Abstract
We report the high-resolution crystal structures of an extensively simplified variant of bovine pancreatic trypsin inhibitor containing 20 alanines (BPTI-20st) and a reference single-disulfide-bonded variant (BPTI-[5,55]st) at, respectively, 1.39 and 1.09 Å resolutions. The sequence was simplified based on the results of an alanine scanning experiment, as reported previously. The effects of the multiple alanine substitutions on the overall backbone structure were surprisingly small (Cα atom RMSD of 0.53 Å) being limited to small local structural perturbations. Both BPTI variants retained a wild-type level of trypsin inhibitory activity. The side-chain configurations of residues buried in the hydrophobic cores (<30% accessible surface area) were almost perfectly retained in both BPTI-20st and BPTI-[5,55]st, indicating that neither multiple alanine replacements nor the removal of the disulfide bonds affected their precise placements. However, the side chains of three partially buried residues (Q31, R20, and to some extent Y21) and several unburied residues rearranged into alternative dense-packing structures, suggesting some plasticity in their shape complementarity. These results indicate that a protein sequence simplified over its entire length can retain its densely packed, native side-chain structure, and suggest that both the design and fold recognition of natively folded proteins may be easier than previously thought.
Keywords: native structure, protein design, protein folding, protein structure, sequence simplification
The protein folding problem (1), or how a protein structure is encoded in its amino acid sequence, remains fundamentally unsolved. Indeed, an astronomical number of sequences are generated by the combination of the 20 natural amino acids at each site of a sequence, but few of these can fold into native, fully functional protein structures. However, the information contained in natural amino acid sequences is redundant (2, 3); namely, a small fraction of the information can fully determine a protein structure (4, 5). Protein sequence simplification has emerged as a method of choice in deciphering the determinants of protein structures.
Information redundancy can be conceptually classified into class and site redundancy (6). Class redundancy refers to the overlapping physicochemical properties of the amino acids, and it can be removed by designing proteins with a reduced amino acid alphabet (7). Class redundancy has been reported in the Src SH3 domain, for which a functional variant with 70% of its sequence encoded by only five amino acids types was identified by phage display (8). Similarly, simplified variants of chorismate mutase (9) and orotate phosphoribosyl transferase (10), with sequences predominantly encoded by only nine amino acid types, rescued auxotrophic host cells lacking the respective enzymes.
Site redundancy refers to the occurrence of several residues in a protein's amino acid sequence that do not contribute to the formation of the structure, except that their backbone atoms act as linker residues in the amino acid sequence. Alanine is often used as a natural representative of such linker residues because its side chain is small enough to avoid steric clashes and does not introduce excessive flexibility into the backbone structure (11). Site redundancy was first demonstrated experimentally in the T4 lysozyme sequence, in which 10 consecutive residues at positions 40–49 were substituted with alanines with very little effect, if any, on its structure and function (12). Simplified variants of the Arc repressor, containing up to 11 alanine substitutions in 53 residues, were also shown by circular dichroism to be folded correctly and retained a wild-type level of DNA-binding activity (13). In line with these developments, we recently reported that bovine pancreatic trypsin inhibitor (BPTI) variants with up to 47% alanines could fold into a native BPTI-like structure (6), and that their thermodynamics were, in most aspects, very similar to those of natural globular proteins (14).
These reports, based on functional or spectroscopic assays, strongly suggest that proteins with simplified sequences can remain folded and retain some or all of their functions. However, very few if any high-resolution structures of simplified proteins are presently available, and when available, they report either modest simplifications of short sequence segments (12) or modifications to a small number of residues located in the protein core (15, 16). Especially, no high-resolution structure of a protein simplified over its entire sequence is yet available. Such high-resolution structures could truly address the question of the sequence determinants of protein structures by providing a precise picture of how side-chain packing is affected by extensive sequence simplification.
Here, we report the high-resolution crystal structure of an extensively simplified BPTI in which over one-third of the residues are alanines, spread over its entire sequence. BPTI-21 (containing 21 alanines) was first designed by simultaneously substituting the least destabilizing BPTI-[5,55] residues to alanines (6), according to an alanine scanning experiment, in which each residue was individually changed to alanine and the stability of the mutated BPTIs were experimentally measured (17). Among the substitution sites, three residues are fully buried and two are partially buried. In the present study, we stabilized BPTI-21 by introducing an A14G substitution (18) that yielded BPTI-20st. As a control for the native structure, we determined the structure of a stabilized BPTI-[5,55] variant (BPTI-[5,55]st; BPTI-[5,55] with the A14G mutation), which is a single-disulfide-bonded BPTI variant regarded as a reference native molecule in several protein folding experiments (19). The effects of multiple alanine substitutions were surprisingly few. The overall structure of BPTI-20st was very similar to those of BPTI-[5,55]st and the wild-type BPTI, and the side-chain configurations of the deeply buried residues were almost perfectly retained. Alternative or new side-chain configurations were observed for a few partially buried residues.
Results and Discussion
Structures of the Simplified BPTIs.
The space group of BPTI-20st and BPTI-[5,55]st was P212121 and P1211 with four and two molecules per unit cell, respectively. High-resolution anisotropic models of BPTI-20st and BPTI-[5,55]st were determined at resolutions of 1.39 and 1.09 Å, respectively, and with crystallographic R factors of 0.1565 and 0.1811 (Table 1). All residue side chains had excellent electron density maps, in both BPTI-20st and BPTI-[5,55]st. The overall backbone fold of both BPTI-20st and BPTI-[5,55]st was very similar to that of wild-type BPTI (Fig. 1A), with overall backbone root mean square deviations (RMSDs) of 0.55 and 0.47 Å, respectively (when compared with the 5PTI structure), whereas the RMSDs among wild-type structures are between 0.40 and ∼0.44 Å, reflecting experimental error arising from variable crystallization conditions [see supporting information (SI) Table S1; also see Figs. S1–S5 and Tables S2 and S3]. These structural observations were corroborated by the full trypsin inhibition activity of the simplified sequence (Fig. S2).
Table 1.
Statistics after final refinement of simplified BPTIs
| BPTI-20st | BPTI-[5,55]st | |
|---|---|---|
| Space group | P212121 | P1211 |
| No. of molecules in the asymmetric unit | Four molecules | Two molecules |
| Matthews coefficient | 2.14 | 2.17 |
| Solvent content, % | 42.50 | 43.32 |
| Resolution in refinement, Å | 44.76∼1.39 | 53.91∼1.09 |
| No. of reflections used | 38,836 | 40,917 |
| R factor | 0.1565 | 0.1811 |
| Rfree | 0.2052 | 0.2090 |
| B factor,* Å2 | ||
| Protein atoms | 15.16 | 11.84 |
| Main chain | 11.06 | 10.77 |
| Side chain | 18.44 | 12.82 |
| Ramachandran plot statistics† | ||
| Residues in most favored region, % | 90.8 (98.2) | 92.2 (95.5) |
| Planarity, % | 100 | 100 |
| RMS Deviation from ideality† | ||
| Bond lengths/bad length, Å/% | 0.008∼0.009/(0.0) | 0.007∼0.015/(0.0) |
| Bond angles/bad angles, °/% | 1.06∼2.79/(1.7‡) | 0.96∼2.89/(0.0) |
*Probe used in the refinement process, 5PTI, is solved at a resolution of 1.0 Å with the main-chain and side-chain crystallographic B factors of 12.2 and 14.5, respectively.
†The qualities of the models were assessed by the SFCHECK and PROCHECK (35, 36) modules in the CCP4 suite. Figures in parentheses refer to data obtained using MolProbity (37).
‡The bad bond angle reported by MolProbity is that of A58, the C-terminal residue of BPTI-20st.
Fig. 1.
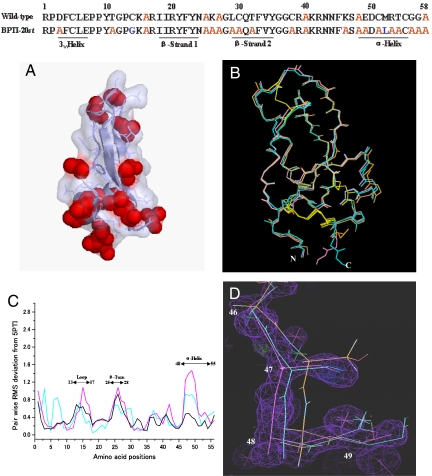
Sequences and structures of the wild-type and simplified BPTIs. At the top are shown the sequences of the wild-type BPTI and the simplified BPTI-20st. Alanines in BPTI-20st are shown in red. C14G was introduced to stabilize the BPTI variants, and M52L was introduced for avoiding cleavage by cyanogen bromide during the purification protocol. BPTI-[5,55]st is a single disulfide bonded variant with the C14G, C30A, C38A, and C51A mutations. The BPTI-20st sequence contains a total of 14 alanine substitutions, of which 3 are buried and 2 are partially buried. (A) Surface representation of BPTI-20st. The alanine residues are shown with red spheres, and the other residues are represented with light blue transparent surfaces. The backbone structure of BPTI-20st is shown with a ribbon model. (B) Wire models of 5PTI (orange), BPTI-20st (pink), and BPTI-[5,55]st (cyan), superimposed using their Cα atoms. N and C indicate the N and C termini, respectively. (C) Pairwise deviations from 5PTI. The distances between the Cα atoms of 6PTI, BPTI-[5,55]st, and BPTI-20st are shown, respectively, in black, cyan, and violet. The three most perturbed regions are indicated by black arrowed lines with residue numbers. The average RMSD between Cα atoms of the wild-type structures and BPTI-[5,55]st and BPTI-20st were 0.47 and 0.55 Å, respectively (see details in Table S1). (D) Structural perturbation observed in the α-helix region with the electron-density map of BPTI-20st. Color codes are the same as defined in B.
Small, local structural perturbations in the 13–17 loop, 25–28 β-turn, and 47–54 α-helix regions were clearly demonstrated by calculating the distances between the Cα atoms of the simplified BPTI and those of the wild-type BPTIs (Fig. 1C). Structural perturbations in the 13–17 and 25–28 regions are also observed in several wild-type structures, suggesting that these loop regions are flexible and tend to be distorted. In contrast, no distortion is observed in the 47–54 α-helix region of the wild-type structures, except for the 4PTI wild-type structure (Fig. S3 a and b), which suggests that the distortion in the α-helix region, although it can occasionally occur in the wild-type structure, is essentially attributable to the multiple alanine substitutions. Furthermore, when separately fitted, the 47–54 α-helix structures of the simplified BPTI could be superimposed almost perfectly onto that of the wild type, indicating that the entire α-helix moved toward the 29–35 β-strand without affecting the α-helix structure, per se (Fig. S3c).
Side-Chain Packing in the Simplified BPTIs.
Overall, the side-chain geometries of both BPTI-20st and BPTI-[5,55]st were similar to that of the wild-type BPTI (RMSD between heavy atoms, 0.75–1.06 Å; the RMSDs of residues substituted with alanines were calculated by using their Cβ atoms). However, a classification of the side-chain configuration changes according to their side-chain accessible surface area (ASA) values yielded some interesting insights into how the protein core is packed.
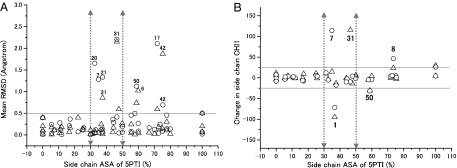
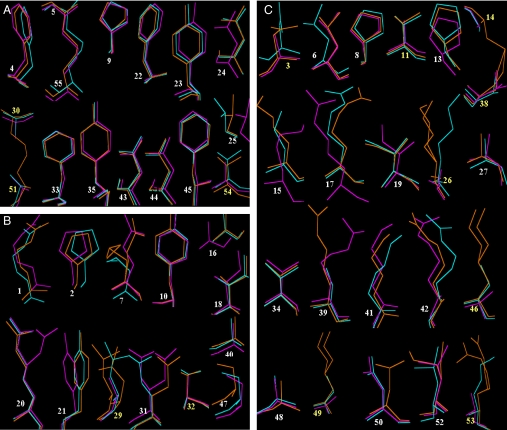
First, the side-chain configurations of all of the buried residues (ASA 0–30%) were surprisingly well conserved in both BPTI-20st and BPTI-[5,55]st. The side-chain RMSDs and χ1 deviations were 0.01–0.39 Å and 0.24–11.2°, respectively (Figs. 2 and 3). These variations are fully within the values observed among the wild-type BPTI structures and can thus be essentially attributed to differences in the crystallization conditions. This extreme conservation is all the more surprising because three buried residues (C30, C51, and T54) were substituted with alanines without apparently disturbing the side-chain packing of the other buried residues. It is also worth noting that the buried structural water molecules conserved in the wild-type BPTI structures were almost perfectly conserved in both simplified BPTIs (Fig. S5 and Table S2).
Fig. 2.
Comparison of the side-chain structures. In both panels, chain 1 of BPTI-20st and chain 1 of BPTI-[5,55]st were used for the calculation, but similar results were obtained independently of the chain used. Residues showing large variations are numbered. (A) Side-chain displacement in RMSD versus side-chain ASA. The side-chain displacement was calculated as the heavy-atoms RMSD minus the backbone atoms RMSD between BPTI-[5,55]st and 5PTI (triangles) or BPTI-20st and 5PTI (circles). The horizontal continuous line represents an RMSD value of 0.5 Å, which can be considered as the experimental error. Arrowed dotted vertical lines are drawn between the buried (ASA < 30%) and partially buried (30% < ASA < 50%) regions, and between the partially buried and surface-exposed (50% < ASA) regions. The residues' ASAs are calculated by using 5PTI. (B) χ1 Deviation versus side-chain ASA. Continuous gray lines represent a χ1 deviation (−25° to +25°) considered as the experimental error.
Fig. 3.
Side-chain conformations. Side-chain structures of buried (ASA 0∼30%) (A), partially buried (ASA 30∼50%) (B), and surface-exposed (ASA 50∼100%) residues (C). Chain 1 of BPTI-[5,55]st and chain 3 of BPTI-20st are superimposed onto that of 5PTI. Dual conformations observed in 5PTI are shown (E7, L29, C38, K26, K46, E49, and R53). Residues that are glycine in the wild sequences (G12, G28, G36, G37, G56, and G57) and the C-terminal A58 are not shown. Multiple alanine substitution sites are shown by yellow identities. Color codes are the same as in Fig. 1B. Significant conformational variability among the wild-type structures is observed for R1 and E7, indicating that the structural variations visible in image C are related to differences in crystallization conditions (see Figs. S3 and S4 for comparison among wild-type structures).
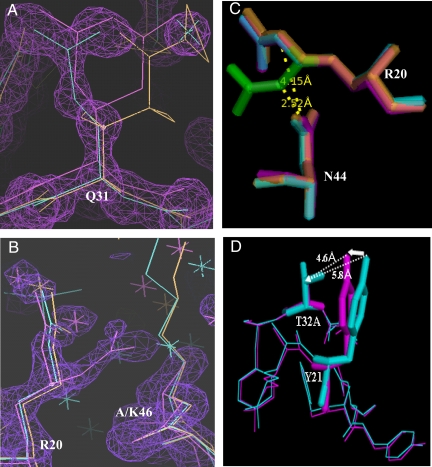
Three partially buried residues (ASA 30–50%; R20, Y21, and Q31) were observed in alternative configurations (Fig. 3) that are absent from any wild-type structures (Fig. S4) and may thus be attributed either to the removal of the disulfide bonds or to the multiple alanine substitutions, rather than to differences in the crystallization conditions. Q31 and R20 were observed in dual configurations in one of the four BPTI-20st chains (Fig. 4). Substitutions of buried and partially buried residues to alanine could have created small packing defects due to the removal of the side-chain atoms beyond Cβ. However, the BPTI-20st structure indicates that all potential packing defects were filled by tiny displacement of the nearby side-chain atoms. For example, potential defects arising from the T32A and T54A substitutions were filled by small displacements of the nearby Y21 and F4 side chains, respectively (Fig. 4D and Table S3). Furthermore, in all four BPTI-20st chains, the Y21 side chain was displaced by 1.2–1.4 Å (Fig. 4D), which enabled the formation of a hydrogen bond between its OH group and a water molecule (HOH5271). This interaction does not occur in either the wild-type or the BPTI-[5,55]st structures. Interestingly, these changes had no effect on the melting temperatures (Tm) of the simplified BPTIs (M.M.I., et al., unpublished work).
Fig. 4.
Details of alternate side-chain conformations. (A) The side chain of Q31 beyond Cβ was in an alternate conformation in both BPTI-20st and BPTI-[5,55]st, which is likely to originate from the neighboring C30AC51A substitutions. (B) An alternate conformation was also observed for the side chain of R20 beyond Cδ in BPTI-20st. The electron-density map of chain 3 of BPTI-20st is shown. The alternate structure was probably enabled by the K46A substitution, which provided both space for the R20 side chain and removed electrostatic repulsion between K46 and R20. (C) Intermolecular contact between the Nε atom of R20 and O1δ of N44. In the alternate conformation (green sticks), the Nε atom of R20 is hydrogen-bonded to the O1δ of N44 (2.52 Å), whereas they are far apart in the original structure (>4 Å). (D) Alternate conformation of Y21 side chain. A potential cavity created upon the T32A substitution is compensated for by displacement of the Y21 side chain.
Finally, the side-chain configurations of surface residues (ASA 50–100%) were quite variable (Figs. 2 and 3), but such structural variations are also observed among the wild-type BPTI structures (Fig. S4). For example, the variations in the side-chain configurations of L6, E7, R17, R42, and D50, as assessed by RMSDs, were also observed in the different wild-type structures (Fig. S4). Thus, most configurational changes in the surface residues are probably unrelated to the multiple alanine substitutions.
Effect of Sequence Simplification on Core Packing.
We simplified the sequences based on the individual residue's contribution to the BPTI structure stability (6), as assessed by an alanine scanning experiment (17). A priori, we imagined that the replacement of large side chains to the small alanine side chain's methyl group might provide spaces facilitating alternate side-chain conformations of nearby residues resulting into either an alternative densely packed protein interior or a molten globule-like protein interior (20, 21). However, our observations indicate that the replacement of three buried residues [C30, C51, and T54 (Fig. 3)] and two partially buried residues (L29 and T32) does not significantly affect the precise side-chain shape complementarity in the protein core, i.e., in a jigsaw-puzzle-like manner (22), except for the few local perturbations reported above. This observation is consistent with an alanine scanning experiment in which the substitution of any of the six core residues (F4, F22, Y23, F33, Y35, and F45) caused BPTI-[5,55] to unfold (17), probably by creating large packing defects that could not be accommodated in the finely interdigitized side-chain structure. A similar view is supported by α-helical coiled-coil structures that can switch from dimers to trimers and tetramers upon the substitution of a few core residues (23), as well as by a three-helix fold that can switch to an α/β fold by changing as few as 12% of the amino acids (24).
Previous reports suggest that the cores of some large proteins are more resilient to extensive repacking. For example, the high-resolution structure analyses of T4 lysozyme in which 10 residues in the core were simultaneously mutated to methionine residues (15) and of a staphylococcal nuclease with six core residues mutated to leucine (16), indicating that they retain native structures, suggest some plasticity in the formation of the cores of large proteins. In BPTI, which is a small protein, we observed such flexibility, at a more modest level, for some partially buried residues of the BPTI-20st.
Possible Implications for Protein Design and Fold Recognition.
The response to the packing defects introduced by extensive sequence simplification seems to involve only the local displacement of backbone atoms, accompanied by minimal side-chain configuration changes, but without any major rearrangement of the buried side-chain configurations. As an ideal model for small proteins, we may hypothesize that each backbone structure has one unique corresponding side-chain packing arrangement of the core residues, and that all sequences producing a similar backbone structure, to within a few sub-angstroms (e.g., 0.5–1.0 Å), will have either fully conserved buried residues or a small number of mutations that are easily accommodated by minimal side-chain or local backbone packing rearrangements. This hypothesis implies that fold recognition (25, 26) and de novo design (27–29) of native proteins can be simplified by restricting any detailed analysis to the small number of deeply buried residues of the template structure, because the physicochemical constraints at the remaining residues appears to be minor.
Conclusions
We report the high-resolution structural analysis of an extensively simplified BPTI variant, in which over one-third of the residues are alanines. The overall backbone and side-chain configurations were well retained. In particular, the side-chain configurations of deeply buried residues were fully conserved in both BPTI-20st and BPTI-[5,55]st. In contrast, there was some degree of plasticity in the conformations of a few partially buried residues, whose side chains were rearranged into novel, densely packed structures. Thus, some residues (mostly buried ones) seem to interact strongly and act like a jigsaw puzzle model, whereas some others (mostly partially buried residues) are able to rearrange into alternative side-chain conformations for accommodating structural perturbations arising from mutations of nearby residues. Together, these observations indicate that most protein structure determinants occur at a few sites deeply buried in the protein core, at least for small globular proteins.
Materials and Methods
Protein Expression and Purification.
BPTI variants were expressed by using the pMMHA expression vector in the Escherichia coli JM109(DE3)pLysS cell line as inclusion bodies, and purified as described in ref. 14. After cell lysis by sonication, the cysteines were air-oxidized overnight in 6 M GnHCl at room temperature. The fusion partner, a His-tagged TrpΔLE leader, was cleaved by cyanogen bromide treatment of methionine and removed as precipitate upon dialysis in 20 mM phosphate buffer (pH 6.0). Proteins were purified by reverse-phase HPLC, lyophilized, and preserved at −80°C until use. The protein identities were confirmed by electrospray ionization time-of-flight mass spectroscopy.
Crystallization and Data Collection.
Initial crystallization conditions were screened with Hampton reagents by the sitting-drop technique at 20°C. Crystals of BPTI-20st and BPTI-[5,55]st were grown at 20°C in 0.1 M Tris·HCl buffer (pH 8.5) with 30% PEG-4000 and 0.2 M lithium sulfate. The x-ray diffraction data were recorded from single crystals by using a synchrotron beamline at the Photon Factory (Tsukuba, Japan). The data were processed with the HKL2000 program package, using DENZO for the integration and SCALEPACK for the merging and statistical analysis of the diffraction intensities (30).
Structure Determination.
The structures of both BPTI variants were determined by molecular replacement method, using 5PTI (31) as a molecular probe and with the molrep program in the CCP4 program suite (32). The initial refinement was started with 48.2∼2.5 Å resolutions data, and the high-resolution limit was gradually extended to the maximum resolution, leading to the inclusion of all reflections. The structural refinement was performed with SHELXL-97 (33). A step-by-step improvement of the models was achieved through model optimization with the Xfit and followed by subsequent positional refinement. Solvent molecules were automatically added to the models at the lowest accepted occupancies of 0.5. The final structures were determined by including anisotropic displacement parameters in the refinement process. The structures were visualized by using COOT (34) and PyMOL (www.pymol.org).
Supplementary Material
Acknowledgments.
We thank Dr. Akashi Ohtaki and Mr. Kouichi Hashimoto for their help with x-ray diffraction data collection at the Photon Factory, Japan; Dr. Mark B. Swindells for discussions; Mr. Ryoh Yajima and Teppei Ebina for computational help; and Mr. Takayuki Kobayashi for installation of structure refinement software. Y.K. thanks Professor Peter S. Kim for the pMMHA expression vectors constructed while in his laboratory at the Whitehead Research Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 3CI7 (BPTI-20st) and 2ZJX (BPTI-[5,55]st)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0802699105/DCSupplemental.
References
- 1.Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 2.Bowie JU, Reidhaar-Olson JF, Lim WA, Sauer RT. Deciphering the message in protein sequences: Tolerance to amino acid substitutions. Science. 1990;247:1306–1310. doi: 10.1126/science.2315699. [DOI] [PubMed] [Google Scholar]
- 3.Rose GD. Lysozyme among the Lilliputians. Proc Natl Acad Sci USA. 2000;97:526–528. doi: 10.1073/pnas.97.2.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Plaxco KW, Riddle DS, Grantcharova V, Baker D. Simplified proteins: Minimalist solutions to the “protein folding problem”. Curr Opin Struct Biol. 1998;8:80–85. doi: 10.1016/s0959-440x(98)80013-4. [DOI] [PubMed] [Google Scholar]
- 5.Kallenbach N. Breaking open a protein barrel. Proc Natl Acad Sci USA. 2001;98:2958–2960. doi: 10.1073/pnas.071051798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuroda Y, Kim PS. Folding of bovine pancreatic trypsin inhibitor (BPTI) variants in which almost half the residues are alanine. J Mol Biol. 2000;298:493–501. doi: 10.1006/jmbi.2000.3622. [DOI] [PubMed] [Google Scholar]
- 7.Schafmeister CE, LaPorte SL, Miercke LJ, Stroud RM. A designed four helix bundle protein with native-like structure. Nat Struct Biol. 1997;4:1039–1046. doi: 10.1038/nsb1297-1039. [DOI] [PubMed] [Google Scholar]
- 8.Riddle DS, et al. Functional rapidly folding proteins from simplified amino acid sequences. Nat Struct Biol. 1997;4:805–809. doi: 10.1038/nsb1097-805. [DOI] [PubMed] [Google Scholar]
- 9.Walter KU, Vamvaca K, Hilvert D. An active enzyme constructed from a 9-amino acid alphabet. J Biol Chem. 2005;280:37742–37746. doi: 10.1074/jbc.M507210200. [DOI] [PubMed] [Google Scholar]
- 10.Akanuma S, Kigawa T, Yokoyama S. Combinatorial mutagenesis to restrict amino acid usage in an enzyme to a reduced set. Proc Natl Acad Sci USA. 2002;99:13549–13553. doi: 10.1073/pnas.222243999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cunningham BC, Wells JA. High-resolution epitope mapping of hGH-receptor interactions by alanine-scanning mutagenesis. Science. 1989;244:1081–1085. doi: 10.1126/science.2471267. [DOI] [PubMed] [Google Scholar]
- 12.Heinz DW, Baase WA, Matthews BW. Folding and function of a T4 lysozyme containing 10 consecutive alanines illustrate the redundancy of information in an amino acid sequence. Proc Natl Acad Sci USA. 1992;89:3751–3755. doi: 10.1073/pnas.89.9.3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown BM, Sauer RT. Tolerance of Arc repressor to multiple-alanine substitutions. Proc Natl Acad Sci USA. 1999;96:1983–1988. doi: 10.1073/pnas.96.5.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kato A, Yamada M, Nakamura S, Kidokoro S, Kuroda Y. Thermodynamic properties of BPTI variants with highly simplified amino acid sequences. J Mol Biol. 2007;372:737–746. doi: 10.1016/j.jmb.2007.06.066. [DOI] [PubMed] [Google Scholar]
- 15.Gassner NC, Baase WA, Matthews BW. A test of the “jigsaw puzzle” model for protein folding by multiple methionine substitutions within the core of T4 lysozyme. Proc Natl Acad Sci USA. 1996;93:12155–12158. doi: 10.1073/pnas.93.22.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J, Lu Z, Sakon J, Stites WE. Proteins with simplified hydrophobic cores compared to other packing mutants. Biophys Chem. 2004;110:239–248. doi: 10.1016/j.bpc.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 17.Yu MH, Weissman JS, Kim PS. Contribution of individual side-chains to the stability of BPTI examined by alanine-scanning mutagenesis. J Mol Biol. 1995;249:388–397. doi: 10.1006/jmbi.1995.0304. [DOI] [PubMed] [Google Scholar]
- 18.Hagihara Y, et al. Screening for stable mutants with amino acid pairs substituted for the disulfide bond between residues 14 and 38 of bovine pancreatic trypsin inhibitor (BPTI) J Biol Chem. 2002;277:51043–51048. doi: 10.1074/jbc.M208893200. [DOI] [PubMed] [Google Scholar]
- 19.Staley JP, Kim PS. Complete folding of bovine pancreatic trypsin inhibitor with only a single disulfide bond. Proc Natl Acad Sci USA. 1992;89:1519–1523. doi: 10.1073/pnas.89.5.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohgushi M, Wada A. “Molten-globule state”: A compact form of globular proteins with mobile side-chains. FEBS Lett. 1983;164:21–24. doi: 10.1016/0014-5793(83)80010-6. [DOI] [PubMed] [Google Scholar]
- 21.Kuwajima K. The molten globule state as a clue for understanding the folding and cooperativity of globular-protein structure. Proteins. 1989;6:87–103. doi: 10.1002/prot.340060202. [DOI] [PubMed] [Google Scholar]
- 22.Harrison SC, Durbin R. Is there a single pathway for the folding of a polypeptide chain? Proc Natl Acad Sci USA. 1985;82:4028–4030. doi: 10.1073/pnas.82.12.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harbury PB, Zhang T, Kim PS, Alber T. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science. 1993;262:1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- 24.Alexander PA, He Y, Chen Y, Orban J, Bryan PN. The design and characterization of two proteins with 88% sequence identity but different structure and function. Proc Natl Acad Sci USA. 2007;104:11963–11968. doi: 10.1073/pnas.0700922104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fischer D, Rice D, Bowie JU, Eisenberg D. Assigning amino acid sequences to 3-dimensional protein folds. FASEB J. 1996;10:126–136. doi: 10.1096/fasebj.10.1.8566533. [DOI] [PubMed] [Google Scholar]
- 26.Jones DT, Taylor WR, Thornton JM. A new approach to protein fold recognition. Nature. 1992;358:86–89. doi: 10.1038/358086a0. [DOI] [PubMed] [Google Scholar]
- 27.Jin W, Kambara O, Sasakawa H, Tamura A, Takada S. De novo design of foldable proteins with smooth folding funnel: Automated negative design and experimental verification. Structure. 2003;11:581–590. doi: 10.1016/s0969-2126(03)00075-3. [DOI] [PubMed] [Google Scholar]
- 28.Alvizo O, Allen BD, Mayo SL. Computational protein design promises to revolutionize protein engineering. BioTechniques. 2007;42 doi: 10.2144/000112336. 31, 33, 35 passim. [DOI] [PubMed] [Google Scholar]
- 29.Kuhlman B, et al. Design of a novel globular protein fold with atomic-level accuracy. Science. 2003;302:1364–1368. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- 30.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276(Part A):307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 31.Wlodawer A, Walter J, Huber R, Sjolin L. Structure of bovine pancreatic trypsin inhibitor. Results of joint neutron and x-ray refinement of crystal form II. J Mol Biol. 1984;180:301–329. doi: 10.1016/s0022-2836(84)80006-6. [DOI] [PubMed] [Google Scholar]
- 32.Collaborative Computational Project, Number 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 33.Sheldrick GM. SHELXL-97: Program for the Refinement of Crystal Structures. 1997 [Google Scholar]
- 34.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 35.Vaguine AA, Richelle J, Wodak SJ. SFCHECK: A unified set of procedures for evaluating the quality of macromolecular structure-factor data and their agreement with the atomic model. Acta Crystallogr D. 1999;55:191–205. doi: 10.1107/S0907444998006684. [DOI] [PubMed] [Google Scholar]
- 36.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 37.Davis IW, et al. MolProbity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W83. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.