Abstract
Highly specific amplification of complex DNA pools without bias or template-independent products (TIPs) remains a challenge. We have developed a method using phi29 DNA polymerase and trehalose and optimized control of amplification to create micrograms of specific amplicons without TIPs from down to subfemtograms of DNA. With an input of as little as 0.5–2.5 ng of human gDNA or a few cells, the product could be close to native DNA in locus representation. The amplicons from 5 and 0.5 ng of DNA faithfully demonstrated all previously known heterozygous segmental duplications and deletions (3 Mb to 18 kb) located on chromosome 22 and even a homozygous deletion smaller than 1 kb with high-resolution chromosome-wide comparative genomic hybridization. With 550k Infinium BeadChip SNP typing, the >99.7% accuracy was compared favorably with results on unamplified DNA. Importantly, underrepresentation of chromosome termini that occurred with GenomiPhi v2 was greatly rescued with the present procedure, and the call rate and accuracy of SNP typing were also improved for the amplicons with a 0.5-ng, partially degraded DNA input. In addition, the amplification proceeded logarithmically in terms of total yield before saturation; the intact cells was amplified >50 times more efficiently than an equivalent amount of extracted DNA; and the locus imbalance for amplicons with 0.1 ng or lower input of DNA was variable, whereas for higher input it was largely reproducible. This procedure facilitates genomic analysis with single cells or other traces of DNA, and generates products suitable for analysis by massively parallel sequencing as well as microarray hybridization.
Keywords: copy number variation, single nucleotide polymorphism, comparative genomic hybridization, multiple displacement amplification
Whole-genome amplification (WGA) plays an important role in genomic research, especially where the amount of the DNA is limited (1–3). Multiple displacement amplification (MDA)-based methods (4) exhibit significantly higher sequence fidelity, less allele and locus bias, higher sensitivity, and higher efficiency than PCR-based methods (1–6). In MDA, ϕ29 DNA polymerase and random exonuclease-resistant primers are used, and DNA is amplified in a reaction maintained at 30°C over a 16- to 18-h period or, in recent modifications of the protocol, for as short a time as 1.5–2 h (7). ϕ29 DNA polymerase can synthesize DNA strands that are up to 100 kb in length with an average length greater than 10 kb (2). Given these advantages, in recent years, MDA WGA has been applied extensively in genomics.
The current MDA methods generate template independent product (TIP). This TIP synthesis is largely oligonucleotide-derived, but exogenous DNA contamination can also contribute (8–13). When the input is limited, such as with a subnanogram amount of template DNA or a limited number of cells, TIPs are very abundant, often representing 70–75% of the total yield (10, 14–17). Therefore, TIPs significantly impair some of the applications of the amplicon (18–20).
Recently, several efforts have been made to eliminate TIP and improve the specificity of MDA (1, 9, 11, 13, 19, 21–23). The outstanding examples include steps for strict control of experimental procedures to avoid exogenous DNA contamination (11), and minimization of the reaction volume (600 to 60 nl) (9, 19) or the reaction time (7). These efforts have succeeded in reducing TIPs but have been difficult to apply in routine practical operation and/or do not completely eliminate TIPs. For example, the most recent Gv2 (GenomiPhi Gv2; GE Healthcare Life Sciences) method results in up to 10 ng/μl TIP (7, 17, 18), whereas the nanoliter reaction produces on average amplicons containing 53–62% TIP when a single cell is used as a template (19).
Another serious limitation is that significant locus and allele biases, or genomic distortion further reduce the quality of MDA amplicons (4, 8, 11, 16, 19, 24, 25). In the investigations mentioned above, a substantial locus dropout was seen, while some loci showed preferential amplification. Even with a larger input (10–100 ng) (24, 25) or with a recently updated protocol (18), the amplicon showed significant locus bias as well as allele dropout. Even the new formats of MDA typically recovered only ≈75% of the genome from a single bacterial cell, based on deep sequencing (11, 19). This underrepresentation or dropout was shown to involve a distinct subset of genomic loci (18, 20, 26), but an observation to the contrary was also reported (11, 16). Amplification bias, according to our observations, occurs often with TIP, but also without TIP.
An improved WGA procedure could facilitate genomic studies of a single or a limited number of cells, and archive samples for sequencing, genomic segmental copy number variation (CNV) analysis, and SNP typing (1, 2, 19, 26–28). We describe here a relatively simple MDA approach for WGA. This approach, labeled as Wpa (whole-pool amplification), provides highly specific, unbiased, and hypersensitive amplification of very small amounts of entire genomes or complex DNA pools.
Results
Proper Concentration of Trehalose in the Reaction Eliminated TIP and Suppressed Locus Bias.
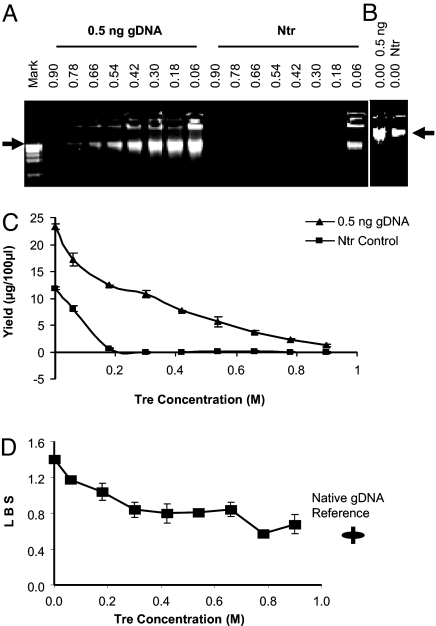
To validate the yield of specific products and TIP, different amounts of gDNA were amplified in parallel with an Ntr control (N9-Ntr) by using a series of concentrations of Tre. A set of amplicons with 0.5 ng of gDNA input vs. Ntr with the general amplification procedure is shown [Fig. 1 A–C and supporting information (SI) Fig. S1]. The absence of products in Ntr when the concentration of Tre was >45–50 μl (or 0.54–0.6 M) was reconfirmed by the absence of product even after a second round of amplification. We routinely took N9-Ntr negativity as the indicator of the absence of TIP. If there was significant exogenous contamination, verified by R9-Ntr, we treated reagents, tips, and tubes and their cups with UV light in a tissue culture hood for decontamination [adapted from a previous method (11)].
Fig. 1.
At appropriate Tre concentrations, TIPs were eliminated, and the locus representation of the amplicon was improved. Extracted gDNA (0.5 ng) vs. Ntr control was tested. Tre preparation lot T808 was used. The numbers above refer the final concentration of Tre. Arrow: 12 kb. (A) Tre concentration was varied from 0.06 to 0.9 M in a 100-μl reaction. Mark: 1-kb DNA ladder (Invitrogen). (B) Without Tre. (C) Product yields at different Tre concentrations, duplicate measurements by PicoGreen assay (two times). (D) LBS (SI Text) of products at different Tre concentrations, and LBS of reference gDNA. The measurement was repeated (two times) for each amplicon, and multiply repeated (nine times) for the reference in different independent tests.
Up to a certain point, the locus representation was improved with higher Tre concentrations in the reaction system (Fig. 1D; Tre preparation lot T808 was used). When the Tre in reaction was >0.42 M, and no TIP was detectable, the locus bias usually was very low and almost constant. Only when Tre was below a certain threshold (<0.3 M) was locus bias greatly increased. However, when Tre was too high, this resulted in a very low yield, and the locus representation could also be distorted. We chose a Tre concentration that was low enough to enable a significant yield of amplified template DNA in a limited time, yet high enough to suppress any TIP. To ensure safe and robust operation, this concentration was usually between 45 and 70 μl × 1.2 M (or 0.54–0.84 M final concentration) in a 100-μl reaction, depending on the preparation of Tre. Even without Tre, the yield for N9-Ntr was apparently less than that of specific product (Fig. 1B), which was different from the yields with the original MDA (12). This is presumably due to the optimized concentration of other reagents in our reaction system. In addition, the Wpa-40°C, which resulted in a specific amplification of DNA without TIP, also obviously improved the locus representation and call rate compared with the original MDA procedure, Repli-g 625S (Molecular Staging) (data not shown). However, the 40°C amplification was not as uniform as the 30°C procedure described above (see below for details).
Estimation of Efficiency and Sensitivity of Amplification.
A set of serially diluted gDNAs was used for amplification with the general procedure. For the 16-h reaction, when the input was reduced from 87.5 ng to 20 pg, the yield of amplicons was reduced, but the log2 value of the fold amplification increased linearly with an increasing log2 value of the input (Fig. S2 and Table S1, where a different preparation lot of Tre, T913, was used). Thus, there was ample yield of product without any TIP under these conditions, and the window of time for TIP free amplification was much wider than with other methods.
A second round of amplification was performed by using 10 ng of purified first-round amplicon as input. We used four amounts of first-round amplified product (12.5, 2.5, 0.5, and 0.1 ng of gDNA) and obtained a yield in the second round of amplification of between 90 and 40 μg (Table S1), which corresponded to a 4,000- to 9,000-fold additional amplification, whereas in the second round Ntr was still undetectable. Interestingly, reamplification with a higher original input in the first round resulted in a higher yield in the second round of amplification, despite the fact that the measured inputs for second-round amplifications were actually the same. This may indicate that the product quality with higher original input was in some way better than that with lower original input. If the whole product from the first-round amplification had been used as the input for a second round of reamplification, then the total yield would have been milligrams of DNA.
As a validation test with the 80-μl trace DNA amplification procedure and real-time monitoring (Fig. 2), human gDNA was diluted by serial 10-fold steps from 10 ng down to 0.01 fg, and 0.1-ng to 0.01-fg input were analyzed. Usually, with 0.1-ng to 1-pg input, ≈4 μg of purified product was obtained when the reaction was allowed to continue for 20 h. With this protocol and these levels of input DNA, the reaction had gone to completion by 20 h. For 0.1-pg to 0.01-fg input, in different repetitions, the fluorescence signal varied: In some tests, the amplicons from all different inputs reached the same saturation yield; in other tests, the amplicons with different input reached different plateaus, ranging approximately from 100% to 25% of the signal of that with 0.1-ng input (i.e., ≈1–4 μg), while the fluorescent signal with positive DNA input was significantly higher than the Ntr signal, which was at the baseline level. In general, the reaction is suggested to be ended before the Ntr signal started to rise up from the baseline. The yield variation was presumably associated with the denaturing treatment and the Tre concentration applied. Another important phenomenon was that after the amplicon reached its plateau, prolonging the reaction sometimes reduced the yield when the original input was relatively high, which could be the result of 3′ → 5′ exonuclease activity of phi29 DNA polymerase when the dNTPs/primers were exhausted. Therefore, the reaction should be ended at an optimal time.
Fig. 2.
SYBR Green real-time monitoring of the amplification reaction (80 μl) with trace gDNA inputs. The reaction was ended at 26 h. RFU, relative fluorescence unit; Ct, cycle threshold. (A) Time course of amplification. Input was ranged from 100 pg to 0.01 fg of extracted gDNA, intact cells, and Ntr control. The variation among cells a, b, and c may reflect the variation in their actual cell numbers or in preparing the amplification reaction. (B) The log2 input in femtograms showed a linear relationship with the Ct from 0.1 ng to 0.01 fg (eight orders of magnitude). Signal was collected every cycle (15 min).
The time of reaction to produce a certain level of product varied linearly with the log value of input (Fig. 2B). This together with the other analysis (Fig. S2 C and D) revealed a logarithmic amplification process (in term of total yield) before the reaction reagents were exhausted. Importantly, the monitor curve showed that, at short reaction times, the yield from intact cells (a single to a few cells; average 2.8 cells; see below) amplified with a 80-μl reaction procedure was significantly higher (>50-fold) than the expected yield based on their DNA amount (≈17 pg) (Fig. 2A), showing that intact cells are better templates for amplification than the corresponding amount of purified and presumably relatively more fragmented gDNA, and short DNA fragments are well known to be not efficiently amplifiable with phi29 polymerase-based isothermal amplification.
Minimized Locus Bias in Amplicons from Trace Inputs.
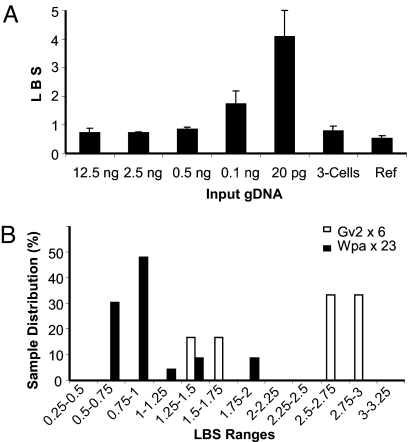
The locus bias (termed locus bias score, LBS; defined as the standard deviation of a panel of eight loci for their locus-by-locus-based qPCR ΔCt with a reference DNA as the control; see details in SI Text) increased as the amount of input DNA dropped (Fig. 3A and Table S2), but this change was not significant when the input was >0.5 ng. However, with 0.1 ng or less input, the LBS rose sharply. When we mildly denatured these trace inputs with the procedure for trace initial DNA based on our optimization tests, the locus representation was partially improved. Use of freshly prepared buffers A and N and reduced denaturing time were very crucial, and a higher concentration of Tre was also important to minimizing the locus bias.
Fig. 3.
LBS of amplicons with different initial DNA inputs. (A) Different amounts of purified gDNA amplified with the general amplification procedure of Wpa (see Table S2 for detailed data). Each sample was repeated twice for qPCR LBS assessment (SI Text), and the mean was taken. (B) Direct amplification from a single to a few cells (2.8 ± 1.9 cells) with Wpa vs. Gv2. In total, 23 and 6 samples were amplified with Wpa and Gv2, respectively.
Nevertheless, the 20-pg and in most cases also the 0.1-ng input-derived amplicon did not show a uniform locus representation. By comparing the ΔCt of each locus between two repetitions of amplification (Table S3), we found that the locus bias for low input (apparently 0.1 ng or less) was not consistent between the two repeats in different loci, rather than locus-specific. This suggested that the locus bias for low inputs was mainly due to random stochastic effects during the sampling of input DNA for amplification. The different concentrations of input DNA were obtained by serial dilution and random sampling from a pool of DNA, during which stochastic effects and perhaps DNA damage or selective loss occurred. Obviously, intact cells would not have such a locus-by-locus bias because of the integrity of a genome of a cell except for certain sites where the intact DNA is randomly discontinuous. Considering that under certain conditions the number of copies for each locus in sampling from the original DNA pool would have approximately Poisson (“sample amount”/“genomic molecular weight”) distribution, where the sample amount is the amount taken for the test, we propose that the amount of input DNA required to obtain a satisfactory locus representation could be proportionally reduced with a smaller genomic size or less complex pool of DNAs.
High Fidelity and Efficiency of WGA from a Single Cell to a Few Cells.
White blood cells with an average of 2.8 ± 1.9 cells per μl (SI Text) were amplified with the general reaction mixture for 16 h. As shown (Fig. 3B), 23 samples of cells were amplified. The amplicons revealed an LBS of 0.92 ± 0.38 in total. However, the LBS of 83% of the samples was 0.77 ± 0.166. For the remaining17% samples, the LBS was 1.63 ± 0.31. Therefore, most of our amplification products resulting from DNA from an average of 2.8 cells gave excellent quality close to the uniformity seen with an input of 2.5 ng of gDNA (equivalent to ≈400 copies of a diploid human genome; Table S2). The remaining 17% samples of cells showed a higher LBS, approximately the LBS of 0.1 ng of gDNA-derived amplicons, which may suggest that these samples were somehow damaged or lost at one point or another (i.e., during cell sorting or other steps).
These results were superior to those we obtained with any other available WGA kits. In comparison, the amplification of six samples of cells with the Gv2 resulted in an LBS of 2.35 ± 0.76 (Fig. 3B). Our early tests (data not shown) with 0.5 ng of extracted gDNA demonstrated that the Gv2 produced the least locus bias among several MDA protocols (Gv2 < Repli-g UltraFast mini < Repli-g 625S), consistent with another report (18). Recent reports further showed that Gv2 gave a better quality of amplicon than GenomiPlex (18, 29). In contrast, previously, at least hundreds to 1,000/1,500 cells were required as starting materials in MDA to obtain reliable SNP and CNV results (13, 18).
Faithful Locus Representation Enabled the Detection of Heterozygous or Small Segmental CNV with Low Amounts of Template.
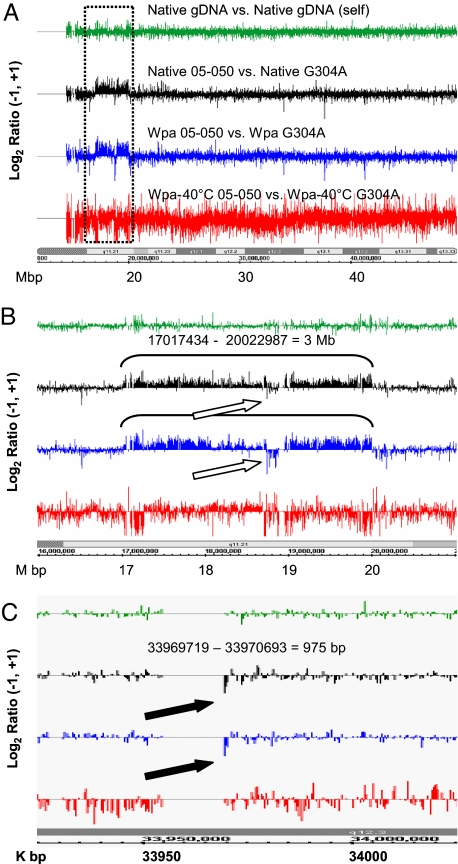
High-resolution comparative genomic hybridization (HR-CGH) with amplicon derived from 5 ng of input gDNA detected all previously known chromosomal segmental aberrations in chromosome 22 in samples from two different probands and was indistinguishable from the HR-CGH result with native gDNA from the same probands (Fig. 4 and Figs. S3 and S4). The break points were also precisely demonstrated. These include a heterozygous genomic segmental duplication [three copies; 3 Mb in size; sample 05-050 (Fig. 4)] and two different heterozygous deletions [one copy; 1.4 Mb and 18 kb, respectively; sample 04-018 (Fig. S4)], all of which are located in or bounded by regions of low copy repeats (LCRs). In addition, a previous known homozygous deletion of 975 bp (in 04-018 and 05-050) was again accurately demonstrated (05-050 data partially degraded shown in Fig. 4C), although sometimes (04-018) the data were a little noisier than with unamplified DNA (Fig. S4D). In contrast, the Wpa-40°C resulted in abundant signal noise and failed in detection of these copy number aberrations (Fig. 4 and Figs. S3 and S4). Impressively, HR-CGH with 0.5 ng of gDNA-derived amplicons via Wpa also clearly detected the known CNVs, although noisier (Fig. S4). The amplicons derived from 0.1 ng of gDNA via Wpa could not unambiguously show CNVs because of higher variability of signals, but the CNVs' patterns were mostly well maintained (Fig. S4 for 04-018). We did also notice some locus imbalance in the amplicon; however, this was minimized, was reproducible when the input was above a certain threshold amount, and could be well compensated for if the same amplified reference sample was applied in parallel as shown above.
Fig. 4.
Amplicon hybridization on a chromosome 22 tiling array reproduced the known chromosomal segment aberrations seen with a native DNA (sample 05-050). The NimbleGen data from arrays were analyzed with NimbleScan 2.3 at CGH DNACopy function option, in which the two channels of signals (Cy3, test sample; Cy5, control) were normalized with Qspline-fit normalization (a minimized nonlinear normalization) and sliding with specified windows. The output gff files [log2 ratio of signals of Cy3 vs. Cy5 (Table S4)] were displayed in Integrated Genome Browser (IGB) (Affymetrix). The “Native gDNA vs. Native gDNA self” (green) is an unamplified 04-018 gDNA against itself. The native DNA set (black) is native 05-050 vs. native G304A. All other Cy3/Cy5 sets are the amplicon of test sample (05-050) vs. amplicon of normal DNA control (G304A). Blue and red refer to the amplification procedure described in this article, Wpa and Wpa-40°C. The labels at the bottom are the chromosome 22 bands and coordinates. (A) The whole chromosome 22 long arm at 2-kb sliding window. The dotted rectangle is the segment zoomed in B. (B) Zoom-in view of the region labeled in A covering a segment with a known heterozygous duplication (3.0 Mb) and analyzed with a 2-kb sliding window. Open arrows indicate reversed signals due to cross-hybridization of the repeat sequence LCR, as analyzed earlier (37). (C) Expansion of a confirmed 975-bp homologous deletion (filled arrows) with a 400-bp sliding window.
Whole-Genome SNP Typing Array Revealed a Minimal Level of Allelic Imbalance and Minimal Sequence Drop-Off in the Chromosome Termini.
As seen in Table S5, the call rates (97.30–99.07%) and accuracy or concordance (>99.85% for the SNPs called in both amplicon and natural reference) for 5-ng-derived amplicons with both Wpa and Gv2 were close to each other and close to native gDNA (call rate of 98.3–99.75%). These call rates were better than a recent report (amplicon 95.9% vs. unamplified 98.5%), in which the early kit Repli-g 625S was applied, and regenotyping was performed when the performance was low and duplicate samples were filtered for the highest call rate (30). The genotyping accuracy of Wpa was actually in the same range as the variation in technical replicates with similar SNP typing arrays (99.87% and 99.88%, replicated Affymetrix array, or between Affymetrix and Illumina arrays) (31). Importantly, the genotyping concordances for amplicons generated from 0.5 ng with Wpa (99.88% and 99.69%) were also close to the technical replicates. In this case, the call rates of Wpa were slightly reduced compared with that with 5 ng of input, but the call rate for the partially degraded sample, 04-018, was modestly improved over Gv2 (92.06% vs. 90.53%).
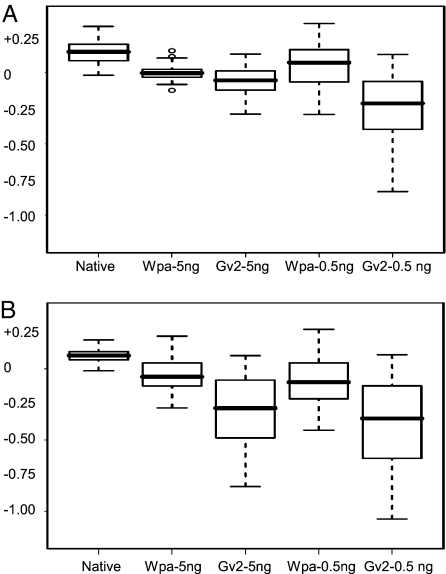
We found that Wpa also showed some amplification nonuniformity among different locations, resulting in some “artificial CNVs” similar to Gv2 (as shown in Fig. S5 and Table S6), with the outputs obtained by taking unamplified gDNAs as their reference. This imbalance, however, was consistent and reproducible for each method but different between Wpa and Gv2. These artificial CNVs can be efficiently cancelled if pairwise amplified test and reference are compared, as observed in CGH results (Fig. 4 and Fig. S4), and also supported by others (32). It is interesting to note that the representation of chromosomal terminal sequences was greatly improved with Wpa compared with Gv2 (Fig. 5), and that some of these regions were significantly underamplified or even lost with Gv2 (Fig. S5 and Tables S6 and S7), as also independently reported recently (32). This occurred especially in the terminal 3–5 Mb and sometimes extended to 10 Mb in many chromosome termini, and was particularly serious when low levels of DNA or degraded DNA was used as input. An analysis for 5 Mb termini is shown (Table S5 calculated all involved SNPs as a cohort; Fig. 5 and Tables S6 and S7 were the result for each chromosome terminus). Importantly, the SNP typing was also greatly improved, as outstandingly exemplified by the amplicons of 0.5-ng input for the partially degraded 04-018, with Wpa vs. Gv2 call rate of 91.9% vs. 84.45% and accuracy of 99.57% vs. 98.62%. The result also showed that the underrepresentation of these terminal regions in Gv2 was not absolutely associated with the distance-to-end but was possibly a sequence-related issue.
Fig. 5.
The mean LRR of 5-Mb termini for all chromosomes applicable, shown in boxplot: minimum, first quartile, median, third quartile and maximum. Outliers are shown as open circles. The log R ratio (LRR) utilizes a collection of multiple native gDNAs as a reference, and it reflects a ratio of amplicon test vs. native reference. This calculation is based on the LRR of all SNPs within 5 Mb of 39 chromosome termini that are represented on the array (Tables S6 and S7). (A) For sample 05-050. (B) For sample 04-018, which was slightly degraded.
Discussion
We demonstrated that an appropriate concentration of Tre combined with other optimized reaction conditions can completely and robustly eliminate the production of endogenous TIP, without minimizing the reaction volume. The product yield increases logarithmically within a certain range (before saturation), and with higher levels of amplification for lower DNA inputs. With Ntr and with the reaction performed with a real-time monitor in the presence of SYBR Green, any exogenous DNA contamination can be easily detected, and the quality and the specificity of the amplicon can be fully controlled. The reaction can be monitored and terminated when there is a satisfactory yield, and exogenous contamination-dependent amplification, if any, is not apparent. In our experience, specific amplification of subnanagra input of DNA or a very small number of intact cells is robust and can be extended to subpicogram or even femtogram DNA if any possible exogenous contamination is firmly excluded.
Importantly, the method described here also substantially improves the locus and allelic representation of the amplicons. When >0.5 ng of purified gDNA or a single to a few intact cells were used as input for amplification, the locus representation of the amplicon was well preserved. The heterozygous segmental copy number changes that occur within or are bounded by stretches of LCRs, which are known to interfere with MDA (4, 5, 18), were clearly detected on genomic tiling arrays. However, this powerful detection may partially be attributed to the pairwise application of amplified test sample and control on the array, because locus bias did exist more or less in the amplicon but was reproducible and consistent when the input was high enough. In contrast, when input was extremely low, the bias was largely a random imbalance from stochastic sampling or random sequences or DNA damage or loss. Importantly, the underamplification or loss of sequences in chromosome termini that occurs with other kits we tested was greatly rescued by this new approach. This is the first WGA method producing amplicons that faithfully detect a heterozygous CNV or subkilobase CNV directly on an oligonucleotide array. In addition, genome-wide SNP typing on array doubly confirmed this locus coverage and allelic representation for 5 ng of input. The 0.5 ng of input-derived amplicons also demonstrated perfect typing accuracy for SNPs, although with a slightly lower call rate. Previously, at least 10, 50, or even 100 ng of good quality DNA was recommended as input for WGA for a faithful amplification (24–26), and the WGA products were only used with PCR fragment arrays (BAC/PAC, etc.), clone arrays, or SNP-CGH (17, 20, 26) that gave much lower resolution in CNV detection.
Tre is a unique, naturally occurring disaccharide that contains glucose molecules bound in an α,α-1,1 linkage. Tre was used to increase the specificity of reverse transcription priming (33). Tre increases the ability of phi29 DNA polymerase to carry out a reaction at 40°C and also to maintain its activity at least for 26 h at 30°C, possibly longer. In addition, Tre was found to decrease the melting temperature, which promotes PCR amplification of GC-rich sequence (34). This may partially explain the rescuing of the loss of the sequences in chromosomal terminal regions with the current procedure compared with Gv2. Furthermore, we propose that Tre, and possibly some other disaccharides, may primarily work as a crowding agent. This is supported by the report that a larger chaperone DNA or a typical molecular crowder, polyethylene glycol 400, reduced the allele bias for subnanograms of DNA (35), and that Tre affected the swelling properties of a polymer, poly(ethylene oxide), in a spectroscopy study (36).
Because of its various attributes—zero background amplification, hypersensitivity, faithful representation of the original starting pool, flexibility in reaction volume and time, and initial DNA volume—the present method for whole DNA pool amplification expedites the use of archived DNA samples, laser-dissected or needle-biopsy samples, and single or a small number of cells for genomic and/or expression profiling (1, 2). The products from this amplification might be applied in massively parallel sequencing, as well as in microarray analysis. Examples of the potential fields of application include whole-genome sequencing of a single microorganism, SNP and CNV analysis, transcriptome analysis, dynamics analysis for timing of DNA replication of different genomic regions, and functionally isolated DNA pools such as ChIP, CpG island methylation, and DNase I-hypersensitive site-selected DNA when the original sample is quantitatively not enough for a desired high-throughput analysis.
Materials and Methods
Procedure for Wpa.
The procedure was a substantially revised phi29 DNA polymerase-based isothermal (30°C) DNA amplification. A critical factor was the employment of Tre [d-(+)-trehalose dehydrate, T5251; Sigma] at a high concentration, which under optimal conditions efficiently suppresses TIP and enables uniform amplification throughout the genome. A SYBR Green real-time monitor was adapted to determine the optimal time for ending the reaction particularly when a trace amount of input DNA is amplified. An optimal sample denature was presented to maximize DNA denaturation while minimizing damage. The total reaction volume was usually 100 μl but could be varied. 9-mer oligonucleotide with random four nucleotides or a random mixture of A and G (recorded as N9 and R9, respectively) and with 2× phosphorothioate modified nucleotides on the 3′-end were applied for amplification or controls.
General procedure for amplification of extracted DNA.
First, 1 μl of freshly prepared buffer A (400 mM KOH, 10 mM EDTA) was placed on the bottom of a precooled 200-μl thin-wall PCR tube, followed by 1 μl of DNA (1–5 ng). After 3 min of denaturation (one more minute was applied for every five more nanograms of DNA applied), 1 μl of buffer N [200 mM HCl, 300 mM Tris·HCl (pH 7.5)] was added and mixed, 5 μl of 1.2 M Tre was immediately added, and the solution was left on ice. Then, a master mixture was made on ice with the following components for one sample (the total volume of each component was multiplied with “Y.Y,” where “Y” was the number of samples to be amplified): 19.8 μl of distilled water (or sufficient volume to make up final 92 μl in total), 10 μl of 10× RXN buffer [500 mM Tris·HCl (pH 7.5), 100 mM NaCl, 100 mM MgCl, 100 mM (NH4)2SO4, 50 mM DTT], a 1.5-μl mixture of 4× dNTPs (each 25 mM), 1.2 μl of 1 nmol/μl N9, 1 μl of 100 ng/μl BSA (NEB), and 57.5 μl of 1.2 M Tre (the volume was adjustable; see below). These components were stirred well and spun down before 1 μl per reaction phi29 DNA polymerase (1 μg/μl; Amersham Pharmacia) was added. A 92-μl aliquot of the master mixture was delivered to each sample, stirred well, spun down again, and incubated at 30°C for 16 h, if not specified otherwise. Finally, the tubes were heated at 70°C for 20 min to stop the reaction. Quantification and purification (see SI Text) were then done. Tre was diluted in distilled water and sterilized with a 0.22-μm filter (Millipore). Aliquots were put at −20°C for long-time storage or at room temperature for 1-week use. There was a wide window of optimal concentration for different preparations of Tre, but the range was somewhat variable (0.54–0.84 M) and therefore was calibrated each time when a new preparation of Tre was made. The non-template-reaction (Ntr) containing N9 and R9, purchased from the Yale DNA Synthesis Laboratory, were separately used as quality control for evaluation of amplification specificity and exogenous contamination.
DNA amplification procedure for trace amounts of DNA or intact cells.
For a <0.5-ng input of extracted gDNA, an 8-fold water dilution of both buffer A and buffer N was used. The denaturation time with diluted buffer A was 8 min. When the sample was <0.1 ng, the time was decreased by ≈1 min for every order of magnitude decrease in the amount of DNA input. For <10 intact cells, 1 μl of cell lysis solution (400 mM KOH, 10 mM EDTA, 100 mM DTT) was added to the 1 μl of cells on ice, mixed gently, and kept on ice for 5 min before 1 μl of buffer N was added. A longer time of lysis was needed for more cells (≈10, 15, and 20 min for 20, 50, and 100 cells). In all cases, 5 μl of 1.2 M Tre was added immediately afterward. The samples then were processed by the general procedure, including the same master mixture, as described above. Alternatively, an 80-μl reaction volume was used for picogram to femtogram starting templates, with the same concentration of Tre, and with the same amount of the other components—thus, their concentrations were increased by 20%. A real-time monitoring procedure for the amplification process was adapted (11). Briefly, 0.1× SYBR Green I (Molecular Probes) was added to the mixture, and the reaction was run on the Bio-Rad qPCR system MyiQ (iQ5) with iCycler. Signals were collected every 15 min. The thermal program was set at 30°C for 16–26 h but was ended when the signal reached a plateau or when the Ntr signal started to rise, if not otherwise specified.
CGH on Chromosome 22 Genomic Tiling Array.
The gDNA samples 05-050 and 04-018 (slightly degraded), for which a series of CNVs were verified (37, 38), and a normalized gDNA control pool from seven male individuals (G304A; Promega) were analyzed with HR-CGH on NimbleGen chromosome 22 oligonucleotide microarrays using sequences from National Center for Biotechnology Information build 35, as described (37). Starting amounts of 5, 0.5, and 0.1 ng, respectively, of purified gDNA were amplified with the Wpa general procedure, or Wpa-40°C that shared all conditions with Wpa procedure except for phi29 added to a 5-min prewarmed reaction mixture and run at 40°C. Four independent amplicons were pooled together to generate sufficient quantity and hybridized to a chromosome 22 array by using the same protocol and the same amount of purified DNA as the native control DNA, and the signal was digitized with the NimbleScan 2.3 software suite, analyzed with its CGH DNACopy function, and displayed with the Integrated Genome Browser from Affymetrix (http://genoviz.sourceforge.net/).
Allele Bias Evaluation on Whole-Genome SNP Genotyping Arrays.
The amplificons of 5 and 0.5 ng of genomic DNA 04-018 and 05-050 were generated with this Wpa and Gv2, and the four replicates of each type were pooled. SNP genotyping of amplified DNAs and their native unamplified gDNA was performed with the HumanHap550-Duo Genotyping BeadChip (Illumina), containing 561,466 SNPs, at the Yale Keck Biotechnology Resource Laboratory according to the Illumina Infinium II Assay protocol.
Supplementary Material
Acknowledgments.
We thank Beverly S. Emanuel for providing genomic DNA samples, Efim Golub for cloning and producing TthSSB mutant protein, and Ruth Halaban and Yasuhiro Nakayama for providing DNA or cells in trial. We thank Drs. Roger Lasken and Haig H. Kazazian for their critical review. This work was supported by National Institute of Health Grants R01 AG23111 and P50 HG02357-01. X.P. was supported in part by the National Cancer Institute Specialized Program of Research Excellence in Skin Cancer Grant 1 P50 CA121974 and the National Institute of Arthritis and Musculoskeletal and Skin Disease Grant 5 P30 AR41942-14. V.S. was supported by the National Institutes of Health core grant P30 DK072442-03.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0808028105/DCSupplemental.
References
- 1.Hutchison CA, III, Venter JC. Single-cell genomics. Nat Biotechnol. 2006;24:657–658. doi: 10.1038/nbt0606-657. [DOI] [PubMed] [Google Scholar]
- 2.Lasken RS. Single-cell genomic sequencing using multiple displacement amplification. Curr Opin Microbiol. 2007;10:510–516. doi: 10.1016/j.mib.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 3.Hughes S, Arneson N, Done S, Squire J. The use of whole genome amplification in the study of human disease. Prog Biophys Mol Biol. 2005;88:173–189. doi: 10.1016/j.pbiomolbio.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 4.Dean FB, et al. Comprehensive human genome amplification using multiple displacement amplification. Proc Natl Acad Sci USA. 2002;99:5261–5266. doi: 10.1073/pnas.082089499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paez JG, et al. Genome coverage and sequence fidelity of phi29 polymerase-based multiple strand displacement whole genome amplification. Nucleic Acids Res. 2004;32:e71. doi: 10.1093/nar/gnh069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hosono S, et al. Unbiased whole-genome amplification directly from clinical samples. Genome Res. 2003;13:954–964. doi: 10.1101/gr.816903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spits C, et al. Whole-genome multiple displacement amplification from single cells. Nat Protoc. 2006;1:1965–1970. doi: 10.1038/nprot.2006.326. [DOI] [PubMed] [Google Scholar]
- 8.Holbrook JF, Stabley D, Sol-Church K. Exploring whole genome amplification as a DNA recovery tool for molecular genetic studies. J Biomol Tech. 2005;16:125–133. [PMC free article] [PubMed] [Google Scholar]
- 9.Hutchison CA, III, Smith HO, Pfannkoch C, Venter JC. Cell-free cloning using phi29 DNA polymerase. Proc Natl Acad Sci USA. 2005;102:17332–17336. doi: 10.1073/pnas.0508809102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spits C, et al. Optimization and evaluation of single-cell whole-genome multiple displacement amplification. Hum Mutat. 2006;27:496–503. doi: 10.1002/humu.20324. [DOI] [PubMed] [Google Scholar]
- 11.Zhang K, et al. Sequencing genomes from single cells by polymerase cloning. Nature Biotechnol. 2006;24:680–686. doi: 10.1038/nbt1214. [DOI] [PubMed] [Google Scholar]
- 12.Dean FB, Nelson JR, Giesler TL, Lasken RS. Rapid amplification of plasmid and phage DNA using Phi 29 DNA polymerase and multiply-primed rolling circle amplification. Genome Res. 2001;11:1095–1099. doi: 10.1101/gr.180501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lage JM, et al. Whole genome analysis of genetic alterations in small DNA samples using hyperbranched strand displacement amplification and array-CGH. Genome Res. 2003;13:294–307. doi: 10.1101/gr.377203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang Z, Zhang X, Deka R, Jin L. Genome amplification of single sperm using multiple displacement amplification. Nucleic Acids Res. 2005;33:e91. doi: 10.1093/nar/gni089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Handyside AH, et al. Isothermal whole genome amplification from single and small numbers of cells: A new era for preimplantation genetic diagnosis of inherited disease. Mol Hum Reprod. 2004;10:767–772. doi: 10.1093/molehr/gah101. [DOI] [PubMed] [Google Scholar]
- 16.Raghunathan A, et al. Genomic DNA amplification from a single bacterium. Appl Environ Microbiol. 2005;71:3342–3347. doi: 10.1128/AEM.71.6.3342-3347.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le Caignec C, et al. Single-cell chromosomal imbalances detection by array CGH. Nucleic Acids Res. 2006;34:e68. doi: 10.1093/nar/gkl336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iwamoto K, et al. Detection of chromosomal structural alterations in single cells by SNP arrays: A systematic survey of amplification bias and optimized workflow. PLoS ONE. 2007;2:e1306. doi: 10.1371/journal.pone.0001306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marcy Y, et al. Nanoliter reactors improve multiple displacement amplification of genomes from single cells. PLoS Genet. 2007;3:1702–1708. doi: 10.1371/journal.pgen.0030155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bredel M, et al. Amplification of whole tumor genomes and gene-by-gene mapping of genomic aberrations from limited sources of fresh-frozen and paraffin-embedded DNA. J Mol Diagn. 2005;7:171–182. doi: 10.1016/S1525-1578(10)60543-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang G, et al. Balanced-PCR amplification allows unbiased identification of genomic copy changes in minute cell and tissue samples. Nucleic Acids Res. 2004;32:e76. doi: 10.1093/nar/gnh070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brukner I, Paquin B, Belouchi M, Labuda D, Krajinovic M. Self-priming arrest by modified random oligonucleotides facilitates the quality control of whole genome amplification. Anal Biochem. 2005;339:345–347. doi: 10.1016/j.ab.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 23.Inoue J, Shigemori Y, Mikawa T. Improvements of rolling circle amplification (RCA) efficiency and accuracy using Thermus thermophilus SSB mutant protein. Nucleic Acids Res. 2006;34:e69. doi: 10.1093/nar/gkl350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bergen AW, Qi Y, Haque KA, Welch RA, Chanock SJ. Effects of DNA mass on multiple displacement whole genome amplification and genotyping performance. BMC Biotechnol. 2005;5:24. doi: 10.1186/1472-6750-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lasken RS, Egholm M. Whole genome amplification: Abundant supplies of DNA from precious samples or clinical specimens. Trends Biotechnol. 2003;21:531–535. doi: 10.1016/j.tibtech.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 26.Arriola E, et al. Evaluation of Phi29-based whole-genome amplification for microarray-based comparative genomic hybridisation. Lab Invest. 2007;87:75–83. doi: 10.1038/labinvest.3700495. [DOI] [PubMed] [Google Scholar]
- 27.Lovmar L, Syvanen AC. Multiple displacement amplification to create a long-lasting source of DNA for genetic studies. Hum Mutat. 2006;27:603–614. doi: 10.1002/humu.20341. [DOI] [PubMed] [Google Scholar]
- 28.Kvist T, Ahring BK, Lasken RS, Westermann P. Specific single-cell isolation and genomic amplification of uncultured microorganisms. Appl Microbiol Biotechnol. 2007;74:926–935. doi: 10.1007/s00253-006-0725-7. [DOI] [PubMed] [Google Scholar]
- 29.Ballantyne KN, van Oorschot RA, Mitchell RJ. Comparison of two whole genome amplification methods for STR genotyping of LCN and degraded DNA samples. Forensic Sci Int. 2007;166:35–41. doi: 10.1016/j.forsciint.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 30.Teo YY, et al. Whole genome-amplified DNA: Insights and imputation. Nat Methods. 2008;5:279–280. doi: 10.1038/nmeth0408-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levy S, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pugh TJ, et al. Impact of whole genome amplification on analysis of copy number variants. Nucleic Acids Res. 2008;36:e80. doi: 10.1093/nar/gkn378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carninci P, et al. Thermostabilization and thermoactivation of thermolabile enzymes by trehalose and its application for the synthesis of full length cDNA. Proc Natl Acad Sci USA. 1998;95:520–524. doi: 10.1073/pnas.95.2.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spiess AN, Mueller N, Ivell R. Trehalose is a potent PCR enhancer: Lowering of DNA melting temperature and thermal stabilization of taq polymerase by the disaccharide trehalose. Clin Chem. 2004;50:1256–1259. doi: 10.1373/clinchem.2004.031336. [DOI] [PubMed] [Google Scholar]
- 35.Ballantyne KN, van Oorschot RA, Mitchell RJ, Koukoulas I. Molecular crowding increases the amplification success of multiple displacement amplification and short tandem repeat genotyping. Anal Biochem. 2006;355:298–303. doi: 10.1016/j.ab.2006.04.039. [DOI] [PubMed] [Google Scholar]
- 36.Magazù S, et al. Experimental simulation of macromolecules in trehalose aqueous solutions: A photon correlation spectroscopy study. J Chem Phys. 1999;111:9086–9092. [Google Scholar]
- 37.Urban AE, et al. High-resolution mapping of DNA copy alterations in human chromosome 22 using high-density tiling oligonucleotide arrays. Proc Natl Acad Sci USA. 2006;103:4534–4539. doi: 10.1073/pnas.0511340103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Korbel JO, et al. Systematic prediction and validation of breakpoints associated with copy-number variants in the human genome. Proc Natl Acad Sci USA. 2007;104:10110–10115. doi: 10.1073/pnas.0703834104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.