Abstract
Neural precursor cells (NPCs) differentiate into neurons, astrocytes, and oligodendrocytes in response to intrinsic and extrinsic changes. Notch signals maintain undifferentiated NPCs, but the mechanisms underlying the neuronal differentiation are largely unknown. We show that SIRT1, an NAD+-dependent histone deacetylase, modulates neuronal differentiation. SIRT1 was found in the cytoplasm of embryonic and adult NPCs and was transiently localized in the nucleus in response to differentiation stimulus. SIRT1 started to translocate into the nucleus within 10 min after the transfer of NPCs into differentiation conditions, stayed in the nucleus, and then gradually retranslocated to the cytoplasm after several hours. The number of neurospheres that generated Tuj1+ neurons was significantly decreased by pharmacological inhibitors of SIRT1, dominant-negative SIRT1 and SIRT1-siRNA, whereas overexpression of SIRT1, but not that of cytoplasm-localized mutant SIRT1, enhanced neuronal differentiation and decreased Hes1 expression. Expression of SIRT1-siRNA impaired neuronal differentiation and migration of NPCs into the cortical plate in the embryonic brain. Nuclear receptor corepressor (N-CoR), which has been reported to bind SIRT1, promoted neuronal differentiation and synergistically increased the number of Tuj1+ neurons with SIRT1, and both bound the Hes1 promoter region in differentiating NPCs. Hes1 transactivation by Notch1 was inhibited by SIRT1 and/or N-CoR. Our study indicated that SIRT1 is a player of repressing Notch1-Hes1 signaling pathway, and its transient translocation into the nucleus may have a role in the differentiation of NPCs.
Keywords: Hes1, N-CoR, neural precursor cell
Self-renewing multipotential stem cells are present in the ventricular zone (VZ) of mammalian embryonic brain (1). During development, neural precursor cells (NPCs) primary undergo extensive self-renewal and then generate only neurons first, followed by the sequential genesis of astrocytes and oligodendrocytes (2). Notch signaling, triggered by the interaction between the Notch receptor and its ligands, increases the number of NPCs and prevents their differentiation into neurons (3). Upon activation, the Notch intracellular domain (Notch-ICD) is released and translocates into the nucleus, where it forms a complex with RBP-J/CSL/CBF1 and induces expression of downstream genes such as Hes1. Hes1, a repressor-type basic helix–loop–helix (bHLH) transcriptional factor, is expressed at high levels in the VZ, and the level decreases as neural differentiation proceeds. Misexpression of Hes1 in the embryonic brain prevents differentiation and maintains NPCs, and conversely, in the Hes1-deficient brain, NPCs prematurely differentiate into neurons (4). Attenuation of the Notch signaling seems to be an important issue for neuronal differentiation, however the mechanisms modulating Notch activity are not clear.
SIRT1 is an NAD+-dependent class III histone deacetylase (HDAC) that deacetylates histones and transcription factors (5–7). It deacetylates and modulates p53 and forkhead transcription factors (FOXOs) thereby promoting cell survival. SIRT1 also participates in cell metabolism (6, 7). It deacetylates and activates the transcriptional coactivator PGC-1α, which induces gluconeogenesis and mitochondrial oxidative phosphorylation, and may contribute to longevity in calorie restriction (6–8). In adipocytes, SIRT1 partners with nuclear receptor corepressor (N-CoR) and silencing mediator of retinoid and thyroid hormone receptors (SMRT) to repress peroxisome proliferator-activated receptor-γ (PPAR-γ) and thereby suppresses fat accumulation (9). N-CoR was initially defined as a regulator of nuclear receptor-mediated transcriptional repression. It has since been shown to interact with HDACs and act as a corepressor for many transcription factors including RBP-J (10). Interestingly, NPCs from N-CoR gene-disrupted mice display spontaneous differentiation into astroglia-like cells (11).
SIRT1 is highly expressed during embryogenesis (12). SIRT1-deficient mice exhibit severe abnormalities, including small body, exencephaly, and retinal and heart defects, and they only infrequently survive postnatally (13, 14). Actually, SIRT1 affects differentiation of adipocytes and muscle cells by inhibiting PPAR-γ (9) and MyoD (15), respectively. We found that SIRT1's activity was regulated by nucleocytoplasmic shuttling and that its subcellular localization changed after differentiation in C2C12 myoblast cells (16).
Here, we found that cytoplasmic SIRT1 in embryonic NPCs was transiently translocated into the nucleus by differentiation conditions, suppressed Hes1 expression and increased the number of Tuj1+ neurons. Our findings indicate that SIRT1 is important for neuronal differentiation and its spatial regulation may be critical for suppressing Notch function.
Results
SIRT1 Expression in Neural Precursor Cells.
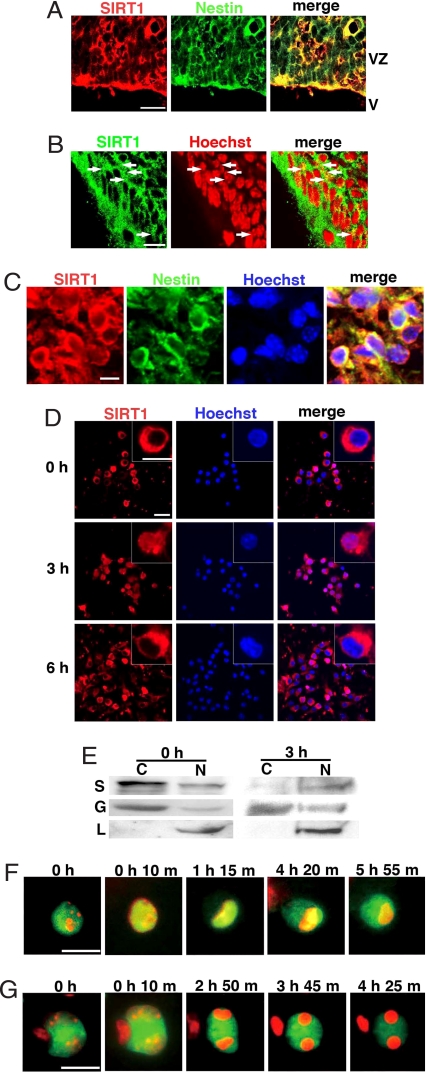
SIRT1 is highly expressed in the mouse embryo (12). We examined its distribution in the embryonic brain [supporting information (SI) Text]. SIRT1 was strongly expressed in the VZ and subventricular zone (SVZ) and almost all of the SIRT1+ cells expressed nestin, a marker of NPCs (Fig. 1A). Although SIRT1 of some cells was found to localize in the nucleus, most cells showed cytoplasmic staining of SIRT1 (Fig. 1B). When NPCs were cultured from striatum of embryonic day (E)14.5 brain, cultured NPCs also expressed SIRT1 in the cytoplasm (Fig. 1C). In the adult brain, SIRT1 immunoreactivity was found in the vicinity of the anterior lateral ventricles, and almost all of the SIRT1+ cells expressed nestin (Fig. S1 A and B). In contrast, the SIRT1 expression was weak in other regions. Nestin-positive cells in the subgranular zone of the hippocampal dentate gyrus showed few expression of SIRT1 (Fig. S1C). NeuN+ neurons in the gray matter weakly expressed SIRT1 in their cytoplasm, but astrocytes and oligodendrocytes were not stained by the SIRT1 antibody (data not shown). When we labeled dividing NPCs with BrdU, BrdU+ cells were labeled by the SIRT1 antibody (Fig. S1D). NPCs in the adult brain are classified into three types; i.e., type B, type C, and type A cells (17, 18). SIRT1 immunoreactivity was detected in the most primitive GFAP+ type B cells, intermediate NG2+/PSA-NCAM− type C-like cells (Fig. S1 E and F) and some of PSA-NCAM+ type A cells (Fig. S1G). SIRT1 was also expressed in S100B+ ependymal cells (Fig. S1 E–G).
Fig. 1.
Nuclear translocation of SIRT1 in NPCs. (A) Immunostaining of E14.5 mouse brain with anti-SIRT1 (red) and anti-nestin (green) antibodies. VZ, ventricular zone; V, ventriculus. (Scale bar: 20 μm.) (B) SIRT1 immunostaining of E16.5 mouse brain. Nuclei are shown in red as a pseudocolor. Some cells express nuclear SIRT1 (arrows). (Scale bar: 50 μm.) (C) Immunostaining of an undifferentiated neurosphere. (Scale bar: 20 μm.) (D) Dissociated neurosphere cells were transferred into differentiation conditions, fixed immediately (0 h), 3 h, and 6 h after the transfer and then immunostained. (Scale bars: 20 μm and 10 μm, Inset.) (E) Cytoplasmic (C) and nuclear (N) fractions were prepared from cells before (0 h) or 3 h after transfer to differentiation conditions and then subjected to Western blot analysis. GAPDH (G) and lamin (L) are cytoplasmic and nuclear markers, respectively. S, SIRT1. (F and G) Time-lapse image analyses of SIRT1-EGFP (F) and SIRT1mtNLS-EGFP (G). Nuclei are shown in red. In G, a cluster of three cells, two of which express SIRT1mtNLS-EGFP, is shown. (Scale bars: 10 μm.)
Transient Nuclear Translocation of SIRT1 by Differentiation Conditions.
SIRT1 in C2C12 cells localizes in the nucleus and translocates into the cytoplasm after differentiation (16). When cultured NPCs were transferred into differentiation conditions and fixed immediately after transfer, SIRT1 was predominantly located in the cytoplasm (Fig. 1D). After a 3-h incubation in differentiation conditions, SIRT1 was predominantly expressed in the nucleus (Fig. 1D). Unexpectedly, the SIRT1 immunoreactivity in many cells again localized in the cytoplasm after a 6-h incubation (Fig. 1D).
The subcellular distribution of SIRT1 was further investigated by Western blot analysis. SIRT1 was detected mainly in the cytoplasmic fraction of the undifferentiated cells (0 h), but dominantly in the nuclear fraction of the cells that had been under differentiation conditions for 3 h (Fig. 1E).
Cultured NPCs were electroporated with SIRT1-fused GFP (SIRT1-EGFP) and monitored by time-lapse microscopy (Fig. 1F and Movie S1). At first, SIRT1-EGFP was mainly expressed in the cytoplasm of a NPC. SIRT1-EGFP started to translocate into the nucleus within 10 min after the transfer into the differentiation conditions, and it was mostly localized to the nucleus by 1 h. SIRT1-EGFP stayed in the nucleus for the next 3 h and then gradually retranslocated to the cytoplasm (Fig. 1F and Movie S1). When SIRT1mtNLS-EGFP, a mutant SIRT1 that consistently localizes to the cytoplasm (16), was expressed in NPCs, it did not show nuclear translocation (Fig. 1G and Movie S2).
Decrease of Neuronal Differentiation by Inhibition of SIRT1.
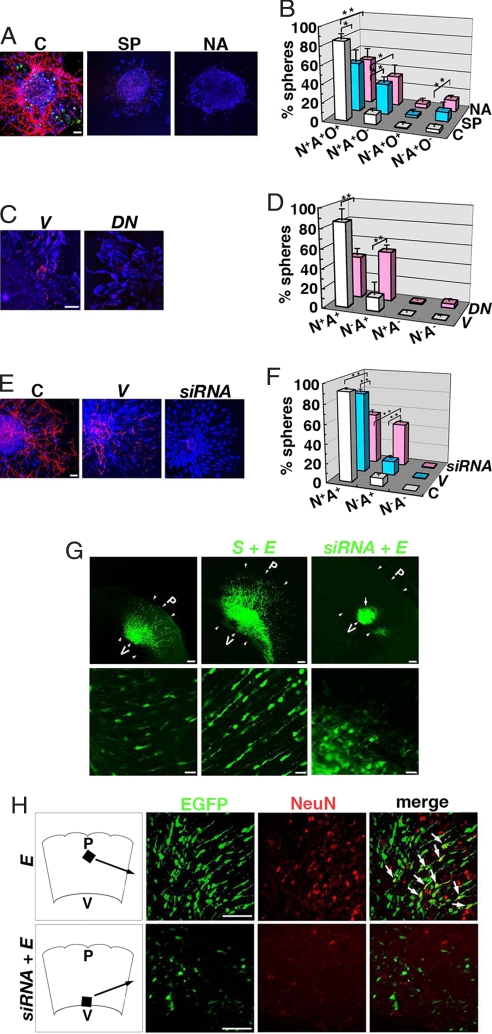
When neurospheres were cultured in the differentiation conditions for 5 days, 84.7 ± 2.8% of the spheres gave rise to Tuj1+, GFAP+, and O4+ cells, indicating that most of spheres differentiate into neurons, astrocytes, and oligodendrocytes (Fig. 2 A and B). In contrast, fewer neurospheres cultured in the presence of the pharmacological SIRT1 inhibitors gave rise to Tuj1+ and O4+ cells after differentiation, and the percentage of spheres that yielded all three types of cells fell to 53.5 ± 7.6% and 49.8 ± 5.9% in the presence of splitomicin and nicotinamide, respectively (Fig. 2 A and B). Sirtinol, another SIRT1 inhibitor, also im-paired the differentiation into neurons and oligodendrocytes (data not shown). At the morphological level, the Tuj1+ neurons that differentiated in the presence of SIRT inhibitors had fewer and shorter neurites than those from control spheres (Fig. 2A; data not shown). The number of spheres containing GFAP+ cells was not altered by the SIRT1 inhibitors: After differentiation, GFAP+ cells were found in nearly 90% of the spheres treated with SIRT1 inhibitors, which was comparable with the proportion of control spheres containing astrocytes (Fig. 2 A and B; data not shown). We could not detect decrease of the number of spheres and apparent apoptosis by SIRT1 inhibitors (data not shown).
Fig. 2.
Inhibition of SIRT1 impairs neuronal differentiation. (A) Neurospheres were cultured in the absence (C) or presence of SIRT1 inhibitors (SP, splitomicin; NA, nicotinamide) and stained with anti-Tuj1 (neurons, red), anti-GFAP (astrocytes, blue), and anti-O4 (oligodendrocytes, green) antibodies. (Scale bar: 50 μm.) (B) The number of neurospheres containing Tuj1+ (N), GFAP+ (A), and/or O4+ cells (O) was counted. *, P < 0.05; **, P < 0.01. (C) NPCs were electroporated with control vector (V) or SIRT1H355Y (DN) and immunostained after differentiation. (Scale bar: 50 μm.) (D and F) The number of spheres containing Tuj1+ (N) and/or GFAP+ (A) cells was counted. **, P < 0.01. (E) NPCs were untreated (C) or infected with lentivirus vector (V) or SIRT1-siRNA lentivirus (siRNA) and immunostained after differentiation. (Scale bar: 50 μm.) (G) E14 brain was electroporated with EGFP (E), SIRT1-EGFP with EGFP (S + E), or SIRT1-siRNA with EGFP (siRNA + E) and examined at E17. Arrowheads indicate tangential thickness of the brain section. P, pia; V, ventriculus. (Scale bars: Upper, 100 μm; Lower, 20 μm.) (H) NeuN immunostaining of E17 brains. Black arrows indicate the localization of the sections. White arrows indicate EGFP+/NeuN+ cells. (Scale bars: 50 μm.)
When SIRT1H355Y, a dominant-negative SIRT1, was expressed in NPCs (Fig. S2), only 42.9 ± 3.5% of the spheres contained Tuj1+ and GFAP+ cells after differentiation (Fig. 2 C and D). On the other hand, 85.9 ± 6.4% of control spheres contained both these cell types. The expression of SIRT1H355Y did not affect the differentiation into GFAP+ cells; the percentage of neurospheres giving rise to GFAP+ cells was 98.0% for mock-transfected and 94.0% for SIRT1H355Y-transfected cells.
The effect of siRNA for SIRT1 on NPCs was also examined. After the differentiation of neurospheres infected with control and SIRT1-siRNA lentivirus (Fig. S3), the percentage of neurospheres giving rise to both Tuj1+ and GFAP+ cells dropped from 83.5 ± 2.4% (control) to 53.8 ± 2.1% (SIRT1-siRNA) (Fig. 2 E and F), which reflected a significant decrease in the number of spheres that generated Tuj1+ neurons. On the other hand, 87.5% of the spheres infected with SIRT1-siRNA lentivirus contained GFAP+ cells, which was comparable to the percentage of GFAP+ spheres infected with the control virus. The Tuj1+ cells differentiated from neurospheres infected with the SIRT1-siRNA lentivirus were rounder and had shorter neurites than those from control neurospheres (Fig. 2E).
To assess the function of SIRT1 in vivo, we used in utero electroporation. SIRT1-EGFP or SIRT1-siRNA was introduced with EGFP plasmid in the E14 brain and then the EGFP-labeled cells were examined at E17. In the control brain, EGFP+ cells had migrated from the VZ to the intermediate zone (IZ) and cortical plate (CP) (Fig. 2G) and some of them were labeled with an anti-NeuN antibody (Fig. 2H). Many EGFP+ cells also migrated from the VZ to the IZ and CP when SIRT1-EGFP was overexpressed. In contrast, when SIRT1-siRNA was coexpressed with EGFP, the normal bipolar orientation of the migrating EGFP+ cells was abolished (Fig. 2G). Instead, round EGFP+ cells clustered in the SVZ and IZ of the SIRT1-siRNA-expressing brains, and these cells had few or short processes. Furthermore, these cells were not stained with an anti-NeuN antibody (Fig. 2H).
Promotion of Neuronal Differentiation by Nuclear SIRT1.
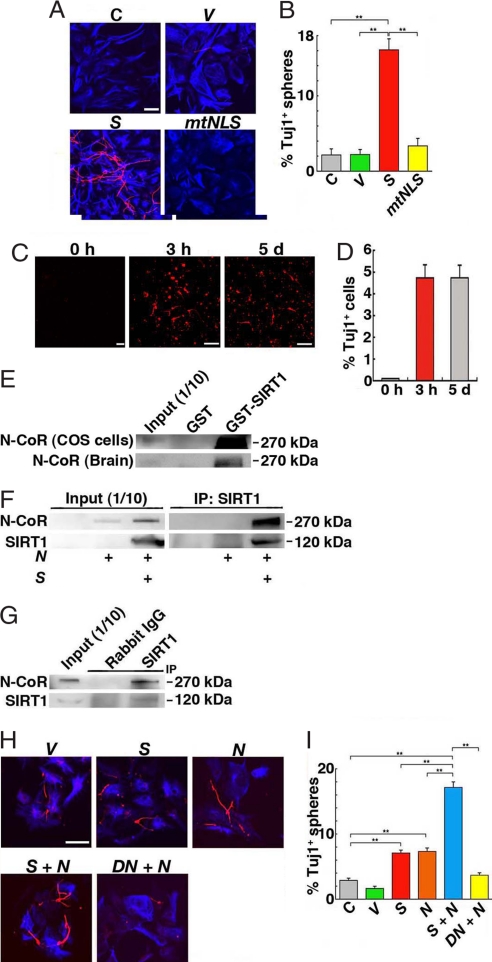
To investigate the effect of SIRT1 overexpression on cultured NPCs, SIRT1 was electroporated in dissociated small spheres, which rarely give rise to Tuj1+ neurons under differentiation conditions. After differentiation, 2.1 ± 0.8% and 2.2 ± 0.7% of mock-transfected and empty vector-transfected spheres contained Tuj1+ cells (Fig. 3 A and B). The overexpression of SIRT1 consistently increased the number of spheres containing neurons to 16.1 ± 1.5% (Fig. 3 A and B). On the other hand, only 3.4 ± 1.0% of spheres that overexpressed the cytoplasm-localized mutant SIRT1, SIRT1mtNLS, generated neurons. These results indicate that SIRT1 promotes neuronal differentiation by its nuclear translocation.
Fig. 3.
SIRT1 and N-CoR promotes neuronal differentiation. (A) NPCs were untreated (C) or transfected with vector (V), SIRT1 (S) or SIRT1mtNLS (mtNLS) and then immunostained with anti-Tuj1 (red) and anti-GFAP (blue) antibodies after differentiation. (Scale bar: 50 μm.) (B) The number of spheres containing Tuj1+ cells was counted. More than 20 visual fields were counted for each experiment. **, P < 0.01. (C) NPCs were cultured in the differentiation conditions for 3 h and then cultured in MHM medium containing EGF and bFGF for 5 days (3 h). For control, NPCs were cultured in the differentiation conditions for 5 days (5 d). Cells were immunostained with an anti-Tuj1 antibody. (Scale bars: 50 μm.) (D) Tuj1+ cells were counted. (E) GST-SIRT1 binds N-CoR from COS cells and fetal brain. (F and G) Immunoprecipitation of N-CoR with anti-SIRT1 antibody. (F) N-CoR in COS cells is coimmunoprecipitated with SIRT1. (G) N-CoR from E16.5 embryonic brain cells cultured in differentiation conditions for 3 h is immunoprecipitated by anti-SIRT1 antibody. (H and I) SIRT1 and N-CoR induce neuronal differentiation. NPCs transfected with the indicated plasmids (V, control vector; S, SIRT1; N, N-CoR; DN, SIRT1H355Y) were analyzed. (Scale bar: 50 μm.) (I) The number of spheres containing Tuj1+ cells was counted. **, P < 0.01.
SIRT1 stayed in the nucleus only for a few hours under the differentiation conditions (Fig. 1 D and F). If SIRT1 has a role in the neuronal differentiation, incubation of NPCs for 3 h in the differentiation conditions would be sufficient to yield Tuj1+ cells. Dissociated NPCs were incubated in differentiation conditions for 3 h, transferred to the medium containing EGF and basic fibroblast growth factor (bFGF) that was used to maintain NPCs and then incubated for 5 days. The number of Tuj1+ cells differentiated from the spheres exposed to the differentiation medium for 3 h was comparable with that of cells cultured in the same medium for 5 days (Fig. 3 C and D). When NPCs were cultured with BrdU, the number of BrdU+ cells was significantly reduced by a 3-h incubation in differentiation conditions (Fig. S4). Thus, 3-h exposure of NPCs to the differentiation conditions might induce cell-cycle exit and be sufficient in the neuronal differentiation.
Leukemia inhibitory factor (LIF) and bone morphogenetic protein (BMP) promote the differentiation of NPCs into GFAP+ cells. We examined the effect of LIF and BMP4. LIF and BMP4 promoted astrocyte differentiation and the number of GFAP+ cells was not reduced by the overexpression of SIRT1-EGFP (Fig. S5). When SIRT1-EGFP was overexpressed, similar number of Tuj1+ cells was differentiated in the presence or absence of LIF and BMP4. However, the ratio of Tuj1+ cells to GFAP+ cells was significantly reduced by these signaling molecules (Fig. S5).
Cooperation of SIRT1 and N-CoR in Neuronal Differentiation.
Nuclear SIRT1 may interact with some transcription factor or repressor and modify gene expression. We focused on the corepressor N-CoR (9). N-CoR mRNA was expressed in both undifferentiated and differentiating NPCs (Fig. S6). Furthermore, export of nuclear N-CoR into the cytoplasm by ciliary neurotrophic factor (CNTF) induces astrocyte differentiation and NPCs of N-CoR knockout mice spontaneously differentiate into astrocyte-like cells (11). N-CoR overexpressed in COS cells and that of fetal brains were bound to the GST fusion protein of SIRT1 (Fig. 3E). Immunoprecipitation experiments also showed that SIRT1 bound N-CoR in COS cells and embryonic brain cells (Fig. 3 F and G).
We examined the effect of overexpression of N-CoR in NPCs. The overexpression of N-CoR alone in dissociated small neurospheres significantly increased the number of spheres containing Tuj1+ cells (7.3 ± 0.5%) to a level comparable to that obtained by the overexpression of SIRT1 alone (7.1 ± 0.4%) (Fig. 3 H and I). The coexpression of SIRT1 and N-CoR further increased the number of neurospheres that yielded Tuj1+ cells to 17.1 ± 0.9%. In contrast, the coexpression of SIRT1H355Y with N-CoR significantly reduced the number of spheres that gave rise to neurons, to only 4% (Fig. 3 H and I). These results suggest that N-CoR and SIRT1 cooperatively promote neuronal differentiation.
Repression of Hes1 by SIRT1 and N-CoR.
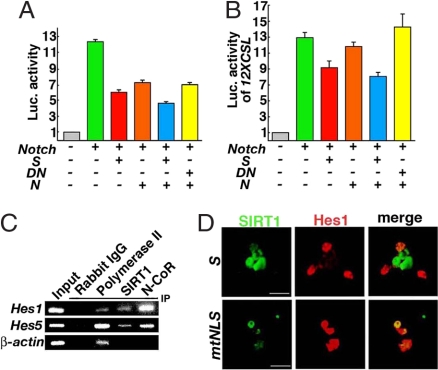
N-CoR represses the transactivation of Hes1 (10). Hes1 mRNA expression in NPCs was significantly reduced after the differentiation (Fig. S7). Therefore, we investigated whether SIRT1 and N-CoR might function together to repress Hes1 transcription. Notch1-ICD induced Hes1 promoter activity in HEK293 cells (Fig. 4A). The coexpression of either SIRT1 or N-CoR with Notch1-ICD significantly repressed the Hes1 promoter activity, although Renilla luciferase, an internal control, was also suppressed by SIRT1 and N-CoR. The coexpression of SIRT1 and N-CoR with Notch1-ICD further repressed the promoter activity (Fig. 4A). The coexpression of SIRT1H355Y with N-CoR resulted in a transcription level similar to that seen with SIRT1 alone or N-CoR alone. Repression of Notch-mediated transcriptional activation by SIRT1 and N-CoR was further investigated by using 12XCSL promoter, which responds only to RBP-J-mediated activation by Notch1-ICD (19). Induction of 12XCSL luciferase activity by Notch1-ICD was repressed by SIRT1 (Fig. 4B). Although N-CoR weakly suppressed 12XCSL luciferase, coexpression of SIRT1 with N-CoR further decreased 12XCSL activity induced by Notch1-ICD. The coexpression of dominant-negative SIRT1H355Y and N-CoR with Notch1-ICD resulted in a transcription level similar to that of Notch1-ICD alone (Fig. 4B).
Fig. 4.
Suppression of Hes1 by SIRT1. (A) Effects of SIRT1 (S), N-CoR (N), and SIRT1H355Y (DN) on the Hes1 promoter activity induced by Notch-ICD (Notch). (B) Effects of SIRT1 (S), N-CoR (N), and SIRT1H355Y (DN) on the 12XCSL1 promoter activity induced by Notch-ICD (Notch). (C) Binding of SIRT1 and N-CoR on the Hes1 and Hes5 promoter regions in differentiating NPCs. (D) SIRT1 suppresses Hes1 expression. NPCs electroporated with SIRT1-EGFP (S) or SIRT1mtNLS-EGFP (mtNLS) were cultured for 24 h in differentiation conditions and then immunostained with an anti-Hes1 antibody. (Scale bars: 20 μm.)
A ChIP assay was used to find whether SIRT1 and N-CoR bound the Hes1 gene in NPCs. SIRT1 and N-CoR both were detected in the promoter region of the Hes1 gene of NPCs cultured in differentiation conditions for 24 h (Fig. 4C). Furthermore, when SIRT1-EGFP was overexpressed in cultured NPCs, Hes1 immunoreactivity was reduced (Fig. 4D). In contrast, the forced expression of SIRT1mtNLS-EGFP, which consistently resides in the cytoplasm, failed to suppress Hes1 expression (Fig. 4D). Hes5 is also a critical mediator of Notch and RBP-J. The promoter region of the Hes5 gene bound SIRT1 and N-CoR (Fig. 4C). Similar to Hes1, Hes5 expression was also reduced in the cells overexpressed with SIRT1-EGFP (Fig. S8).
Discussion
The Notch signal and the repressor-type bHLH transcriptional factors maintain stem cells and prevent neuronal differentiation (3, 4). Inhibitory bHLH transcription factors repress activator-type bHLH genes such as Mash, Math, and Neurogenin (4). When the expression levels of inhibitory bHLH transcription factors are reduced, neuronal differentiation commences spontaneously (3, 4). In this study, we demonstrated that SIRT1 transiently translocated into the nucleus under differentiation conditions. We found that some cells in the embryonic brain expressed SIRT1 in the nucleus, which might indicate the translocation of SIRT1 in vivo (Fig. 1B). Our results indicated that SIRT1 repressed Hes1 and Hes5 expression by directly repressing RBP-J activity promoted by Notch1-ICD. Knockdown of RBP-J promotes the conversion of neural stem cells into intermediate neural progenitors (INPs) that generate mostly Tuj1+ neurons after withdrawal of bFGF (20). Because INPs show attenuated RBP-J signaling, transient translocation of SIRT1 may have a role on conversion of NPCs into INPs.
In Drosophila, the unequal distribution of Numb affects neural cell fate (21). Numb segregates preferentially into the neuronal daughter cell during asymmetric divisions where it inhibits Notch signaling, although the function of Numb homologs in mammalian NPCs is still under investigation (22, 23). If an extracellular signal recruits SIRT1 into the nucleus, it is a candidate for a non-cell-autonomous signal in the neural differentiation. When SIRT1 was inhibited, differentiated Tuj1+ neurons often had fewer or shorter neurites than those in control spheres (Fig. 2 A, C, and E). NPCs expressing SIRT1-siRNA were round and had short or no neurites in the embryonic mouse brain (Fig. 2G). Neurite outgrowth is inhibited by the overexpression of Notch and promoted by the expression of Numb (24). The inhibition of SIRT1 may increase Notch activity in Tuj1+ neurons, thereby altering the neuronal morphology. However, in vivo genetic manipulation of Notch1 induced dendritic arborization and branching of hippocampal neurons (25). Thus, additional function of SIRT1 on neurite outgrowth may exist.
Promotion of neuronal differentiation by the overexpression of SIRT1-EGFP was still found in the presence of LIF and BMP in cultured NPCs. However, the ratio of the number of Tuj1+ cells to that of GFAP+ cells reduced by LIF plus BMP4 (Fig. S5). Further study is necessary to elucidate the functional role of SIRT1 on the signaling cascade of astrocyte differentiation.
N-CoR was shown to repress astrocyte differentiation (11). In the present study, N-CoR interacts with SIRT1, inhibited Hes1 transactivation (Fig. 4) and promoted neuronal differentiation (Fig. 3 E and F). These results indicate that N-CoR has an important role on neuronal differentiation. CNTF recruits N-CoR into the cytoplasm by inducing phosphorylation of N-CoR (11). SIRT1 may translocate into the cytoplasm with N-CoR by CNTF.
SIRT1 was previously reported to inhibit the differentiation of cells (9, 15). Class I and class II HDAC family inhibited or promoted differentiation in a tissue-specific manner. Valproic acid, an inhibitor of class I and class II HDACs, inhibits the differentiation of preadipocyte cells (26), whereas it promotes the neuronal differentiation of adult hippocampal neural progenitors (27). HDAC1 represses MyoD and thereby inhibits myogenesis (28), but it induces differentiation of NPCs during zebrafish neurogenesis (29).
Recently, Prozorovski et al. (30) showed that oxidative conditions repressed Mash1 expression in neonates and cultured NPCs from E17.5 embryos and indicated that the repression of Mash1 was mediated by the complex of Hes1 and SIRT1. They suggested that SIRT1 participated in oxidation-mediated suppression of neurogenesis. Most of Mash1+ cells in the neonate SVZ differentiate into oligodendrocytes and olfactory interneurons (31), which have not been examined in our experiments. Although cell lineages of Mash1+ cells were not determined by Prozorovski et al., SIRT1 might play a distinct role on differentiation under specific conditions.
Methods
Animals.
The Animal Welfare Guidelines of Sapporo Medical University were followed in all of the studies and experiments using mice. ddY and ICR mice were obtained from Nihon SLC.
Neurosphere.
Neurospheres were cultured in media hormone mix (MHM) medium with EGF (20 ng/ml) and bFGF (20 ng/ml) (32). For differentiation, cells were plated onto poly-l-ornithine-coated microcoverslips and incubated for 5 days in growth factor-free MHM medium containing 1% FBS (differentiation conditions).
Nucleofection.
Dissociated NPCs were suspended in 100 μl of mouse neural stem cell nucleofector solution (Amaxa) with plasmid DNA (5 μg) and electroporated.
In Vivo Electroporation.
The methods for in utero electroporation were described previously (33). Briefly, pregnant mice were deeply anesthetized on E14, and the uterine horns were exposed. Plasmid DNA was injected with pEGFP-N1 (BD Biosciences) to each embryonic brain in the uterus and electroporated with a square-pulse electroporator (Nepa Gene). Electronic pulses (40 V; 50-msec duration) were applied five times at intervals of 950 msec. The uterine horns were then placed back into the abdominal cavity to continue development. Embryos were examined at E17.
ChIP Assay.
The EZ ChIP kit (UBI) was used. Sheared DNA samples prepared from NPCs cultured under differentiation conditions for 24 h were immunoprecipitated and were analyzed by PCR.
Supplementary Material
Acknowledgments.
We thank T. Seki, S. Ishii, R. M. Evans, J. Milbrandt, D. L. Turner, and U. Lendahl for kindly providing materials. We also thank T. Bando and S. Takeuchi for excellent technical assistance in the in utero electroporation and siRNA experiments, respectively. We are grateful to the Nikon Imaging Center at Hokkaido University for their help with the time-lapse study. This work was supported in part by Grants-in-Aid for Scientific Research 13035038 and 15659062 and a National Project “Knowledge Cluster Initiative” (2nd stage, “Sapporo Biocluster Bio-S”) from the Ministry of Education, Culture, Sports, Science and Technology of Japan and by grants from The Special Fund for Medical Research from Sapporo Medical University, the Sapporo Medical University Foundation for Promotion of Medical Science, and Northern Advancement Center for Science and Technology.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0800612105/DCSupplemental.
References
- 1.Davis AA, Temple S. A self-renewing multipotential stem cell in embryonic rat cerebral cortex. Nature. 1994;372:263–266. doi: 10.1038/372263a0. [DOI] [PubMed] [Google Scholar]
- 2.Miller FD, Gauthier AS. Timing is everything: Making neurons versus glia in the developing cortex. Neuron. 2007;54:357–369. doi: 10.1016/j.neuron.2007.04.019. [DOI] [PubMed] [Google Scholar]
- 3.Louvi A, Artavanis-Tsakonas S. Notch signalling in vertebrate neural development. Nat Rev Neurosci. 2006;7:93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- 4.Kageyama R, Ohtsuka T, Kobayashi T. The Hes gene family: Repressors and oscillators that orchestrate embryogenesis. Development. 2007;134:1243–1251. doi: 10.1242/dev.000786. [DOI] [PubMed] [Google Scholar]
- 5.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 6.Haigis MC, Guarente LP. Mammalian sirtuins—Emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20:2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- 7.Longo VD, Kennedy BK. Sirtuins in aging and age-related disease. Cell. 2006;126:257–268. doi: 10.1016/j.cell.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 8.Rodgers JT, et al. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 9.Picard F, et al. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kao HY, et al. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev. 1998;12:2269–2277. doi: 10.1101/gad.12.15.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hermanson O, Jepsen K, Rosenfeld MG. N-CoR controls differentiation of neural stem cells into astrocytes. Nature. 2002;419:934–939. doi: 10.1038/nature01156. [DOI] [PubMed] [Google Scholar]
- 12.Sakamoto J, Miura T, Shimamoto K, Horio Y. Predominant expression of Sir2alpha, an NAD-dependent histone deacetylase, in the embryonic mouse heart and brain. FEBS Lett. 2004;556:281–286. doi: 10.1016/s0014-5793(03)01444-3. [DOI] [PubMed] [Google Scholar]
- 13.Cheng HL, et al. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci USA. 2003;100:10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McBurney MW, et al. The mammalian SIR2alpha protein has a role in embryogenesis and gametogenesis. Mol Cell Biol. 2003;23:38–54. doi: 10.1128/MCB.23.1.38-54.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fulco M, et al. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol Cell. 2003;12:51–62. doi: 10.1016/s1097-2765(03)00226-0. [DOI] [PubMed] [Google Scholar]
- 16.Tanno M, Sakamoto J, Miura T, Shimamoto K, Horio Y. Nucleocytoplasmic shuttling of the NAD+-dependent histone deacetylase SIRT1. J Biol Chem. 2007;282:6823–6832. doi: 10.1074/jbc.M609554200. [DOI] [PubMed] [Google Scholar]
- 17.Aguirre AA, Chittajallu R, Belachew S, Gallo V. NG2-expressing cells in the subventricular zone are type C-like cells and contribute to interneuron generation in the postnatal hippocampus. J Cell Biol. 2004;165:575–589. doi: 10.1083/jcb.200311141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seki T, Arai Y. Highly polysialylated neural cell adhesion molecule (NCAM-H) is expressed by newly generated granule cells in the dentate gyrus of the adult rat. J Neurosci. 1993;13:2351–2358. doi: 10.1523/JNEUROSCI.13-06-02351.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kato H, et al. Involvement of RBP-J in biological functions of mouse Notch1 and its derivatives. Development. 1997;124:4133–4141. doi: 10.1242/dev.124.20.4133. [DOI] [PubMed] [Google Scholar]
- 20.Mizutani K, Yoon K, Dang L, Tokunaga A, Gaiano N. Differential Notch signalling distinguishes neural stem cells from intermediate progenitors. Nature. 2007;449:351–355. doi: 10.1038/nature06090. [DOI] [PubMed] [Google Scholar]
- 21.Rhyu MS, Jan LY, Jan YN. Asymmetric distribution of numb protein during division of the sensory organ precursor cell confers distinct fates to daughter cells. Cell. 1994;76:477–491. doi: 10.1016/0092-8674(94)90112-0. [DOI] [PubMed] [Google Scholar]
- 22.Yoon K, Gaiano N. Notch signaling in the mammalian central nervous system: Insights from mouse mutants. Nat Neurosci. 2005;8:709–715. doi: 10.1038/nn1475. [DOI] [PubMed] [Google Scholar]
- 23.Knoblich JA. Mechanisms of asymmetric stem cell division. Cell. 2008;132:583–597. doi: 10.1016/j.cell.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 24.Sestan N, Artavanis-Tsakonas S, Rakic P. Contact-dependent inhibition of cortical neurite growth mediated by notch signaling. Science. 1999;286:741–746. doi: 10.1126/science.286.5440.741. [DOI] [PubMed] [Google Scholar]
- 25.Breunig JJ, Silbereis J, Vaccarino FM, Sestan N, Rakic P. Notch regulates cell fate and dendrite morphology of newborn neurons in the postnatal dentate gyrus. Proc Natl Acad Sci USA. 2007;104:20558–20563. doi: 10.1073/pnas.0710156104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lagace DC, Nachtigal MW. Inhibition of histone deacetylase activity by valproic acid blocks adipogenesis. J Biol Chem. 2004;279:18851–18860. doi: 10.1074/jbc.M312795200. [DOI] [PubMed] [Google Scholar]
- 27.Hsieh J, Nakashima K, Kuwabara T, Mejia E, Gage FH. Histone deacetylase inhibition-mediated neuronal differentiation of multipotent adult neural progenitor cells. Proc Natl Acad Sci USA. 2004;101:16659–16664. doi: 10.1073/pnas.0407643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mal A, Sturniolo M, Schiltz RL, Ghosh MK, Harter ML. A role for histone deacetylase HDAC1 in modulating the transcriptional activity of MyoD: Inhibition of the myogenic program. EMBO J. 2001;20:1739–1753. doi: 10.1093/emboj/20.7.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cunliffe VT. Histone deacetylase 1 is required to repress Notch target gene expression during zebrafish neurogenesis and to maintain the production of motoneurones in response to hedgehog signalling. Development. 2004;131:2983–2995. doi: 10.1242/dev.01166. [DOI] [PubMed] [Google Scholar]
- 30.Prozorovski T, et al. Sirt1 contributes critically to the redox-dependent fate of neural progenitors. Nat Cell Biol. 2008;10:385–394. doi: 10.1038/ncb1700. [DOI] [PubMed] [Google Scholar]
- 31.Parras CM, et al. Mash1 specifies neurons and oligodendrocytes in the postnatal brain. EMBO J. 2004;23:4495–4505. doi: 10.1038/sj.emboj.7600447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shimazaki T, Shingo T, Weiss S. The ciliary neurotrophic factor/leukemia inhibitory factor/gp130 receptor complex operates in the maintenance of mammalian forebrain neural stem cells. J Neurosci. 2001;21:7642–7653. doi: 10.1523/JNEUROSCI.21-19-07642.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagano T, Morikubo S, Sato M. Filamin A and FILIP (Filamin A-Interacting Protein) regulate cell polarity and motility in neocortical subventricular and intermediate zones during radial migration. J Neurosci. 2004;24:9648–9657. doi: 10.1523/JNEUROSCI.2363-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.