Abstract
Egress of lipoprotein-derived cholesterol from lysosomes requires two lysosomal proteins, polytopic membrane-bound Niemann–Pick C1 (NPC1) and soluble Niemann–Pick C2 (NPC2). The reason for this dual requirement is unknown. Previously, we showed that the soluble luminal N-terminal domain (NTD) of NPC1 (amino acids 25–264) binds cholesterol. This NTD is designated NPC1(NTD). We and others showed that soluble NPC2 also binds cholesterol. Here, we establish an in vitro assay to measure transfer of [3H]cholesterol between these two proteins and phosphatidylcholine liposomes. Whereas NPC2 rapidly donates or accepts cholesterol from liposomes, NPC1(NTD) acts much more slowly. Bidirectional transfer of cholesterol between NPC1(NTD) and liposomes is accelerated >100-fold by NPC2. A naturally occurring human mutant of NPC2 (Pro120Ser) fails to bind cholesterol and fails to stimulate cholesterol transfer from NPC1(NTD) to liposomes. NPC2 may be essential to deliver or remove cholesterol from NPC1, an interaction that links both proteins to the cholesterol egress process from lysosomes. These findings may explain how mutations in either protein can produce a similar clinical phenotype.
Keywords: cholesterol trafficking, kinetics of cholesterol binding, Niemann–Pick C disease
Although much has been learned about the regulation of cellular cholesterol metabolism, a major question remains: How does lipoprotein-derived cholesterol transfer from its entry point in endosomes/lysosomes to the endoplasmic reticulum (ER), where it performs regulatory functions, or to the plasma membrane, where it plays a crucial structural role? A major source of cellular cholesterol comes from plasma low-density lipoproteins (LDL), which enter cells by receptor-mediated endocytosis after binding to cell-surface LDL receptors (1). After internalization in coated pits and vesicles, LDL particles are delivered to endosomes and lysosomes. On average, each LDL particle contains 1,500 molecules of cholesteryl esters. The esters are hydrolyzed in lysosomes by lysosomal acid lipase (2), and the resulting free cholesterol somehow leaves the lysosome and arrives at the ER, plasma membrane, and other membranes.
A major clue to the mechanism of cholesterol exit from lysosomes was supplied by discoveries in cells from patients with autosomal recessive Niemann–Pick Type C (NPC) disease (3). In affected individuals, unesterified cholesterol, sphingomyelin, and other lipids accumulate in endosomes and lysosomes of many organs, including the brain. Lysosomal cholesterol accumulation can be reproduced in patients' cells in vitro by incubating the cells with LDL (4, 5).
The defect in NPC disease has been traced to mutations in either of two genes, both of whose products are required for cholesterol export from lysosomes. One gene, NPC1, encodes a polytopic membrane protein of 1,278 aa that is located in membranes of late endosomes and lysosomes (6). The second gene, NPC2, encodes a soluble protein of 131 aa that is concentrated in lysosomes and is also excreted in seminal fluid and milk (7). Homozygous loss-of-function mutations in either gene produce the same NPC phenotype, suggesting that both genes function in the same pathway for lysosomal cholesterol export (4).
Of the two NPC proteins, NPC2 is the more well studied. NPC2 has been shown to bind cholesterol and derivatives such as cholesterol sulfate (8–10). X-ray crystallography reveals that cholesterol sulfate binds in such a manner that the iso-octyl side chain of the sterol is deeply buried in a hydrophobic pocket and the sulfate on the 3 position of the A ring is exposed to solvent (11). Introduction of a hydrophilic moiety, such as a hydroxyl group, on the cholesterol side chain prevents binding (10). On the other hand, changing the configuration of the 3-hydroxyl group, as with cholesterol sulfate or epicholesterol, does not reduce binding (10). Because of its ability to bind cholesterol, NPC2 can transfer cholesterol from one liposome to another (12). This transfer is accelerated in the presence of anionic phospholipids such as bis(monooleoylglycero)phosphate (BMP) and phosphatidyl inositol, which are found in lysosomes (12, 13).
Recently, the NPC1 protein has come under study as a result of its purification from rabbit liver membranes (14). The 1,254 aa of the mature protein (after removal of its signal peptide) are organized into 13 putative membrane-spanning helices separated by 12 cytosolic or luminal loops (15). A noteworthy feature is the presence of three large luminal domains. The first luminal domain consists of the N-terminal 240 aa (residues 25–264) whose N terminus lies free in the lumen after cleavage by signal peptidase. We designate this sequence as the N-terminal domain (NTD), i.e., NPC1(NTD), which is the most highly conserved sequence in the full-length protein (6). The second and third luminal domains are loops that connect transmembrane helices 2/3 and 8/9, respectively. Full-length NPC1 was shown to bind cholesterol with saturation kinetics (14), and the binding site was localized to the NPC1(NTD) (10). NPC1(NTD) was prepared as a soluble secreted protein by transfecting cells with a cDNA encoding the signal sequence (amino acids 1–24), followed by amino acids 25–264, followed by an epitope tag (10). In direct contrast to the findings with NPC2, sterol binding to NPC1(NTD) was not blocked by addition of hydroxyl groups to the 24, 25, or 27 positions on the iso-octyl side chain (10). Indeed, these modifications increased the apparent affinity for the protein. On the other hand, binding was abolished when the 3-hydroxyl was switched from the β to the α orientation. These findings led to the suggestion that NPC1(NTD) binds primarily to the A ring, whereas NPC2 binds to the opposite end of the sterol, namely the iso-octyl side chain (10).
The current study addresses the question of how NPC1(NTD) and NPC2 interact in the transfer of cholesterol. In agreement with previous findings (12, 13), we show that the on-rate and off-rate of cholesterol from NPC2 is rapid. In sharp contrast, the on- and off-rates of cholesterol from NPC1(NTD) are slow, even in the presence of liposomal membranes. NPC2 accelerates both the binding and release of cholesterol by NPC1(NTD). These findings suggest that NPC2 functions to transfer cholesterol to and/or from NPC1, thereby explaining the requirement for both proteins in the exit of LDL-cholesterol from lysosomes.
Results
The current studies take advantage of the fact that NPC1(NTD) as well as NPC2 are water-soluble proteins, and therefore transfer assays can be carried out in the absence or presence of very low concentrations of detergents, i.e., 0.004% Nonidet P-40. For this purpose, we isolated recombinant NPC1(NTD) and NPC2 from the culture medium of CHO cells that were transfected with plasmids encoding the proteins with or without C-terminal His or FLAG tags. After purification as described in Methods, NPC1(NTD) gave a single diffuse Coomassie-stained band on SDS/PAGE [supporting information (SI) Fig. S1]. NPC2 gave multiple bands, owing to differing degrees of N-linked glycosylation (10, 16).
To measure cholesterol binding, we incubated the His-tagged proteins with [3H]cholesterol at either 4°C or 37°C in the presence of 0.004% Nonidet P-40. Protein-bound [3H]cholesterol was isolated by nickel chromatography and quantitated by scintillation counting (Fig. 1). To measure dissociation rates, we first incubated the proteins with [3H]cholesterol at 4°C and then isolated the cholesterol–protein complexes by gel filtration as described in Methods. The proteins were then bound to nickel agarose beads, and dissociation was measured by release of [3H]cholesterol into the supernatant. The rates of association and dissociation of [3H]cholesterol to and from NPC1(NTD) were strongly influenced by temperature (Fig. 1 A and C). At 4°C, binding was extremely slow (Fig. 1A), and dissociation was so slow as to be unmeasurable, even after 2 h (Fig. 1C). Both processes were accelerated dramatically at 37°C. In comparison with NPC1(NTD), NPC2 bound and released [3H]cholesterol relatively rapidly at 4°C as well as at 37°C (Fig. 1 B and D). To measure saturation binding at equilibrium for NPC1(NTD), we carried out incubations for 45 min at 37°C and for 20 h at 4°C (Fig. 1E). The calculated Kd for [3H]cholesterol binding was 50 nM at 4°C and 90 nM at 37°C; maximal binding was slightly higher at 37°C. For NPC2, saturation binding at equilibrium was done for 45 min at both 4°C and 37°C (Fig. 1F). The calculated Kd was 90 nM at 4°C and 130 nM at 37°C. The binding of [3H]cholesterol to both proteins was unaffected when the pH was varied from 5.5 to 7.4 at either 4°C or 37°C (data not shown).
Fig. 1.
Kinetics of [3H]cholesterol binding to purified NPC proteins. (A and B) Time course at different temperatures. Each reaction, in a final volume of 80 μl of buffer B (pH 6.5) with 0.004% Nonidet P-40, contained 4 pmol of NPC1(NTD)-LVPRGS-His-8-FLAG (A) or 8 pmol of NPC2-His-10 (B) and 100 nM [3H]cholesterol (132 × 103 dpm/pmol). After incubation for indicated time at 4°C (○) or 37°C (●), the amount of bound [3H]cholesterol was measured with the Ni-NTA-agarose binding assay. Each value is the average of duplicate assays and represents total binding after subtraction of a blank value (0.01–0.02 pmol per tube). (C and D) Dissociation of previously bound [3H]cholesterol from NPC proteins at different temperatures. [3H]Cholesterol complexed to NPC1(NTD)-LVPRGS-His-8-FLAG (C) or NPC2-His-10 (D) at 4°C was isolated by gel filtration. Fractions containing NPC1 or NPC2 complexed to [3H]cholesterol were pooled (3 ml), diluted 7-fold with buffer B (pH 6.5) containing 0.004% Nonidet P-40 and 10 μM unlabeled cholesterol, and incubated at 4°C (○) or 37°C (●). At the indicated time, a 1-ml-aliquot of the pooled 21-ml sample was transferred to a tube containing 600 μl of Ni-NTA-agarose beads. After incubation for 3 min at 4°C, the beads were centrifuged at 600g for 1 min, after which the supernatant was assayed for radioactivity. Each value is the average of duplicate assays and represents the percentage of [3H]cholesterol remaining bound to the beads relative to zero-time value. The “100% initial binding” values at zero time for NPC1(NTD) and NPC2 were 1.6 and 0.24 pmol per tube, respectively. (E and F) Saturation curves for equilibrium binding at different temperatures. This experiment was carried out as in A and B except that [3H]cholesterol concentration varied as indicated, and incubation time at 4°C was 20 h for NPC1(NTD) and 45 min for NPC2 and at 37°C, 45 min for both proteins.
Next, we conducted protein-to-protein cholesterol transfer assays in which [3H]cholesterol was transferred from a donor NPC protein that lacks a His tag to an acceptor NPC protein that possesses a His tag. To prepare the donors, we incubated NPC1(NTD) or NPC2 with [3H]cholesterol at 4°C and isolated each sterol-protein complex by gel filtration (Fig. S2). The transfer reactions were conducted either at 4°C or at 37°C (Fig. 2). After incubation, the reaction mixture was subjected to nickel agarose chromatography, and the amount of [3H]cholesterol transferred to the indicated His-tagged NPC protein was measured by scintillation counting of the imidazole-eluted fraction. No donor protein was detected in the eluted fraction as determined by immunoblot analysis (data not shown). At 4°C, untagged NPC1(NTD) rapidly transferred [3H]cholesterol to His-tagged NPC2 (Fig. 2A, filled circles). The measured values are probably an underestimate of true transfer because of the relatively long time required to process the sample, i.e., ≈8–10 min. Remarkably, at 4°C, untagged NPC1(NTD) did not transfer [3H]cholesterol to His-tagged NPC1(NTD) (Fig. 2A, open circles). On the other hand, even at 4°C, untagged NPC2 rapidly transferred [3H]cholesterol to tagged versions of both NPC1(NTD) and NPC2 (Fig. 2B). The differences between NPC1(NTD) and NPC2 persisted when the concentrations of the acceptor protein were varied (Fig. 2 E and F). At 4°C, transfer of [3H]cholesterol from NPC1(NTD) to NPC2 showed a maximum at pH 5.5, which reflects the pH of endosomes/lysosomes (Fig. 2G, filled circles). NPC1(NTD) did not transfer to itself at any pH (Fig. 2G, open circles). When NPC2 was the donor, the transfer rates were relatively constant over the pH range of 5.5 to 8.5 (Fig. 2H).
Fig. 2.
Transfer of [3H]cholesterol from donor NPC protein to acceptor NPC protein. Schematic diagrams of the two transfer assays are shown at the top of each column. (A–D) Time course at different temperatures. Each reaction, in a final volume of 100 μl of buffer B (pH 5.5) with 0.004% Nonidet P-40, contained ≈40 pmol of NPC1(NTD)-LVPR (A and C) or NPC2-FLAG (B and D) each complexed to [3H]cholesterol (1.3–1.9 pmol; 132 × 103 dpm/pmol) and 200 pmol of NPC1(NTD)-LVPRGS-His-8-FLAG (○) or NPC2-His-10 (●). After incubation for indicated time at 4°C (A and B) or 37°C (C and D), the amount of [3H]cholesterol transferred to the indicated acceptor His-tagged NPC protein was measured with the Ni-NTA-agarose cholesterol transfer assay. Each value represents the percentage of [3H]cholesterol transferred. The 100% values for transfer from donor were 1.9 pmol (A), 1.5 (B and C), and 1.3 (D). (E and F) Transfer at 4°C as a function of the amount of acceptor NPC protein. This experiment was carried out as in A–D except that the amount of acceptor NPC protein varied as indicated and time of incubation at 4°C was 30 min. The 100% values for transfer of [3H]cholesterol from NPC1(NTD) and NPC2 were 2.0 and 1.2 pmol, respectively. (G and H) Transfer at 4°C as a function of pH. Each reaction in a final volume of 300 μl of buffer A, B, or E with 0.004% Nonidet P-40 at the indicated pH, contained ≈30 pmol of NPC1(NTD)-LVPR (G) or NPC2-FLAG (H) each complexed to [3H]cholesterol (1.6 and 1.0 pmol, respectively) and 200 pmol of either NPC1(NTD)-LVPRGS-His-8-FLAG (○) or NPC2-His-10 (●). After incubation for 30 min at 4°C, the amount of [3H]cholesterol transferred to the indicated acceptor His-tagged NPC was measured as described above except samples were diluted with 1.1 ml of buffer A with 0.004% Nonidet P-40. Each value is the average of duplicate assays and represents the percentage of [3H]cholesterol transferred. The 100% values for transfer from NPC1(NTD) and NPC2 were 1.6 and 1.0 pmol, respectively.
At 37°C, NPC1(NTD) was able to transfer [3H]cholesterol to the His-tagged version of itself (Fig. 2C, open circles), but even at this temperature the transfer to NPC2 was much faster (Fig. 2C, filled circles). Again, NPC2 transferred its [3H]cholesterol rapidly either to NPC1(NTD) or NPC2 at 37°C (Fig. 2D).
To measure protein-to-liposome transfer, we prepared unilamellar phosphatidylcholine (PC) liposomes by sonication as described in Methods. The donors were His-tagged NPC1(NTD) or NPC2 that had been complexed with [3H]cholesterol and isolated by gel filtration. After incubation with liposomes, the reaction mixture was applied to a nickel agarose column. The liposomes (labeled with Texas red dye) appeared in the flow-through fractions, whereas the His-tagged NPC proteins appeared in the imidazole-eluted fractions (Fig. S3). We measured the amount of [3H]cholesterol transferred from donor NPC proteins to liposomes by scintillation counting of the flow-through fraction. To assure that transfer of [3H]cholesterol was liposome dependent, we conducted parallel incubations in the absence of liposomes, and the amount of [3H]cholesterol in the flow-through was measured. These values were subtracted from those in the liposome-containing incubations to measure liposome-dependent transfer. At 4°C, NPC1(NTD) failed to transfer [3H]cholesterol to liposomes (Fig. 3A, open circles). Transfer to liposomes was enhanced dramatically (>100-fold) when NPC2 was included in the incubation (Fig. 3A, filled circles). As the temperature was increased from 4°C to 22°C to 37°C, there was a progressive increase in the ability of NPC1(NTD) to transfer [3H]cholesterol to liposomes (Fig. S4A). This rate was enhanced dramatically when NPC2 was present (Fig. 3 A and C; Fig. S4A). When NPC2 was the donor, [3H]cholesterol was transferred rapidly to liposomes at all temperatures, and the addition of NPC1(NTD) had no significant effect (Fig. 3 B and D and Fig. S4B). To confirm that the [3H]cholesterol in the nickel agarose flow-through had indeed been transferred to liposomes, we isolated the liposomes by flotation through a discontinuous sucrose gradient in an ultracentrifuge. All of the [3H]cholesterol comigrated with the Texas red dye-labeled liposomes (data not shown).
Fig. 3.
Transfer of [3H]cholesterol from donor NPC protein to acceptor PC liposomes. Schematic diagrams of the two transfer assays are shown at the top of each column. (A–D) Time course at different temperatures. Each reaction, in a final volume of 200 μl of buffer B (pH 5.5), contained ≈30 pmol of NPC1(NTD)-LVPRGS-His-8-FLAG (A and C) or NPC2-His-10 (B and D) each complexed to [3H]cholesterol (1.0 and 0.3 pmol, respectively; 132 × 103 dpm/pmol) and 60 μg of PC liposomes labeled with Texas red dye in the absence (○) or presence (●) of 100 pmol NPC2-His-10 (A and C) or NPC1(NTD)-LVPRGS-His-8-FLAG (B and D). After incubation for the indicated time at 4°C (A and B) or 37°C (C and D), the amount of [3H]cholesterol transferred to liposomes was measured in the Ni-NTA-agarose cholesterol transfer assay. Each value is the average of duplicate assays and represents the percentage of [3H]cholesterol transferred to liposomes. The 100% values for transfer from NPC1(NTD) and NPC2 were 1.0 and 0.3 pmol, respectively. Blank values in the absence of liposomes (1–4%) were subtracted. (E and F) Transfer at 4°C as a function of the amount of liposomes. This experiment was carried out as in A–D except that the amount of liposomes varied as indicated, and the time of incubation at 4°C was 30 min. The 100% values for transfer of [3H]cholesterol from NPC1(NTD) and NPC2 were 0.4 and 1.7 pmol, respectively. Blank values in the absence of liposomes (2–5%) were subtracted. (G and H) Transfer at 4°C as a function of pH. The conditions for this experiment are the same as those in E and F except that the final volume of the reaction was 300 μl in buffer A, B, or E at the indicated pH, and the acceptor was 60 μg of PC liposomes in the absence (○) or presence (●) of 100 pmol of NPC2-His-10 (G) or 100 pmol NPC1(NTD)-LVPRGS-His-8-FLAG (H). The 100% values for transfer of [3H]cholesterol from NPC1(NTD) and NPC2 were 0.8 and 0.5 pmol, respectively. Blank values in absence of liposomes (1–5%) were subtracted.
Fig. 3E shows the transfer of [3H]cholesterol from NPC1(NTD) to liposomes as a function of the concentration of liposomes. Transfer required the presence of NPC2 at all liposome concentrations, whereas transfer from NPC2 to liposomes did not require NPC1(NTD) (Fig. 3F). The transfer reactions showed only minor effects of pH in the range of 4.5–8.5 (Fig. 3 G and H).
To confirm that the transfer activity of NPC2 required cholesterol binding, we exploited a naturally occurring missense mutation in NPC2(P120S). Homozygosity for this mutation was observed in a patient with NPC2 disease (17). Immunofluorescence of the patient's fibroblasts showed that the mutant protein reaches lysosomes, suggesting that the protein folds sufficiently to escape the quality-control system of the ER. However, the mutant protein is unable to carry out its function in facilitating cholesterol egress from lysosomes (17). To study NPC2(P120S), we prepared a cDNA encoding the mutant protein and introduced it into CHO cells by transfection. The protein was purified from the culture medium like native NPC2. SDS/PAGE, gel filtration, and circular dichroism spectroscopy (wavelength scan at 4°C) showed that the mutant protein behaved similarly to the wild type (Fig. 4A Inset and data not shown). The human P120S mutation replaces a residue that lies at the edge of the hydrophobic cholesterol-binding pocket as indicated by the crystal structure of the bovine NPC2 (11). Indeed, as shown in Fig. 4A, the P120S mutant failed to bind [3H]cholesterol under our assay conditions. The mutant NPC2 was also severely defective in facilitating the transfer of [3H]cholesterol from NPC1(NTD) to liposomes (Fig. 4B). These data suggest that NPC2 must bind cholesterol to facilitate transfer of cholesterol from NPC1(NTD) to liposomes.
Fig. 4.
Mutant NPC2 fails to transfer [3H]cholesterol from NPC1(NTD) to acceptor PC liposomes. (A) Saturation curves for equilibrium binding of [3H]cholesterol for wild-type and mutant NPC2. Binding reactions were carried out as described in Fig. 1F except that each reaction was incubated for 2 h at 4°C and contained 8 pmol of wild-type (●) or P120S mutant (○) version of NPC2-His-10. Each value is the average of duplicate assays and represents binding after subtraction of blank values (0.01–0.11 pmol). (Inset) Coomassie Brilliant blue R-250 stain of wild-type and mutant NPC2 proteins after electrophoresis on 13% SDS/PAGE (3 μg of each protein loaded on gel). (B) Transfer of [3H]cholesterol from donor NPC1(NTD) to acceptor liposomes as a function of varying concentrations of wild-type or mutant NPC2. Assays were carried out as in Fig. 3 except that each reaction was carried out for 10 min at 4°C and contained ≈50 pmol of NPC1(NTD)-LVPRGS-His-8-FLAG complexed to [3H]cholesterol (1.2 pmol; 132 × 103 dpm/pmol), 60 μg of PC liposomes, and the indicated concentration of wild-type (●) or P120S mutant (○) version of NPC2-His-10. Each value is the average of duplicate assays and represents the percentage of [3H]cholesterol transferred to liposomes. The 100% value for transfer from NPC1(NTD) was 1.2 pmol. Blank values in the absence of NPC2 protein (8%) were subtracted.
Fig. 5 shows the results of the reciprocal assay, namely, the transfer of [3H]cholesterol from PC liposomes to NPC1(NTD) or NPC2. At 4°C, there was little transfer to NPC1(NTD), and transfer was stimulated markedly by NPC2 (Fig. 5A). As the temperature was increased from 4°C to 22°C to 37°C, there was a progressive increase in the ability of NPC1(NTD) to accept [3H]cholesterol from liposomes (data not shown). This rate was enhanced markedly at 4°C and 37°C when NPC2 was added (Fig. 5 A and C). Here again, NPC2 showed no requirement for NPC1(NTD). NPC2 rapidly accepted [3H]cholesterol from liposomes at all temperatures, and there was no effect of adding NPC1(NTD) (Fig. 5 B and D).
Fig. 5.
Transfer of [3H]cholesterol from donor PC liposomes to acceptor NPC protein. Schematic diagrams of the two transfer assays are shown at the top of each column. (A–D) Time course at different temperatures. Each reaction, in a final volume of 200 μl of buffer B (pH 5.5), contained 17 μg of PC liposomes containing 540 pmol of [3H]cholesterol (930 dpm/pmol) labeled with Texas red dye and 100 pmol of NPC1(NTD)-LVPRGS-His-8-FLAG (A and C) or NPC2-His-10 (B and D) in the presence (●) or absence (○) of 100 pmol of either NPC2-FLAG (A and C) or NPC1(NTD)-LVPR (B and D). After incubation for the indicated time at 4°C (A and B) or 37°C (C and D), the amount of [3H]cholesterol transferred to acceptor His-tagged NPC protein was measured in the Ni-NTA-agarose cholesterol transfer assay. Each value is the average of duplicate assays and represents the amount of [3H]cholesterol transferred from liposomes to the indicated acceptor His-tagged NPC protein. Blank values in the absence of His-tagged NPC protein (0.2–0.3 pmol) were subtracted. (E and F) Transfer at 4°C as a function of the concentration of NPC protein. This experiment was carried out as in A–D except that the amount of acceptor NPC1(NTD)-LVPRGS-His-8-FLAG (E) or acceptor NPC2-FLAG (F) varied as indicated, and the time of incubation at 4°C was 30 min. Each value is the average of duplicate assays and represents the amount of [3H]cholesterol transferred from liposomes to the indicated acceptor NPC protein. Blank values in the absence of His-tagged NPC protein (0.2–0.4 pmol) were subtracted. (G and H) Transfer at 4°C as a function of pH. The conditions for this experiment are the same as those in Fig. 3 G and H except that the donor was 17 μg of PC:[3H]cholesterol liposomes (930 dpm/fmol), and the acceptor was 100 pmol of NPC1(NTD)-LVPRGS-His-8-FLAG (G) or NPC2-His-10 (H) in the absence (○) or presence (●) of 100 pmol of either NPC2-FLAG (G) or NPC1(NTD)-LVPR(H). Each value is the average of duplicate assays and represents the amount of [3H]cholesterol transferred from liposomes to the indicated His-tagged NPC protein. Blank values in the absence of His-tagged NPC protein (0.2–0.3 pmol) were subtracted.
Fig. 5E shows the transfer of [3H]cholesterol from PC liposomes to NPC1(NTD) as a function of increasing concentrations of NPC1(NTD) in the absence or presence of NPC2. As seen with the protein-to-liposome transfer, little transfer occurred unless NPC2 was present. Similarly, when the acceptor was NPC2, it alone effected transfer of [3H]cholesterol from liposomes to itself, and this transfer was not influenced by NPC1(NTD) (Fig. 5F). Interestingly, the liposome-to-protein assay was pH dependent at 4°C. In the presence of NPC2, NPC1(NTD) accepted cholesterol optimally at pH 5.5 (Fig. 5G). NPC2 accepted cholesterol optimally at pH 4.5 (Fig. 5H).
Discussion
The present studies establish conditions for study of the transfer of cholesterol between the water-soluble NTD of NPC1 and water-soluble NPC2 and between both proteins and liposomes. NPC1 and NPC2 are both required in order for lipoprotein-derived cholesterol to exit from endosomes and lysosomes. Consistent with this common function, NPC2 and NPC1 can both bind cholesterol, the latter in the luminal NTD (10, 14). Despite these similarities, the cholesterol binding reactions exhibit major differences, as shown in Fig. 1. NPC2 behaves like a typical receptor. Binding is rapid and reversible at both 4°C and 37°C. NPC1(NTD) behaves quite differently. The protein binds cholesterol extremely slowly at 4°C. The rate of binding is accelerated at least 140-fold at 37°C. Once bound to NPC1(NTD), cholesterol does not dissociate from NPC1(NTD) at 4°C over at least 2 h. At 37°C the dissociation is much faster. Because of these reciprocal changes, the equilibrium dissociation constant for NPC1(NTD) is similar at the two temperatures (50 nM and 90 nM at 4°C and 37°C, respectively).
The above data suggest that the binding site on NPC1(NTD) exists in a relatively closed conformation, although less so at 37°C than at 4°C. Remarkably, this site on NPC1(NTD) can be opened by NPC2. When NPC2 is used as a donor to transfer cholesterol to NPC1(NTD), the transfer is rapid both at 4°C and 37°C (Fig. 2B). Indeed, NPC2 transfers cholesterol to NPC1(NTD) as rapidly as it transfers cholesterol to unoccupied NPC2 molecules. NPC2 can also rapidly remove cholesterol from NPC1(NTD) (Fig. 2 A and C) even under conditions where cholesterol has not dissociated from NPC1(NTD) (Fig. 1C).
Although the physical basis for the temperature effect on NPC1(NTD) is not yet known, the ability to slow the reaction at 4°C experimentally permits detailed study of the kinetics using the relatively slow bead-trapping assay. In the cell, NPC1 operates at 37°C where it has an intrinsic ability to bind and release cholesterol. However, even at 37°C, the binding and release reactions are markedly accelerated by NPC2, suggesting that NPC2 performs this function in the living cell.
Because of its ability to open the binding site on NPC1(NTD), NPC2 facilitates the transfer of cholesterol from NPC1(NTD) to PC liposomes (Fig. 3) and from liposomes to NPC1(NTD) (Fig. 5). Genetic evidence for this function of NPC2 was provided by the studies in Fig. 4 in which a naturally occurring NPC2 mutant (P120S) failed both to bind [3H]cholesterol and to facilitate its transfer from NPC1(NTD) to lysosomes. The precise mechanism for the facilitated transfer reaction cannot be ascertained with the relatively slow bead-trapping assay, which requires several minutes of adherence and washing before the bound [3H]cholesterol can be quantified. It seems likely that the process is catalytic, i.e., NPC2 accepts or donates a cholesterol molecule in a hit-and-run fashion. Consistent with this hypothesis, we were unable to isolate a stable complex between untagged versions of NPC1(NTD) and NPC2 and His-tagged versions of these proteins as determined by nickel agarose chromatography (data not shown).
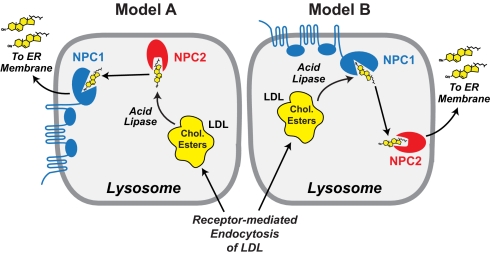
Within cells, cholesterol egress from lysosomes is unidirectional, i.e., from LDL to lysosomal membranes to ER. To establish this unidirectional flux, NPC1 and NPC2 could act in one of two possible orders, as illustrated in Fig. 6. In both models, NPC2 performs a shuttling function with respect to NPC1. In Model B, acid lipase interacts with the membranous domain of NPC1 and releases cholesterol directly to the NTD of NPC1, which then transfers it to NPC2. Given the relative simplicity of NPC2's structure, insertion of cholesterol into the lysosomal membrane, from which it travels to the ER, would likely require NPC2 to interact with a lysosomal cholesterol transporter, as yet unidentified. In Model A, acid lipase liberates cholesterol which is then bound by NPC2 and delivered to the NTD of NPC1. NPC1 then inserts the cholesterol into lysosomal membranes, from which it is transferred to the ER. In this case, membrane insertion is likely mediated by the complex membranous domain of NPC1. The current in vitro data do not permit an experimental distinction between these two models.
Fig. 6.
Alternative models for the transfer of cholesterol from LDL to lysosomal membranes. The interpretations of Model A and Model B are explained in Discussion.
Another important unanswered question concerns the role of the lysosomal anionic phospholipids such as bio(monooleoylglycerol)phosphate and phosphatidyl inositol that have been reported to accelerate the transfer of cholesterol from NPC2 to liposomes (12, 13). Do either or both of these two lipids influence the rate and/or direction of transfer between the two NPC proteins? The answers to these questions may be forthcoming now that it is possible to study the cholesterol transfer reactions with soluble forms of both NPC proteins in a test tube.
Methods
[3H]Cholesterol Binding Assay.
This assay was previously described (10). In brief, each reaction contained, in a final volume of 80 μl, the indicated buffer, varying concentrations of [3H]cholesterol (132 × 103 dpm/pmol; delivered in ethanol at a final concentration of 2%), 1 μg of BSA, and varying amounts of purified human NPC1 or NPC2. After incubation for the indicated time at 4°C or 37°C, the mixture in buffer A was loaded onto a column packed with 0.3 ml of Ni-NTA-agarose beads (Qiagen) that had been preequilibrated with buffer A with 0.004% (wt/vol) Nonidet P-40 and then washed with 6 ml of the same buffer. (Before Ni chromatography, mixtures in buffer B were diluted 5-fold with buffer A containing 0.004% Nonidet P-40). Protein-bound [3H]cholesterol was eluted with 1 ml of buffer A containing 250 mM imidazole and 0.004% Nonidet P-40 and quantified by scintillation counting.
The [3H]Cholesterol Transfer Assays.
Three types of transfer assays were performed: (i) movement of [3H]cholesterol from a donor protein (NPC1(NTD) or NPC2) to an acceptor protein (NPC1(NTD) or NPC2); (ii) movement of [3H]cholesterol from NPC proteins to liposomes; and (iii) movement of [3H]cholesterol from liposomes to NPC proteins. In the NPC protein-to-NPC protein transfer assays, each reaction contained, in a final volume of 100 μl of buffer B (pH 5.5) supplemented with 0.004% Nonidet P-40, 0.4–2 pmol of [3H]cholesterol complexed to ≈40 pmol of a donor protein, either NPC1(NTD)-LVPR or NPC2-FLAG. The donor proteins did not contain a His tag. The assay mixture contained varying amounts of an acceptor protein (either NPC1(NTD)-LVPRGS-His-8-FLAG or NPC2-His-10), both of which contained His tags. After incubation for 30 min at 4°C or 37°C, each reaction mixture was diluted with 500 μl of buffer A supplemented with 0.004% Nonidet P-40 and loaded onto a 2-ml column (Bio-Rad) packed with 0.3 ml of Ni-NTA-agarose beads (Qiagen) that had been preequilibrated with buffer A with 0.004% Nonidet P-40. Each column was washed at 4°C with 3 ml of buffer A with 0.004% Nonidet P-40, after which the His-tagged acceptor proteins were eluted with 1 ml of buffer A containing 0.004% Nonidet P-40 and 250 mM imidazole. The amount of [3H]cholesterol transferred to each His-tagged acceptor protein was quantified by scintillation counting of the eluate as described (18).
In the NPC protein-to-liposome transfer assays, each reaction contained, in a final volume of 200 μl of buffer B, 0.3–1.7 pmol of [3H]cholesterol complexed to 30–50 pmol of either NPC1(NTD)-LVPRGS-His-8-Flag or NPC2-His-10, and varying amounts of acceptor PC liposomes in the absence or presence of the other His-tagged NPC protein. After incubation for 30 min at 4°C or 37°C, each mixture was processed as described above except that reactions were diluted with 750 μl of buffer A, and none of the buffers contained Nonidet P-40. The amount of [3H]cholesterol transferred to liposomes was quantified by scintillation counting of the flow-through.
In the [3H]cholesterol/liposome-to-NPC protein transfer assays, each reaction contained, in a final volume of 200 μl of buffer B, 17 μg of PC:[3H]cholesterol liposomes (930 dpm/pmol), and increasing amounts of an acceptor protein, either NPC1(NTD)-LVPRGS-His-8-FLAG or NPC2-His-10, in the absence or presence of the other non-His-tagged NPC protein. After incubation for 30 min at 4°C, each mixture was processed as described above except that reactions were diluted with 750 μl of buffer A, and none of the buffers contained Nonidet P-40. The amount of [3H]cholesterol transferred to the respective His-tagged NPC protein was quantified by scintillation counting of the eluate.
Other Methods.
Materials, buffers, plasmids, purification of NPC proteins, isolation of complexes of [3H]cholesterol bound to NPC proteins, preparation of liposomes, and immunoblot analysis are described in SI Text.
Supplementary Material
Acknowledgments.
We thank Shomanike Head and Lisa Beatty for invaluable help with tissue culture. Dorothy Goddard and Bethany Cartwright provided excellent technical assistance. This work was supported by National Institutes of Health Grant HL20948 and a grant from the Perot Family Foundation. R.E.I and M.L.W. are supported by Medical Scientist Training Program Grant 5T32 GM08014. R.E.I. is also supported by the Ara Parseghian Medical Foundation.
Footnotes
The authors declare no conflict of interest.
See Commentary on page 15223.
This article contains supporting information online at www.pnas.org/cgi/content/full/0807328105/DCSupplemental.
References
- 1.Brown MS, Goldstein JL. A receptor-mediated pathway for cholesterol homeostasis. Science. 1986;232:34–47. doi: 10.1126/science.3513311. [DOI] [PubMed] [Google Scholar]
- 2.Goldstein JL, Dana SE, Faust JR, Beaudet AL, Brown MS. Role of lysosomal acid lipase in the metabolism of plasma low density lipoprotein: Observations in cultured fibroblasts from a patient with cholesteryl ester storage disease. J Biol Chem. 1975;250:8487–8495. [PubMed] [Google Scholar]
- 3.Pentchev PG, Vanier MT, Suzuki K, Patterson MC. In: The Metabolic and Molecular Basis of Inherited Disease. Scriver CR, Beaudet AL, Sly WS, Valle D, editors. New York: McGraw–Hill; 1995. pp. 2625–2639. [Google Scholar]
- 4.Pentchev PG. Niemann–Pick C research from mouse to gene. Biochim Biophys Acta. 2004;1685:3–7. doi: 10.1016/j.bbalip.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 5.Li AC, Tanaka RD, Callaway K, Fogelman AM, Edwards PA. Localization of 3-hydroxy-3-methylglutaryl CoA reductase and 3-hydroxy-3-methlyglutaryl CoA synthase in the rat liver and intestine is affected by cholestyramine and mevinolin. J Lipid Res. 1988;29:781–796. [PubMed] [Google Scholar]
- 6.Carstea ED, et al. Niemann–Pick C1 disease gene: Homology to mediators of cholesterol homeostasis. Science. 1997;277:228–231. doi: 10.1126/science.277.5323.228. [DOI] [PubMed] [Google Scholar]
- 7.Naureckiene S, et al. Identification of HE1 as the second gene of Niemann–Pick C disease. Science. 2000;290:2298–2301. doi: 10.1126/science.290.5500.2298. [DOI] [PubMed] [Google Scholar]
- 8.Ko DC, Binkley J, Sidow A, Scott MP. The integrity of a cholesterol-binding pocket in Niemann–Pick C2 protein is necessary to control lysosome cholesterol levels. Proc Natl Acad Sci USA. 2003;100:2518–2525. doi: 10.1073/pnas.0530027100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friedland N, Liou H-L, Lobel P, Stock AM. Structure of a cholesterol-binding protein deficient in Niemann–Pick type C2 disease. Proc Natl Acad Sci USA. 2003;100:2512–2517. doi: 10.1073/pnas.0437840100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Infante RE, et al. Purified NPC1 protein: II. Localization of sterol binding to a 240-amino acid soluble luminal loop. J Biol Chem. 2008;283:1064–1075. doi: 10.1074/jbc.M707944200. [DOI] [PubMed] [Google Scholar]
- 11.Xu S, Benoff B, Liou H-L, Lobel P, Stock AM. Structural basis of sterol binding by NPC2, a lysosomal protein deficient in Niemann–Pick type C2 disease. J Biol Chem. 2007;282:23525–23531. doi: 10.1074/jbc.M703848200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Babalola JO, et al. Development of an assay for the intermembrane transfer of cholesterol by Niemann–Pick C2 protein. Biol Chem. 2007;388:617–626. doi: 10.1515/BC.2007.063. [DOI] [PubMed] [Google Scholar]
- 13.Cheruku SR, Xu Z, Dutia R, Lobel P, Storch J. Mechanism of cholesterol transfer from the Niemann–Pick type C2 protein to model membranes supports a role in lysosomal cholesterol transport. J Biol Chem. 2006;281:31594–31604. doi: 10.1074/jbc.M602765200. [DOI] [PubMed] [Google Scholar]
- 14.Infante RE, et al. Purified NPC1 protein: I. Binding of cholesterol and oxysterols to a 1278-amino acid membrane protein. J Biol Chem. 2008;283:1052–1063. doi: 10.1074/jbc.M707943200. [DOI] [PubMed] [Google Scholar]
- 15.Davies JP, Ioannou YA. Topological analysis of Niemann–Pick C1 protein reveals that the membrane orientation of the putative sterol-sensing domain is identical to those of 3-hydroxy-3-methylglutaryl-CoA reductase and sterol regulatory element binding protein cleavage-activating protein. J Biol Chem. 2000;275:24367–24374. doi: 10.1074/jbc.M002184200. [DOI] [PubMed] [Google Scholar]
- 16.Chikh K, Vey S, Simonot C, Vanier MT, Millat G. Niemann–Pick type C disease: Importance of N-glycosylation sites for function and cellular location of the NPC2 protein. Mol Gen Metabol. 2004;83:220–230. doi: 10.1016/j.ymgme.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 17.Verot L, et al. Niemann–Pick C disease: Functional characterization of three NPC2 mutations and clinical and molecular update on patients with NPC2. Clin Genet. 2007;71:320–330. doi: 10.1111/j.1399-0004.2007.00782.x. [DOI] [PubMed] [Google Scholar]
- 18.Radhakrishnan A, Sun L-P, Kwon HJ, Brown MS, Goldstein JL. Direct binding of cholesterol to the purified membrane region of SCAP: Mechanism for a sterol-sensing domain. Mol Cell. 2004;15:259–268. doi: 10.1016/j.molcel.2004.06.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.