively selected sites was highly variable in number and position in most patients (Figure 2). Notable exceptions were amino acids at positions 267 and 270 in C2 (numbered according to the reference HIV-2ALI strain) that were under strong positive selection in all patients. Selection at these two sites persisted for at least two years in 9 patients (Figure 2). Because of these two sites, the median number of positively selected codons per sample was higher in the C2 region compared with the other regions (p < 0.005) (n = 18). Finally, using linear regression analysis we found that within each patient an average of 1.0 (SD = 3.8) positively selected site varied per year (adaptation rate).
Figure 2.
Frequency, intensity and distribution of positively selected sites in the C2, V3 and C3 regions along the course of HIV-2 infection. Positively selected codons (obtained with Codeml, model M3) were classified in two categories according to the ω ratio:ω>6, codons under strong selective pressure; 1<ω<6, codons under weak selective pressure. The frequency and distribution of positively selected sites in the C2, V3 and C3 regions are shown in each infection year. Higher frequency positively selected sites are shown in bold letters. Sites were numbered according to the reference HIV-2ALI strain. (na, not available)
Glycosylation of the HIV-2 env C2-C3 region
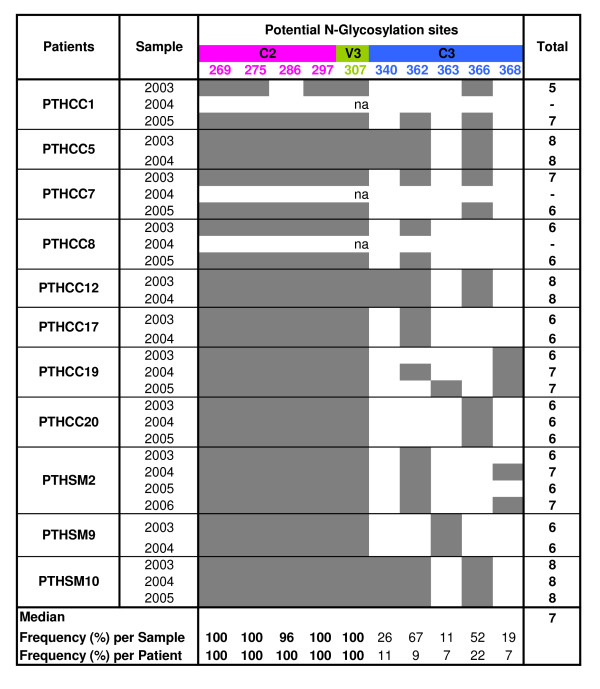
Since the glycosylation pattern of the HIV-1 env gene may influence neutralization escape to the immune system, viral tropism and clinical progression [32,36,54-57], we determined the number of potential N-glycosylation sites in our sequences and examined its variation as a function of time and other parameters analyzed in this study. The number of N-glycosylation sites ranged from 5 to 8 (median, 7) and tended to be conserved along the infection in each patient, the exception being patient PTHCC1 with an increase in two sites over the three years of follow up (Figure 3). The number of glycosylation sites varied significantly between C2, V3 and C3 (p < 0.001), being concentrated particularly in C2 (p < 0.001) (n = 18). At the intra- and inter-patient level, the most conserved N-glycosylation sites were located in C2 and V3. With one exception, all sites that varied over time were located in C3. The number of N-linked glycosylation sites was directly associated with the number of positively selected sites (r2 = 0.301; p = 0.018).
Figure 3.
Frequency and distribution of potential N-glycosylation sites in the C2, V3 and C3 regions along the course of infection. The frequency and distribution of potential N-linked glycosylation sites in the C2, V3 and C3 regions are shown in each infection year. Higher frequency glycosylation sites are shown in bold letters. Sites were numbered according to the reference HIV-2ALI strain. (na, not available)
Molecular evolution of the C2, V3 and C3 regions as a function of the antibody response
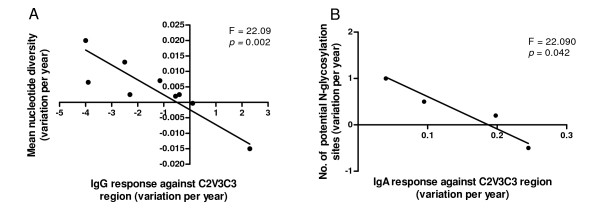
All patients produced IgA antibodies against the C2V3C3 region whereas IgG antibodies were detected in all but two patients, PTHSM9 and PTHSM10 (Table 1). Intra-patient analysis revealed that along the course of the infection the variation of C2V3C3-specific IgG response was inversely associated with the variation of nucleotide diversity (F = 22.09; p = 0.002) as well as with the dN rate (F = 22.800; p = 0.002) and amino acid diversity (Shannon's entropy, F = 23.610; p = 0.002), particularly in the V3 (F = 11.660; p = 0.014) and C3 regions (F = 6.214; p = 0.041) (n = 9) (Figure 4). Variation of the C2V3C3- specific IgA response over time was inversely associated with variation in the number of N-linked glycosylation sites (F = 22.090; p = 0.042; n = 4) which occurred in four patients particularly in the C3 region (Figure 4).
Figure 4.
C2V3C3 sequence evolution along the course of infection as a function of antibody response. Deming regression analysis. (A) Annual variation (slope) of the C2V3C3-IgG response vs Annual variation (slope) of the mean nucleotide diversity; (B) Annual variation (slope) of the C2V3C3-IgA response vs Annual variation (slope) of the number of potential N-glycosylation sites.
Discussion
In this study we have examined, for the first time, the molecular evolution of the envelope C2, V3 and C3 regions during chronic HIV-2 infection and its correlation with the antibody response against the same regions. Our cohort was constituted by long-term infected patients showing, in general, low CD4+ T cell counts and undetectable plasma viremia.
Nucleotide diversity increased with time in all but one patient with values similar to those obtained in an earlier study performed with HIV-2 elite controllers (2.1%, this study, vs 1.7%; p = 0.3440) [24]. This value is also similar to the 2.5% median diversity reported for chronically HIV-1 infected patients [58] and to the 3.0% mean diversity reported for some long-term nonprogressors with low viral load [21].
In phylogenetic analysis we found low quasispecies complexity in most patients, i.e. virus populations from most patients were mostly homogeneous during the follow up period. This was expected since HIV-2 is generally seen as a slowly evolving virus and over a short period of time one would expect to observe few evolutionary changes [22,24,59]. However, in three patients there was evidence for segregation of virus quasispecies according to the year of infection, which implies high rate of evolutionary change and immune selection in these patients [15,60]. Consistent with this, we found that the nucleotide divergence rate varied widely between patients. Moreover, the average nucleotide divergence rate (0.014 substitutions per site per year) was very high when compared to that reported for HIV-2 elite controllers (mean, 0.23%) [24] and for HIV-1 long-term non progressors with low plasma viral load (mean, 0.27%) [21]. Even though we could not detect any association between nucleotide divergence and the number of CD4+ T cells, the higher net divergence observed in our patients might be related to their high immune deterioration, as higher genetic divergence is generally found in HIV-1 rapid progressors compared to slow- or non-progressors [19,20]. In fact, the 0.014 annual divergence rate found in our patients is similar to that found in chronically HIV-1 infected patients (between 1.0% and 1.5% per year) [17,58]. In conclusion, the sampling schedule used in our study, and possibly the fact that we have analyzed the virus present inside the cells and not in the plasma, has enabled us to demonstrate that the evolutionary dynamics of HIV-2 during chronic infection is surprisingly similar to HIV-1. This implies that HIV-2 is actively replicating during chronic infection, possibly in the lymphoid tissue, as in HIV-2 patients the mononuclear cells in the lymph nodes are heavily infected, even more than the mononuclear cells in the peripheral blood [61,62]. Future studies of HIV-2 nucleotide divergence should include also the virus populations present in the lymphoid tissue and other cellular compartments (e.g. GI tract).
Despite the high nucleotide divergence rate, most of the substitutions were of a synonymous nature such that the dN/dS ratio of the C2V3C3 region was always below one and, most importantly, it decreased over time in most patients. These results are in agreement with previous reports that have examined the C2V3C3 region [22,24] and with the observation that, globally, the HIV-2 env gene is under purifying selection [25]. Consistent with previous studies of a cross-sectional nature, we found that C2 and C3, but not V3, were the fastest evolving regions at the nucleotide and amino acid level contributing significantly to the high within-patient nucleotide divergence rate [22,63]. The conservation of the V3 region in vivo implies that in HIV-2, as in HIV-1, this region is submitted to strong structural and conformational constraints which are probably related to its crucial functional roles at the level of coreceptor binding and cell entry [29-32].
It is probable that adaptation to immune pressure is the main driver of the rapid intra-host evolution of the C2 and C3 regions in HIV-2 [15,25,58,60,64-66]. Indeed, we found that most of the amino acids under selection are located in C2, including the two amino acids that are under strongest positive selection in all patients (positions 267 and 270). Moreover, selection at these two sites persisted for at least two years in the majority of the patients which is a clear indication that they are under continued immune pressure in vivo [60,67]. The equivalent amino acids in HIV-1 are not under positive selection [67], are located in the hidden surface of envelope glycoprotein complex [58] and define a cytotoxic T cell epitope [68]. Thus, our results also suggest that the antigenic presentation of the C2, and perhaps the C3 region (see below), in the envelope complex of HIV-2 differs substantially from that of HIV-1.
Glycans on HIV-1 envelope protein play an important role in the folding of the glycoproteins, in infection and in evasion from the host immune response (reviewed in [69]). We found that, as for HIV-1 [51,58], the majority of potential N-glycosylation sites were concentrated in the C2 region. The four N-glycosylation sites in C2 and the site in the beginning of V3 were highly conserved in all patients throughout infection which is strongly indicative of convergent evolution at these glycosylation hotspots and suggests an unexpected constraint on the potential diversity of the HIV-2 envelope [70,71]. The convergent evolution of glycosylation sites may have important implications for both vaccine design and antiviral therapeutic [69].
To try to identify the immune correlates of the molecular evolution of HIV-2 C2, V3 and C3 regions we have looked into all possible associations between the number of CD4+ T cells or the IgA and IgG antibody levels and different parameters that reflect viral molecular evolution. In longitudinal analysis there was no significant association between the number of CD4+ T cells and nucleotide diversity, amino acid entropy, nucleotide divergence, dN/dS ratio and number of positively selected sites. These results are in partial contrast to those of MacNeil et al. [24], who found a direct association between the rates of HIV-2 diversification and rates of CD4+ T cell decline in long-term non progressors followed for a decade in Senegal. The short term follow-up and the associated modest variation in the number of CD4 + T cells might have prevented the detection of this type of association in our patients.
Strikingly, however, there was a close relationship between virus diversification and evolution and C2V3C3-specific antibody response over time. In fact, higher IgG response was significantly associated with lower viral variability at the nucleotide and amino acid levels as well as with lower frequency of nonsynonymous substitutions. These results imply that the anti-C2V3C3 IgG antibodies are effective at reducing viral population size limiting the number of virus escape mutants [72]. This is in striking contrast to the majority of acute and chronic HIV-1 infections where the virus quickly escapes from anti-V3 and anti-C3 autologous neutralizing antibodies [33,73-76]. Consistent with the lower capacity of HIV-2 to escape from C2V3C3- neutralizing antibodies when compared to HIV-1, we found that on average HIV-2 has a five-fold lower adaptation rate in vivo than HIV-1 (1 positively selected site per year vs 5 sites per year) [60,77]. The HIV-2 low adaptation rate may be related to its low replicative capacity and low plasma viral load [12,13,78]. Overall, these results provide support for a crucial role of neutralizing antibody response in the effective containment of viral replication in HIV-2 infection in vivo [36].
Surprisingly, in some patients addition of glycans to the C3 region was associated with a reduction in the IgA immunogenicity of the C2V3C3 region. Envelope-specific plasma IgA antibodies, mostly binding to the gp36 transmembrane glycoprotein, have been found to neutralize HIV-2 [79]. Increasing the number of N-glycans in the envelope gp120 surface glycoprotein, or varying the position of glycosylation sites, has been associated with escape from IgG neutralizing antibody response in simian immunodeficiency virus (SIV) and HIV-1 infection [57,80-82]. Hence, one plausible explanation for the inverse association between IgA response and N-glycosylation is that the C3 envelope region induces IgA neutralizing antibodies to which HIV-2 escapes through the occlusion of the C3 region with N-linked glycans. This may have important implications for vaccine design. Ongoing studies will determine whether C2V3C3- specific IgA antibodies present in these patients effectively neutralize their autologous virus.
Conclusion
The evolutionary dynamics of HIV-2 envelope during chronic and highly suppressed infection is surprisingly similar to HIV-1 implying that the virus is actively replicating in cellular compartments. Convergent evolution of N-glycosylation in C2 and V3, as well as the limited diversification of V3, indicates however that there are important functional constraints to the potential diversity of the HIV-2 envelope. HIV-2 envelope diversification is inversely related to the C2V3C3-specific IgG antibody response over time implying that these antibodies are effective at reducing viral population size, limiting the number of virus escape mutants. The C3 region seems to be a target for IgA antibodies and increasing N-linked glycosylation may prevent HIV-2 envelope recognition by these antibodies. Our results provide new insights into the biology of HIV-2 and its relation with the human host and may have important implications for vaccine design.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
NT designed and coordinated the study. PB performed most of the cloning and sequencing experiments. JMM isolated the viruses and quantified the antibody responses. HB and CR participated in virus isolation and in the sequencing analysis of some patients. MD, FA and FM recruited the patients and were responsible for collecting the blood samples and the clinical data. PG quantified the plasma viremia. CN and PG helped with the interpretation of data and revision of the manuscript. PB and NT preformed statistical analysis. PB and NT interpreted the data and wrote the manuscript. All authors reviewed and accepted the final manuscript.
Supplementary Material
Table 2. Results from sequence and phylogenetic analysis. a dN/dS – ratio of nonsynonymous and synonymous substitutions, obtained with Codeml (model M0). b dN/dS – ratio of nonsynonymous and synonymous substitutions between the first and the last time point, obtained with Codeml (model M0), when applicable. c Sum of Shannon's entropy values at each position in protein alignment. d Number of positively selected codons in the nucleotide alignment, obtained with Codeml (model M3). SD – Standard deviation.
Acknowledgments
Acknowledgements
This work was supported by Fundação para a Ciência e Tecnologia (project POCTI/ESP/48045). Pedro Borrego is supported by a PhD grant from Fundação para a Ciência e Tecnologia.
Contributor Information
Pedro Borrego, Email: pborrego@ff.ul.pt.
José Maria Marcelino, Email: jose.marcelino@ineti.pt.
Cheila Rocha, Email: cheilarocha@ff.ul.pt.
Manuela Doroana, Email: manuela.doroana@hsm.min-saude.pt.
Francisco Antunes, Email: ip231874@sapo.pt.
Fernando Maltez, Email: fmaltez@hccabral.min-saude.pt.
Perpétua Gomes, Email: p.gomes@netcabo.pt.
Carlos Novo, Email: carlos.novo@ineti.pt.
Helena Barroso, Email: mbarroso@ff.ul.pt.
Nuno Taveira, Email: ntaveira@ff.ul.pt.
References
- Hu DJ, Dondero TJ, Rayfield MA, George JR, Schochetman G, Jaffe HW, Luo CC, Kalish ML, Weniger BG, Pau CP, et al. The emerging genetic diversity of HIV. The importance of global surveillance for diagnostics, research, and prevention. JAMA. 1996;275:210–216. doi: 10.1001/jama.275.3.210. [DOI] [PubMed] [Google Scholar]
- Andersson S, Norrgren H, da Silva Z, Biague A, Bamba S, Kwok S, Christopherson C, Biberfeld G, Albert J. Plasma viral load in HIV-1 and HIV-2 singly and dually infected individuals in Guinea-Bissau, West Africa: significantly lower plasma virus set point in HIV-2 infection than in HIV-1 infection. Arch Intern Med. 2000;160:3286–3293. doi: 10.1001/archinte.160.21.3286. [DOI] [PubMed] [Google Scholar]
- Kanki PJ, Travers KU, S MB, Hsieh CC, Marlink RG, Gueye NA, Siby T, Thior I, Hernandez-Avila M, Sankale JL, et al. Slower heterosexual spread of HIV-2 than HIV-1. Lancet. 1994;343:943–946. doi: 10.1016/S0140-6736(94)90065-5. [DOI] [PubMed] [Google Scholar]
- Marlink R, Kanki P, Thior I, Travers K, Eisen G, Siby T, Traore I, Hsieh CC, Dia MC, Gueye EH, et al. Reduced rate of disease development after HIV-2 infection as compared to HIV-1. Science. 1994;265:1587–1590. doi: 10.1126/science.7915856. [DOI] [PubMed] [Google Scholar]
- Whittle H, Morris J, Todd J, Corrah T, Sabally S, Bangali J, Ngom PT, Rolfe M, Wilkins A. HIV-2-infected patients survive longer than HIV-1-infected patients. AIDS. 1994;8:1617–1620. doi: 10.1097/00002030-199411000-00015. [DOI] [PubMed] [Google Scholar]
- Reeves JD, Doms RW. Human immunodeficiency virus type 2. J Gen Virol. 2002;83:1253–1265. doi: 10.1099/0022-1317-83-6-1253. [DOI] [PubMed] [Google Scholar]
- Berry N, Ariyoshi K, Jaffar S, Sabally S, Corrah T, Tedder R, Whittle H. Low peripheral blood viral HIV-2 RNA in individuals with high CD4 percentage differentiates HIV-2 from HIV-1 infection. J Hum Virol. 1998;1:457–468. [PubMed] [Google Scholar]
- Soares R, Foxall R, Albuquerque A, Cortesao C, Garcia M, Victorino RM, Sousa AE. Increased frequency of circulating CCR5+ CD4+ T cells in human immunodeficiency virus type 2 infection. J Virol. 2006;80:12425–12429. doi: 10.1128/JVI.01557-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DE, Llenado RA, Torres JV. Humoral immunity and the evolution of HIV-2. Viral Immunol. 2004;17:436–439. doi: 10.1089/vim.2004.17.436. [DOI] [PubMed] [Google Scholar]
- Berry N, Jaffar S, Schim van der Loeff M, Ariyoshi K, Harding E, N'Gom PT, Dias F, Wilkins A, Ricard D, Aaby P, et al. Low level viremia and high CD4% predict normal survival in a cohort of HIV type-2-infected villagers. AIDS Res Hum Retroviruses. 2002;18:1167–1173. doi: 10.1089/08892220260387904. [DOI] [PubMed] [Google Scholar]
- Lizeng Q, Nilsson C, Sourial S, Andersson S, Larsen O, Aaby P, Ehnlund M, Bjorling E. Potent neutralizing serum immunoglobulin A (IgA) in human immunodeficiency virus type 2-exposed IgG-seronegative individuals. J Virol. 2004;78:7016–7022. doi: 10.1128/JVI.78.13.7016-7022.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNeil A, Sarr AD, Sankale JL, Meloni ST, Mboup S, Kanki P. Direct evidence of lower viral replication rates in vivo in human immunodeficiency virus type 2 (HIV-2) infection than in HIV-1 infection. J Virol. 2007;81:5325–5330. doi: 10.1128/JVI.02625-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaak H, Ende ME van der, Boers PH, Schuitemaker H, Osterhaus AD. In vitro replication capacity of HIV-2 variants from long-term aviremic individuals. Virology. 2006;353:144–154. doi: 10.1016/j.virol.2006.05.029. [DOI] [PubMed] [Google Scholar]
- Levy JA. HIV and the pathogenesis of AIDS. 2. Washington, D.C.: ASM Press; 1998. [Google Scholar]
- Lemey P, Rambaut A, Pybus OG. HIV evolutionary dynamics within and among hosts. AIDS Rev. 2006;8:125–140. [PubMed] [Google Scholar]
- Delwart EL, Pan H, Sheppard HW, Wolpert D, Neumann AU, Korber B, Mullins JI. Slower evolution of human immunodeficiency virus type 1 quasispecies during progression to AIDS. J Virol. 1997;71:7498–7508. doi: 10.1128/jvi.71.10.7498-7508.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankarappa R, Margolick JB, Gange SJ, Rodrigo AG, Upchurch D, Farzadegan H, Gupta P, Rinaldo CR, Learn GH, He X, et al. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J Virol. 1999;73:10489–10502. doi: 10.1128/jvi.73.12.10489-10502.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson S, Perry SM, Bustamante CD, Orive ME, Stearns MN, Kelly JK. A statistical characterization of consistent patterns of human immunodeficiency virus evolution within infected patients. Mol Biol Evol. 2005;22:456–468. doi: 10.1093/molbev/msi029. [DOI] [PubMed] [Google Scholar]
- Castiglione F, Poccia F, D'Offizi G, Bernaschi M. Mutation, fitness, viral diversity, and predictive markers of disease progression in a computational model of HIV type 1 infection. AIDS Res Hum Retroviruses. 2004;20:1314–1323. doi: 10.1089/aid.2004.20.1314. [DOI] [PubMed] [Google Scholar]
- Markham RB, Wang WC, Weisstein AE, Wang Z, Munoz A, Templeton A, Margolick J, Vlahov D, Quinn T, Farzadegan H, Yu XF. Patterns of HIV-1 evolution in individuals with differing rates of CD4 T cell decline. Proc Natl Acad Sci USA. 1998;95:12568–12573. doi: 10.1073/pnas.95.21.12568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello G, Casado C, Sandonis V, Alvaro-Cifuentes T, Dos Santos CA, Garcia S, Rodriguez C, Del Romero J, Pilotto JH, Grinsztejn B, et al. Plasma viral load threshold for sustaining intrahost HIV type 1 evolution. AIDS Res Hum Retroviruses. 2007;23:1242–1250. doi: 10.1089/aid.2007.0074. [DOI] [PubMed] [Google Scholar]
- Barroso H, Taveira N. Evidence for negative selective pressure in HIV-2 evolution in vivo. Infect Genet Evol. 2005;5:239–246. doi: 10.1016/j.meegid.2004.07.008. [DOI] [PubMed] [Google Scholar]
- Sankale JL, de la Tour RS, Renjifo B, Siby T, Mboup S, Marlink RG, Essex ME, Kanki PJ. Intrapatient variability of the human immunodeficiency virus type 2 envelope V3 loop. AIDS Res Hum Retroviruses. 1995;11:617–623. doi: 10.1089/aid.1995.11.617. [DOI] [PubMed] [Google Scholar]
- MacNeil A, Sankale JL, Meloni ST, Sarr AD, Mboup S, Kanki P. Long-term intrapatient viral evolution during HIV-2 infection. J Infect Dis. 2007;195:726–733. doi: 10.1086/511308. [DOI] [PubMed] [Google Scholar]
- Choisy M, Woelk CH, Guegan JF, Robertson DL. Comparative study of adaptive molecular evolution in different human immunodeficiency virus groups and subtypes. J Virol. 2004;78:1962–1970. doi: 10.1128/JVI.78.4.1962-1970.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhail M, Wang B, Lemey P, Beckthold B, Vandamme AM, Gill MJ, Saksena NK. Role of viral evolutionary rate in HIV-1 disease progression in a linked cohort. Retrovirology. 2005;2:41. doi: 10.1186/1742-4690-2-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Gojobori T. Evolutionary mechanisms and population dynamics of the third variable envelope region of HIV within single hosts. Proc Natl Acad Sci USA. 1997;94:1264–1269. doi: 10.1073/pnas.94.4.1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Hoffmann F, He J, He X, Kankasa C, Ruprecht R, West JT, Orti G, Wood C. Evolution of subtype C HIV-1 Env in a slowly progressing Zambian infant. Retrovirology. 2005;2:67. doi: 10.1186/1742-4690-2-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jong JJ, De Ronde A, Keulen W, Tersmette M, Goudsmit J. Minimal requirements for the human immunodeficiency virus type 1 V3 domain to support the syncytium-inducing phenotype: analysis by single amino acid substitution. J Virol. 1992;66:6777–6780. doi: 10.1128/jvi.66.11.6777-6780.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delobel P, Nugeyre MT, Cazabat M, Pasquier C, Marchou B, Massip P, Barre-Sinoussi F, Israel N, Izopet J. Population-based sequencing of the V3 region of env for predicting the coreceptor usage of human immunodeficiency virus type 1 quasispecies. J Clin Microbiol. 2007;45:1572–1580. doi: 10.1128/JCM.02090-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CC, Tang M, Zhang MY, Majeed S, Montabana E, Stanfield RL, Dimitrov DS, Korber B, Sodroski J, Wilson IA, et al. Structure of a V3-containing HIV-1 gp120 core. Science. 2005;310:1025–1028. doi: 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaka Y, Sato A, Miki S, Kawauchi S, Sakaida H, Hori T, Uchiyama T, Adachi A, Hayami M, Fujiwara T, Yoshie O. Small amino acid changes in the V3 loop of human immunodeficiency virus type 2 determines the coreceptor usage for CXCR4 and CCR5. Virology. 1999;264:237–243. doi: 10.1006/viro.1999.0006. [DOI] [PubMed] [Google Scholar]
- Zolla-Pazner S. Improving on nature: focusing the immune response on the V3 loop. Hum Antibodies. 2005;14:69–72. [PubMed] [Google Scholar]
- Bjorling E, Scarlatti G, von Gegerfelt A, Albert J, Biberfeld G, Chiodi F, Norrby E, Fenyo EM. Autologous neutralizing antibodies prevail in HIV-2 but not in HIV-1 infection. Virology. 1993;193:528–530. doi: 10.1006/viro.1993.1160. [DOI] [PubMed] [Google Scholar]
- Andersson S. HIV-2 and the immune response. AIDS Reviews. 2001;3:11–23. [Google Scholar]
- Shi Y, Brandin E, Vincic E, Jansson M, Blaxhult A, Gyllensten K, Moberg L, Brostrom C, Fenyo EM, Albert J. Evolution of human immunodeficiency virus type 2 coreceptor usage, autologous neutralization, envelope sequence and glycosylation. J Gen Virol. 2005;86:3385–3396. doi: 10.1099/vir.0.81259-0. [DOI] [PubMed] [Google Scholar]
- Rodriguez SK, Sarr AD, MacNeil A, Thakore-Meloni S, Gueye-Ndiaye A, Traore I, Dia MC, Mboup S, Kanki PJ. Comparison of heterologous neutralizing antibody responses of human immunodeficiency virus type 1 (HIV-1)- and HIV-2-infected Senegalese patients: distinct patterns of breadth and magnitude distinguish HIV-1 and HIV-2 infections. J Virol. 2007;81:5331–5338. doi: 10.1128/JVI.02789-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano V, Gomes P, Heneine W, Holguin A, Doruana M, Antunes R, Mansinho K, Switzer WM, Araujo C, Shanmugam V, et al. Human immunodeficiency virus type 2 (HIV-2) in Portugal: clinical spectrum, circulating subtypes, virus isolation, and plasma viral load. J Med Virol. 2000;61:111–116. doi: 10.1002/(SICI)1096-9071(200005)61:1<111::AID-JMV18>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Cavaco-Silva P, Taveira NC, Rosado L, Lourenco MH, Moniz-Pereira J, Douglas NW, Daniels RS, Santos-Ferreira MO. Virological and molecular demonstration of human immunodeficiency virus type 2 vertical transmission. J Virol. 1998;72:3418–3422. doi: 10.1128/jvi.72.4.3418-3422.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Maniatis T, Fritsch EF. Molecular cloning: a laboratory manual. 2. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas KB, Nicholas Hugh B., Jr GeneDoc: a tool for editing and annotating multiple sequence alignments. Distributed by the author. 1997.
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP*. Phylogenetic Analysis using Parsimony (*and other Methods). Version 4. Sinauer-Associates, Sunderland, Massachusetts; 1998. [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- Salazar-Gonzalez JF, Bailes E, Pham KT, Salazar MG, Guffey MB, Keele BF, Derdeyn CA, Farmer P, Hunter E, Allen S, et al. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J Virol. 2008;82:3952–3970. doi: 10.1128/JVI.02660-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branchlength Calculator http://www.hiv.lanl.gov/content/sequence/BRANCHLENGTH/branchlength.html
- Pond SL, Frost SD, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- Zhang M, Gaschen B, Blay W, Foley B, Haigwood N, Kuiken C, Korber B. Tracking global patterns of N-linked glycosylation site variation in highly variable viral glycoproteins: HIV, SIV, and HCV envelopes and influenza hemagglutinin. Glycobiology. 2004;14:1229–1246. doi: 10.1093/glycob/cwh106. [DOI] [PubMed] [Google Scholar]
- Korber BT, Kunstman KJ, Patterson BK, Furtado M, McEvilly MM, Levy R, Wolinsky SM. Genetic differences between blood- and brain-derived viral sequences from human immunodeficiency virus type 1-infected patients: evidence of conserved elements in the V3 region of the envelope protein of brain-derived sequences. J Virol. 1994;68:7467–7481. doi: 10.1128/jvi.68.11.7467-7481.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcelino JM, Barroso H, Goncalves F, Silva SM, Novo C, Gomes P, Camacho R, Taveira N. Use of a new dual-antigen enzyme-linked immunosorbent assay to detect and characterize the human antibody response to the human immunodeficiency virus type 2 envelope gp125 and gp36 glycoproteins. J Clin Microbiol. 2006;44:607–611. doi: 10.1128/JCM.44.2.607-611.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botarelli P, Houlden BA, Haigwood NL, Servis C, Montagna D, Abrignani S. N-glycosylation of HIV-gp120 may constrain recognition by T lymphocytes. J Immunol. 1991;147:3128–3132. [PubMed] [Google Scholar]
- Clevestig P, Pramanik L, Leitner T, Ehrnst A. CCR5 use by human immunodeficiency virus type 1 is associated closely with the gp120 V3 loop N-linked glycosylation site. J Gen Virol. 2006;87:607–612. doi: 10.1099/vir.0.81510-0. [DOI] [PubMed] [Google Scholar]
- Pollakis G, Kang S, Kliphuis A, Chalaby MI, Goudsmit J, Paxton WA. N-linked glycosylation of the HIV type-1 gp120 envelope glycoprotein as a major determinant of CCR5 and CXCR4 coreceptor utilization. J Biol Chem. 2001;276:13433–13441. doi: 10.1074/jbc.M009779200. [DOI] [PubMed] [Google Scholar]
- Polzer S, Dittmar MT, Schmitz H, Schreiber M. The N-linked glycan g15 within the V3 loop of the HIV-1 external glycoprotein gp120 affects coreceptor usage, cellular tropism, and neutralization. Virology. 2002;304:70–80. doi: 10.1006/viro.2002.1760. [DOI] [PubMed] [Google Scholar]
- Joos B, Fischer M, Schweizer A, Kuster H, Boni J, Wong JK, Weber R, Trkola A, Gunthard HF. Positive in vivo selection of the HIV-1 envelope protein gp120 occurs at surface-exposed regions. J Infect Dis. 2007;196:313–320. doi: 10.1086/518935. [DOI] [PubMed] [Google Scholar]
- Barroso H, Araujo F, Gomes MH, Mota-Miranda A, Taveira N. Phylogenetic demonstration of two cases of perinatal human immunodeficiency virus type 2 infection diagnosed in adulthood. AIDS Res Hum Retroviruses. 2004;20:1373–1376. doi: 10.1089/aid.2004.20.1373. [DOI] [PubMed] [Google Scholar]
- Rambaut A, Posada D, Crandall KA, Holmes EC. The causes and consequences of HIV evolution. Nat Rev Genet. 2004;5:52–61. doi: 10.1038/nrg1246. [DOI] [PubMed] [Google Scholar]
- Gomes P, Taveira NC, Pereira JM, Antunes F, Ferreira MO, Lourenco MH. Quantitation of human immunodeficiency virus type 2 DNA in peripheral blood mononuclear cells by using a quantitative-competitive PCR assay. J Clin Microbiol. 1999;37:453–456. doi: 10.1128/jcm.37.2.453-456.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobe O, Ariyoshi K, Marchant A, Sabally S, Corrah T, Berry N, Jaffar S, Whittle H. Proviral load and immune function in blood and lymph node during HIV-1 and HIV-2 infection. Clin Exp Immunol. 1999;116:474–478. doi: 10.1046/j.1365-2249.1999.00914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parreira R, Esteves A, Santos C, Piedade J, Venenno T, Canas-Ferreira WF. Genetic variability of human immunodeficiency virus type 2 C2V3 region within and between individuals from Bissau, Guinea-Bissau, West Africa. AIDS Res Hum Retroviruses. 2000;16:1307–1312. doi: 10.1089/08892220050117069. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Kabata Y, Gojobori T. Reevaluation of amino acid variability of the human immunodeficiency virus type 1 gp120 envelope glycoprotein and prediction of new discontinuous epitopes. J Virol. 2000;74:4335–4350. doi: 10.1128/JVI.74.9.4335-4350.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Bielawski JP, Yang Z. Widespread adaptive evolution in the human immunodeficiency virus type 1 genome. J Mol Evol. 2003;57:212–221. doi: 10.1007/s00239-003-2467-9. [DOI] [PubMed] [Google Scholar]
- Lemey P, Kosakovsky Pond SL, Drummond AJ, Pybus OG, Shapiro B, Barroso H, Taveira N, Rambaut A. Synonymous substitution rates predict HIV disease progression as a result of underlying replication dynamics. PLoS Comput Biol. 2007;3:e29. doi: 10.1371/journal.pcbi.0030029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross HA, Rodrigo AG. Immune-mediated positive selection drives human immunodeficiency virus type 1 molecular variation and predicts disease duration. J Virol. 2002;76:11715–11720. doi: 10.1128/JVI.76.22.11715-11720.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber BT M, Brander C, Haynes BF, Koup R, Moore JP, Walker BD, Watkins DI. HIV Molecular Immunology 2006/2007. Los Alamos National Laboratory, Theoretical Biology and Biophysics, Los Alamos, New Mexico. LA-UR 07-4752.; [Google Scholar]
- Scanlan CN, Offer J, Zitzmann N, Dwek RA. Exploiting the defensive sugars of HIV-1 for drug and vaccine design. Nature. 2007;446:1038–1045. doi: 10.1038/nature05818. [DOI] [PubMed] [Google Scholar]
- Fischer PB, Karlsson GB, Butters TD, Dwek RA, Platt FM. N-butyldeoxynojirimycin-mediated inhibition of human immunodeficiency virus entry correlates with changes in antibody recognition of the V1/V2 region of gp120. J Virol. 1996;70:7143–7152. doi: 10.1128/jvi.70.10.7143-7152.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon AF, Lewis FI, Pond SL, Frost SD. Evolutionary interactions between N-linked glycosylation sites in the HIV-1 envelope. PLoS Comput Biol. 2007;3:e11. doi: 10.1371/journal.pcbi.0030011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenfell BT, Pybus OG, Gog JR, Wood JL, Daly JM, Mumford JA, Holmes EC. Unifying the epidemiological and evolutionary dynamics of pathogens. Science. 2004;303:327–332. doi: 10.1126/science.1090727. [DOI] [PubMed] [Google Scholar]
- Zolla-Pazner S. Identifying epitopes of HIV-1 that induce protective antibodies. Nat Rev Immunol. 2004;4:199–210. doi: 10.1038/nri1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richman DD, Wrin T, Little SJ, Petropoulos CJ. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc Natl Acad Sci USA. 2003;100:4144–4149. doi: 10.1073/pnas.0630530100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost SD, Wrin T, Smith DM, Kosakovsky Pond SL, Liu Y, Paxinos E, Chappey C, Galovich J, Beauchaine J, Petropoulos CJ, et al. Neutralizing antibody responses drive the evolution of human immunodeficiency virus type 1 envelope during recent HIV infection. Proc Natl Acad Sci USA. 2005;102:18514–18519. doi: 10.1073/pnas.0504658102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore PL, Gray ES, Choge IA, Ranchobe N, Mlisana K, Abdool Karim SS, Williamson C, Morris L. The c3-v4 region is a major target of autologous neutralizing antibodies in human immunodeficiency virus type 1 subtype C infection. J Virol. 2008;82:1860–1869. doi: 10.1128/JVI.02187-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson S. Adaptation in the env gene of HIV-1 and evolutionary theories of disease progression. Mol Biol Evol. 2003;20:1318–1325. doi: 10.1093/molbev/msg144. [DOI] [PubMed] [Google Scholar]
- Marchant D, Neil SJ, McKnight A. Human immunodeficiency virus types 1 and 2 have different replication kinetics in human primary macrophage culture. J Gen Virol. 2006;87:411–418. doi: 10.1099/vir.0.81391-0. [DOI] [PubMed] [Google Scholar]
- Lizeng Q, Skott P, Sourial S, Nilsson C, Andersson SS, Ehnlund M, Taveira N, Bjorling E. Serum immunoglobulin A (IgA)-mediated immunity in human immunodeficiency virus type 2 (HIV-2) infection. Virology. 2003;308:225–232. doi: 10.1016/S0042-6822(02)00088-0. [DOI] [PubMed] [Google Scholar]
- Chackerian B, Rudensey LM, Overbaugh J. Specific N-linked and O-linked glycosylation modifications in the envelope V1 domain of simian immunodeficiency virus variants that evolve in the host alter recognition by neutralizing antibodies. J Virol. 1997;71:7719–7727. doi: 10.1128/jvi.71.10.7719-7727.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- Sagar M, Wu X, Lee S, Overbaugh J. Human immunodeficiency virus type 1 V1-V2 envelope loop sequences expand and add glycosylation sites over the course of infection, and these modifications affect antibody neutralization sensitivity. J Virol. 2006;80:9586–9598. doi: 10.1128/JVI.00141-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table 2. Results from sequence and phylogenetic analysis. a dN/dS – ratio of nonsynonymous and synonymous substitutions, obtained with Codeml (model M0). b dN/dS – ratio of nonsynonymous and synonymous substitutions between the first and the last time point, obtained with Codeml (model M0), when applicable. c Sum of Shannon's entropy values at each position in protein alignment. d Number of positively selected codons in the nucleotide alignment, obtained with Codeml (model M3). SD – Standard deviation.