Abstract
Exposure of Escherichia coli to alkylating agents activates expression of AidB in addition to DNA repair proteins Ada, AlkA, and AlkB. AidB was recently shown to possess a flavin adenine dinucleotide (FAD) cofactor and to bind to dsDNA, implicating it as a flavin-dependent DNA repair enzyme. However, the molecular mechanism by which AidB acts to reduce the mutagenic effects of specific DNA alkylators is unknown. We present a 1.7-Å crystal structure of AidB, which bears superficial resemblance to the acyl-CoA dehydrogenase superfamily of flavoproteins. The structure reveals a unique quaternary organization and a distinctive FAD active site that provides a rationale for AidB's limited dehydrogenase activity. A highly electropositive C-terminal domain not present in structural homologs was identified by mutational analysis as the DNA binding site. Structural analysis of the DNA and FAD binding sites provides evidence against AidB-catalyzed DNA repair and supports a model in which AidB acts to prevent alkylation damage by protecting DNA and destroying alkylating agents that have yet to reach their DNA target.
Keywords: acyl-CoA dehydrogenase, adaptive response, DNA repair, flavin adenine dinucleotide
Alkylating agents present in the cell and in the environment chemically modify DNA to produce cytotoxic and mutagenic lesions. Alkylation damage to DNA therefore poses a severe threat to the stability of the genome and, in mammals, can lead to heritable diseases and cancer. The cytotoxicity of these lesions is the basis for use of alkylation agents in chemotherapy. As a safeguard against DNA alkylation damage, all organisms have devised multiple DNA repair strategies to remove these modifications and restore DNA to an undamaged state. In addition, bacteria employ an inducible response that serves to protect cells from changing levels of mutagens. In Escherichia coli, exposure to sublethal concentrations of SN1 agents N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) and N-methylnitrosourea (MNU), or SN2 agents such as methyl methanesulfonate (MMS), stimulates the expression of four genes, ada, alkA, alkB, and aidB (reviewed in ref. 1). The activation of these genes confers increased cellular resistance to the mutagenic and cytotoxic effects of these agents and is known as the adaptive response (2).
The DNA repair mechanisms of Ada, AlkA, and AlkB have been structurally and functionally characterized (reviewed in refs. 1 and 3). The multifunctional Ada protein acts both as a methyltransferase to directly demethylate O6-methylguanine and O4-methylthymine and as a transcriptional activator for the other three adaptive response genes. AlkA is a DNA glycosylase that catalyzes the base excision repair of alkylpurines, including cytotoxic 3-methyladenine and mutagenic 1,N6-ethenoadenine, as well as O2-methylated pyrimidines. More recently, AlkB has been identified as an α-ketoglutarate-Fe(II)-dependent DNA dioxygenase to repair 1-methyladenine and 3-methylcytosine lesions by oxidative demethylation.
Despite detailed understanding of Ada, AlkA, and AlkB, the mechanism by which AidB protects against DNA damage in the adaptive response is less well understood. Induction of AidB diminishes the mutagenic effect of MNNG (4). Paradoxically, strains with insertionally inactivated aidB display either increased resistance to MNNG or no change in sensitivity as compared with wild-type E. coli. Sensitivity to other methylating agents in these strains is unaffected (5, 6). On the basis of these observations, it has been suggested that AidB may act as a detoxification enzyme to destroy nitrosoguanidines or their reactive intermediates (4), although no direct evidence of this activity has been observed.
AidB is related in sequence to acyl-CoA dehydrogenases (ACADs), which use a flavin adenine dinucleotide (FAD) to catalyze the α,β-dehydrogenation of acyl-CoA conjugates (4, 7). AidB has been shown recently to bind stoichiometric amounts of redox active FAD, and weak isovaleryl-CoA dehydrogenase (IVD) activity has been detected in vitro from both crude cell extracts and purified preparations (4, 7). However, the visible spectrum of AidB's flavin was unaffected by isovaleryl-CoA, suggesting that fatty acyl-CoAs are not substrates for the enzyme (7). Importantly, AidB was shown to bind dsDNA, and homology modeling predicted the DNA binding site to be located in the C-terminal region of the protein (7). The presence of a redox-active flavin and DNA binding activity led to the suggestion that AidB might catalyze the direct repair of methylated DNA by a dehydrogenase mechanism (7).
Whether AidB acts to reduce mutagenicity by a DNA repair or a detoxification mechanism is currently a matter of speculation. To help resolve this issue, we present a high-resolution crystal structure of AidB together with mutational analysis of DNA binding. AidB adopts a novel homotetrameric architecture decorated with two identical DNA binding surfaces, which reveals that the protein is well equipped to sterically occlude dsDNA from chemical attack. Importantly, the structure is not consistent with a DNA repair function. Alternatively, the unique chemical environment of AidB's putative FAD active site provides a rationale for a possible role in deactivation of alkylating agents.
Results
Overview of the AidB Structure.
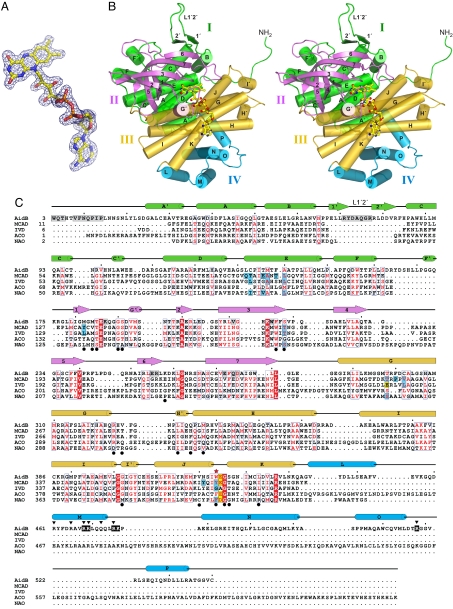
The crystal structure of E. coli AidB was determined with experimental phases obtained by multiple isomorphous replacement with anomalous scattering (MIRAS) [Fig. 1A and supporting information (SI) Table S1]. The model, consisting of one polypeptide chain in the asymmetric unit, has been refined against native x-ray diffraction data to 1.7-Å resolution and to a crystallographic residual of 16.1% (Rfree = 17.9%). As predicted (4, 7), the structure of AidB is representative of the ACAD family of flavoproteins (Fig. 1 B and C) (reviewed in ref. 8). However, AidB contains unique features that distinguish it functionally from the ACAD enzymes. One AidB subunit (Fig. 1B) consists of an N-terminal α-helical domain (domain I, residues 1–179), a seven-stranded β-barrel (domain II, residues 180–285), a central α-helical region (domain III, residues 286–444), and an α-helical domain at its C terminus (domain IV, residues 445–540). Domains I–III collectively constitute the ACAD core fold (Fig. 1B), with rmsd values over all backbone atoms from medium-chain acyl-CoA dehydrogenase (MCAD) and isovaleryl dehydrogenase (IVD) of 1.47 Å and 1.53 Å, respectively (Table S2). Domain IV, however, is not present in the ACAD family (Fig. 1C).
Fig. 1.
The structure of E. coli AidB. (A) Experimental MIRAS electron density at 1.92 Å contoured at 1σ is superimposed on the FAD molecule. Yellow, carbon; blue, nitrogen; red, oxygen; orange, phosphorus. (B) Stereo image of one subunit colored according to domains. The FAD is shown as ball and stick. (C) Structure-based sequence alignment of AidB, medium-chain acyl-CoA dehydrogenase (MCAD, PDB ID code 3MDE) (19), isovaleryl acyl-CoA dehydrogenase (IVD, PDB ID code 1IVH) (18), acyl-CoA oxidase II (ACO, PDB ID code 1IS2) (9), and nitroalkane oxidase (NAO, PDB ID code 2C0U) (26). AidB residues important for tetramerization and DNA binding are highlighted gray and black, respectively, and those predicted to contact DNA are marked with black triangles. ACAD substrate binding and catalytic residues are highlighted blue and yellow, respectively. Black circles denote FAD binding residues, and AidB Trp-424 is marked with a red star.
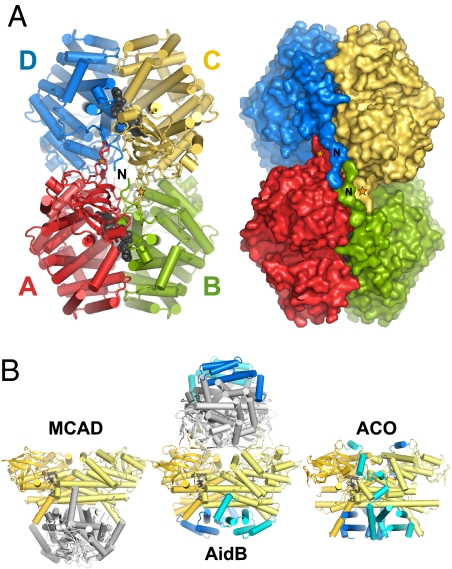
In the crystal structure, AidB forms a tetramer assembled by crystallographic symmetry (Fig. 2A). A second crystal form (Table S1) revealed an identical tetramer from subunits related by noncrystallographic symmetry within the asymmetric unit. Sedimentation velocity ultracentrifugation showed that AidB (60,590 Da per subunit) sediments as a 234-kDa protein, and gel filtration analysis was consistent with a tetramer in solutions as high as 500 mM ionic strength (data not shown). The AidB tetramer is a dimer-of-dimers in which the four subunits A, B, C, and D are related by D2 symmetry to approximate an ellipsoid with dimensions 120 × 94 × 60 Å3 (Fig. 2A). As in ACAD structures, AB and CD dimers are each formed from extensive contacts between domains II and III, with 3,350 Å2 (15% of the total) buried surface area per subunit. Two FAD molecules (one per subunit) are bound at each of these A/B and C/D dimer interfaces (Fig. 2A). Unlike the ACADs, however, AidB has a unique quaternary architecture formed by an extended random coil at the extreme N terminus and a β-hairpin loop (L1′2′) inserted between helices αB and αC that project outward to interlock AB and CD dimers together (Figs. 1C and 2 and Fig. S1).
Fig. 2.
The unique AidB tetramer. (A) Four subunits A, B, C, and D associate with dihedral symmetry as a dimer of dimers. The FAD cofactors at each of the A/B and C/D dimer interfaces are shown as black CPK spheres. N-terminal extensions and L1′2′ loops that form the AB/CD interface unique to AidB are labeled with the letter N and orange stars, respectively. The orientations of the cylinder and surface representations are identical, with the view down one two-fold axis. (B) Variation in subunit organization. MCAD tetramer (Left), AidB tetramer (Center), and ACO dimer (Right) are oriented with respect to the AB dimers (gold and yellow subunits). AidB is rotated 60° about a vertical axis with respect to the orientation in A. The subunits within the CD dimer in the AidB and MCAD tetramers are shaded light and dark gray. C-terminal domains in AidB and ACO are highlighted blue and cyan.
The quaternary structure of AidB is inverted with respect to the ACAD tetramer as a result of the novel N-terminal interface and the additional C-terminal domain IV. In ACADs, the AB/CD dimer interface is formed from helices αH and αK on domain III. In AidB, this interface is occluded by domain IV, which is a truncated version of that found in the dimeric acyl-CoA oxidase II (ACO) (9) (Fig. 2B). Interestingly, AidB helices αM, αN, αO, and αP (domain IV) superimpose remarkably well onto the MCAD symmetry-related helices αG, αH, αI, and αK (domain III) (Fig. S2). Thus, the core AidB dimer is stabilized by an additional C-terminal domain as opposed to the intersubunit interactions found in ACADs. In contrast to the ACO dimer, the N-terminal tetramerization platform on the opposite side of the AidB dimer provides two C-terminal domain surfaces per molecule of AidB.
The FAD Active-Site Cavity.
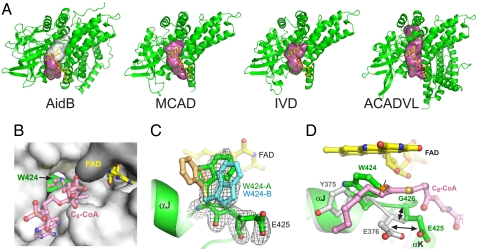
Proteins within the ACAD superfamily share conserved features inside their FAD active sites (8). The FAD binds in an extended conformation at the A/B subunit interface, with the isoalloxazine ring buried within a central cavity that contains catalytic residues (Fig. 3). Immediately adjacent to the FAD binding site is a substrate access channel that leads from the protein exterior into the flavin active site. A conserved phenylalanine/tyrosine residue (e.g., MCAD Tyr-375) forms one side of the active site at the dimethylphenyl side of the flavin ring and helps position the substrate for catalysis (Fig. 3D). The α,β-dehydrogenation/oxidation reactions in ACAD enzymes are typically catalyzed by a conserved glutamate carboxylate (e.g., MCAD Glu-376) and the isoalloxazine N5 nitrogen, which are positioned adjacent to the acyl chain Cα and Cβ carbons, respectively (10).
Fig. 3.
The FAD and putative substrate binding pocket. (A) Comparison of the substrate access cavities (magenta/white surfaces) in AidB, MCAD, IVD, and ACADVL (very long chain acyl-CoA dehydrogenase, PDB ID code 2UXW). The rear of the cavity blocked by Trp-424 in AidB is shown as a white surface. (B) The substrate access channel as viewed from the outside of the protein (gray surface). Yellow sticks, FAD; green, Trp-424. The octanoyl-CoA substrate (pink sticks) from the MCAD structure (PDB ID code 3MDE) is superimposed for reference. (C) Annealed omit electron density (3σ contours) for Trp-424 and Glu-425. The two Trp-424 conformers modeled into the density are shown as green and blue sticks. A hypothetical conformer (tan sticks) in the same position as Tyr-375 in ACAD structures is shown for reference only and was not included in the final structure. (D) Superposition of AidB (green) and MCAD (gray) structures. The AidB flavin is yellow, and the MCAD octanoyl-CoA substrate is pink. The black arrows highlight the shift in Glu-425 position, and the steric clash between Trp-424 and a fatty acyl substrate is shown as an orange starburst.
Several unique features distinguish AidB's FAD cavity from the ACAD active sites despite the conservation of their general properties (Fig. 3). The FAD–protein interactions within domains II and III are largely conserved among AidB and ACADs (Fig. 1C and Fig. S3). In addition, the overall dimensions of the AidB substrate access channel are very similar to those of ACADs (Fig. 3 A and B). However, the AidB channel is blocked at the flavin ring by Trp-424, which is in the same position in the primary sequence as MCAD Tyr-375 (Figs. 1C and 3D). Unbiased annealed omit electron density showed the Trp-424 indole side chain to exist in two predominant conformers, both of which are within van der Waals contact with the re-face of the isoalloxazine ring (Fig. 3C). The indole rings of these conformers are coplanar and are rotated 90° with respect to the conserved Phe/Tyr in the same position in ACADs (e.g., MCAD Tyr-375) (Fig. 3 C and D). Electron density in the position of MCAD Tyr-375 was not observed. This orientation of the indole ring sterically blocks access to the flavin N5 nitrogen and to the rear half of the putative substrate cavity. In addition, the bulky Trp-424 distorts the backbone of the αJ/αK loop and shifts the position of the Glu-425 side chain by ∼5 Å from its normal position in ACAD structures (Fig. 3D). The electron density and higher B-factors of Glu-425 (Fig. 3C) and Asn-421 (data not shown) side chains, both of which contact Trp-424, indicated higher mobility than other side chains in this cavity. Thus, the bulky Trp-424 side chain sterically blocks access to the chemically labile portion of the flavin and to the back half of the channel and significantly restructures the environment of the AidB putative substrate cavity.
DNA Binding by AidB.
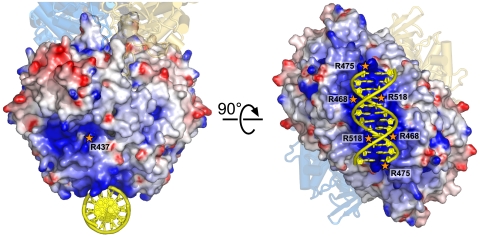
AidB was recently reported to possess DNA binding activity (7) that may be related to its response to DNA-damaging alkylating agents. Interestingly, AidB crystals formed only when DNA was present in the crystallization buffer, although we were unable to locate DNA in the electron density maps. The electrostatic landscape of AidB revealed two positively charged regions of the protein that were suspected to interact with DNA (Fig. 4). At domain IV, pairs of αM and αO helices across the A/B dyad axis of symmetry create a highly electropositive concave groove ∼20 Å wide, perfectly sized to accommodate a DNA duplex. In addition, the mouth of the conserved ACAD acyl-CoA substrate channel is positively charged, and proteolytic cleavage of AidB near this region (Met-194) was shown previously to be protected in the presence of DNA (7). To test these two regions as potential DNA binding sites, we measured the affinity of site-directed mutant proteins for 25mer dsDNA using a fluorescence anisotropy assay (Fig. S4). The wild-type protein binds DNA with a dissociation constant (Kd) of 4.3 ± 0.1 μM (Table 1). Substitution of Arg-518 with glutamine at domain IV reduced DNA binding affinity 30-fold relative to wild type. Likewise, Arg468Gln/Arg469Gln and Arg475Gln/Lys476Asn double mutations in domain IV reduced binding at least 100-fold and 6-fold, respectively. In contrast, glutamine substitution of Arg-437 at the mouth of the ACAD substrate cavity did not significantly affect DNA binding (Table 1).
Fig. 4.
Theoretical model of DNA bound to the C-terminal domain of AidB. The solvent-accessible surface of the AB dimer is colored according to electrostatic potential (red, negative; blue, positive; −7 to +7 kBT). The CD dimer (gold and blue subunits) is shown as a cartoon for reference. The dsDNA dodecamer (yellow) was docked onto the AidB surface using mutational DNA binding data as physical restraints. Mutations tested for DNA binding are labeled with orange stars. Potentials were calculated with DelPhi (27) and rendered with PyMOL (www.pymol.org).
Table 1.
DNA binding activity of wild-type and mutant AidB
| Mutant | Kd, μM | Relative binding |
|---|---|---|
| WT | 4.3 ± 0.1 | 1.00 |
| R437Q | 6.1 ± 1.6 | 0.70 |
| R468Q/R469Q | >300 | <0.01 |
| R475Q/K476N | 26.6 ± 5.2 | 0.16 |
| R518Q | 135.5 ± 35 | 0.03 |
Kd values for a 25mer oligonucleotide duplex were measured by fluorescence anisotropy as described in Experimental Procedures.
Discussion
We report here a high-resolution structure and detailed DNA binding data for AidB, the most enigmatic of the proteins involved in the adaptive response to alkylation agents in E. coli. AidB has been proposed to repair alkylation damaged DNA, similarly to its counterparts Ada, AlkA, and AlkB. Alternatively, AidB's response to E. coli exposure to MNNG, but not MMS or MNU, raises the possibility that AidB acts to protect DNA against alkylation by chemically detoxifying MNNG derivatives themselves (4–6). Although the specific role remains to be determined, the structural features of AidB's unique DNA binding domain, subunit organization, and FAD chemical environment argue for a protective role for AidB. Furthermore, the structure helps to dispel the model that the flavin cofactor is an active participant in the dealkylation of DNA.
Mutational analysis of DNA binding by AidB identified domain IV as the DNA binding site. To illustrate how DNA might engage domain IV, we constructed a model of dsDNA docked onto the molecular surface using the mutagenesis data as restraints (Fig. 4). Our DNA docking model shows that the 40-Å-long positively charged groove will accommodate 12 base pairs of linear duplex DNA, with basic side chains conveniently positioned to interact with the DNA backbone. DNA binding by AidB was highly salt-dependent, consistent with an extended electrostatic binding site. Importantly, there are no protein contacts with the DNA bases in our docking model that would allow for preferential binding of AidB to methylated DNA. This is consistent with previous studies showing that AidB binds methylated and unmethylated plasmid DNA with equal affinity (7) and with results from this laboratory that AidB does not bind oligonucleotides containing abasic sites differently than unmodified DNA (data not shown).
AidB's inverted tetrameric assembly relative to the ACAD superfamily exposes DNA binding surfaces at each end of the tetramer. The lack of positive charge around the sides of the tetramer render it unlikely that the DNA wraps around the protein. Nevertheless, nonspecific DNA binding at the ends of the tetramer suggests that AidB might function to protect naked DNA from attack by methylating agents. A similar role has been observed in the Dps protein, which protects DNA in starved E. coli against oxidative damage (11, 12). Both AidB and Dps are up-regulated during stationary phase and are rpoS-dependent (13, 14). Interestingly, endogenous methylating agents such as nitrosamines are formed as by-products of stationary-phase metabolism (15). This leads to an accumulation of alkylation damage to DNA during stationary phase, as demonstrated by an increase in spontaneous mutation in methyltransferase (ada ogt)-deficient E. coli in nondividing cells (16). Induction of AidB expression, therefore, could serve to prevent endogenous stationary-phase alkylation damage in a manner similar to Dps protection of oxidative damage. Dps protection of DNA is believed to occur by binding DNA duplexes within the pores of hexagonally packed Dps dodecamers (17). Interestingly, both crystal forms of AidB display a similar crystal packing in that the putative DNA binding faces are clustered symmetrically around 25-Å pore that is perfectly sized to accommodate DNA (Fig. S5). These observations, together with the fact that DNA is essential for AidB crystallization, suggest that AidB tetramers cluster around DNA to restrict access by damaging agents.
Our mutational data of AidB's DNA binding activity demonstrate that AidB does not engage DNA at the mouth of the ACAD acyl-CoA substrate binding cavity. This cavity is the only entry point into the FAD active site and was therefore a tantalizing possibility for DNA binding if AidB were to repair damaged DNA by a FAD-dependent mechanism. However, the inability of the Arg437Gln mutation to affect DNA binding (Table 1 and Fig. 4), along with the following structural observations, suggest that this cavity does not engage DNA. First, the positive charge of this region is a conserved feature of ACADs that likely serves to balance the negative charge of the pyrophosphate moiety of CoA. Second, the two positive patches at this cavity and on domain IV are discontinuous, suggesting that DNA does not wrap from one site to the other. Third, the mouth of the active site cavity is separated from the FAD isoalloxazine ring by 13 Å, which would prevent DNA from reaching the active site from the outside of the protein. These observations argue against the notion that AidB repairs DNA by an FAD-dependent mechanism.
The unique features of the FAD pocket provide a structural rationale for AidB's weak isovaleryl dehydrogenase activity and for the absence of flavin spectral perturbation in the presence of isovaleryl-CoA (4, 7). Specifically, AidB differs significantly from the ACAD enzymes in the residues important for positioning the substrates in close proximity to the flavin. Despite the fact that the channel leading to the flavin in AidB is similar in size and shape to the ACAD substrate binding site, the Trp-424 plug restricts access to the rear half of the tunnel and, more importantly, to the isoalloxazine N5 nitrogen involved in the α,β-dehydrogenation reaction (Fig. 3) (10). It has been proposed that the glycine in this position in IVD allows for binding branched acyl chains (8, 18). In addition to the Trp-424 steric block, the 5-Å displacement of the Glu-425 carboxylate from its normal position in ACADs (Fig. 3C) would render the carboxylate ineffective at abstracting the Cα-H hydrogen from the acyl-CoA substrate. Although the FAD cavity is spacious enough for Trp-424 and Glu-425 side chains to shift position, such a large conformational change to accommodate an extended acyl chain is not likely to occur in light of the small active-site changes observed upon acyl-CoA binding in MCAD and isobutyryl-CoA dehydrogenase (19, 20). Thus, it appears that the elongated FAD cavity in AidB is an evolutionary remnant and not a bona fide active site for fatty or amino acid acyl-CoA thioesters.
Does AidB perform catalysis inside the FAD binding pocket? The volume of this cavity (≈290 Å2) is large enough to accommodate a small molecule. Furthermore, the mobility of the Asn-421, Trp-424, and Glu-425 side chains suggests that these residues may bind a substrate. AidB has been proposed to detoxify MNNG or one of its reactive metabolites (4). Unlike MMS or MNU, MNNG is activated by thiols, including glutathione, to produce highly reactive methylation agents (e.g., methyldiazonium ion) (21). Interestingly, AidB's FAD cavity is rich in thiol and methylsulfide groups, a characteristic distinctly different from other structural homologs. In fact, four of 13 solvent-accessible residues lining the AidB pocket are cysteine or methionine, whereas there are no such residues in contact with substrates in MCAD, IVD, ACO, or NAO (Fig. 1C). It is therefore intriguing to speculate that Cys-133, Met-137, Met-182, and Cys-302 may alter the redox potential of AidB's pocket relative to those of other, functionally unrelated enzymes. In particular, Cys-133 and Met-137 sulfur atoms are positioned 5.8 and 4.3 Å from the flavin, respectively, and are oriented toward the mouth of the substrate channel (Fig. S6). Thus, it appears that AidB's putative active site may act as a sink for reactive MNNG derivatives. Coupled with the nonspecific DNA binding function, this suggests that AidB may be the cell's last line of defense to prevent DNA methylation by trapping the compounds before they are able to react with DNA.
Experimental Procedures
Protein Purification and Crystallization.
The E. coli aidB gene was PCR-amplified from genomic DNA and ligated into a modified pET-19b (Novagen) expression vector. AidB protein was overexpressed in E. coli C41 cells for 3 h at 37°C, and selenomethionyl (SeMet)-AidB was produced by using the method described by Van Duyne et al. (22). AidB was isolated from the cell lysate by Ni2+-NTA (Qiagen) affinity chromatography and further purified after cleavage of the His10 tag by heparin-Sepharose chromatography in 50 mM Tris (pH 7.5), 5% glycerol, 2 mM DTT, 0.1 mM EDTA, and a linear gradient of 0.2–1.0 M NaCl. The SeMet-AidB buffer was supplemented with 5 mM methionine and 20 mM DTT after the Ni2+-NTA step.
For crystallization, AidB was concentrated to 10 mg/ml (ε280 = 1.25 mg−1·ml) in 20 mM Hepes (pH 6.8), 250 mM NaCl, 2 mM DTT, and 0.1 mM EDTA. Initial crystallization screens were performed in the presence of 1.2-fold molar excess double-stranded oligonucleotides ranging 10–30 nucleotides in length, from which the highest-quality crystals were obtained with a 25-mer, d(GACTATGCGACAATGCGCCTCCATT)/d(AATGGAGGCGCATTGTCGCATAGTC). Crystals were grown from sitting drop vapor diffusion at 21°C using equal volumes of protein/DNA and reservoir solutions. Native AidB grew from 100 mM Tris (pH 8.0), 50 mM CaCl2, and 4% PEG 2000. Two different crystal forms of SeMet-AidB grew from 50 mM sodium cacodylate (pH 6.5), 40–100 mM magnesium acetate, and 15–27.5% PEG 400 (Se-1) and from 100 mM Mes (pH 6.0), 50 mM CaCl2, and 2% PEG 2000 (Se-2). The mercury derivative was prepared by soaking native AidB crystals for 20 h at 21°C in mother liquor that contained 10% PEG 2000 and 0.5 mM MeHgCl.
X-Ray Data Collection, Phasing, and Structure Refinement.
Native and Se-2 crystals were soaked briefly in mother liquor containing 10% PEG 2000 and 25% ethylene glycol and frozen in liquid nitrogen before data collection, and Se-1 crystals were frozen straight from the mother liquor. Diffraction data (Table S1) were collected at beamlines 22-ID (native) and 21-ID (SeMet, Hg) at the Advanced Photon Source (Argonne, IL) and processed with HKL2000 (23). Experimental phases were determined by MIRAS using mercury (2.5 Å) and Se-1 (1.92 Å) derivatives referenced against the native. Positions of 16 Hg and 21 Se positions were located, and phases were improved by density modification with autoSHARP (24). An atomic model consisting of residues 3–540 in a single polypeptide chain and including side chains was built manually into the 1.92-Å MIRAS electron density map (Fig. 1A) using XtalView/Xfit. Electron density corresponding to DNA could not be identified. Atomic coordinates and B-factors were refined by using REFMAC 5.4 (25). The final model contained 94.6% non-glycine and non-proline residues in the most favored regions, 5.3% in the allowed regions, and 0% in the disallowed regions of the Ramachandran plot. Structure factors and the atomic model have been deposited in the Protein Data Bank under ID code 3DJL.
Mutagenesis and DNA Binding Assays.
Mutations were made in the AidB expression plasmid using the QuikChange Mutagenesis Kit (Stratagene). Mutant proteins were expressed and purified in the same manner as wild-type AidB, and structural integrity was verified by circular dichroism spectroscopy. DNA binding measurements were carried out in 50 mM Hepes (pH 6.8), 200 mM NaCl, 5% glycerol, 2 mM DTT, and 0.1 mM EDTA. Binding was monitored as an increase in fluorescence anisotropy as AidB (0–40 μM) was added to a 50 nM solution of the same 25mer DNA used in crystallization and containing a 6-carboxyfluorescein at the 3′ end of one strand. Polarized fluorescence intensities were measured at excitation and emission wavelengths of 495 and 538 nm, respectively. Dissociation constants (Kd) were derived by fitting a simple two-state binding model to data from three experiments using Kaleidagraph 3.6 (Synergy Software).
Detailed Methods.
Molecular modeling and hydrodynamic measurements are described in SI Text.
Supplementary Material
Acknowledgments.
We thank Jeffry Lary and James Cole at the National Analytical Ultracentrifugation Facility (University of Connecticut, Storrs, CT). Use of the Advanced Photon Source was supported by the U.S. Department of Energy Office of Basic Energy Sciences under Contract No. DE-AC02-06CH11357. Use of LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor for the support of this research program (Grant 085P1000817). This work was funded by the American Cancer Society (RSG-07-063-01-GMC to B.F.E.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3DJL).
This article contains supporting information online at www.pnas.org/cgi/content/full/0806521105/DCSupplemental.
References
- 1.Sedgwick B. Repairing DNA-methylation damage. Nat Rev. 2004;5:148–157. doi: 10.1038/nrm1312. [DOI] [PubMed] [Google Scholar]
- 2.Samson L, Cairns J. A new pathway for DNA repair in Escherichia coli. Nature. 1977;267:281–283. doi: 10.1038/267281a0. [DOI] [PubMed] [Google Scholar]
- 3.Sedgwick B, Bates PA, Paik J, Jacobs SC, Lindahl T. Repair of alkylated DNA: Recent advances. DNA Repair (Amsterdam) 2007;6:429–442. doi: 10.1016/j.dnarep.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 4.Landini P, Hajec LI, Volkert MR. Structure and transcriptional regulation of the Escherichia coli adaptive response gene aidB. J Bacteriol. 1994;176:6583–6589. doi: 10.1128/jb.176.21.6583-6589.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Volkert MR, Nguyen DC. Induction of specific Escherichia coli genes by sublethal treatments with alkylating agents. Proc Natl Acad Sci USA. 1984;81:4110–4114. doi: 10.1073/pnas.81.13.4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Volkert MR, Nguyen DC, Beard KC. Escherichia coli gene induction by alkylation treatment. Genetics. 1986;112:11–26. doi: 10.1093/genetics/112.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rohankhedkar MS, Mulrooney SB, Wedemeyer WJ, Hausinger RP. The AidB component of the Escherichia coli adaptive response to alkylating agents is a flavin-containing, DNA-binding protein. J Bacteriol. 2006;188:223–230. doi: 10.1128/JB.188.1.223-230.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim JJ, Miura R. Acyl-CoA dehydrogenases and acyl-CoA oxidases. Structural basis for mechanistic similarities and differences. Eur J Biochem. 2004;271:483–493. doi: 10.1046/j.1432-1033.2003.03948.x. [DOI] [PubMed] [Google Scholar]
- 9.Nakajima Y, et al. Three-dimensional structure of the flavoenzyme acyl-CoA oxidase-II from rat liver, the peroxisomal counterpart of mitochondrial acyl-CoA dehydrogenase. J Biochem. 2002;131:365–374. doi: 10.1093/oxfordjournals.jbchem.a003111. [DOI] [PubMed] [Google Scholar]
- 10.Ghisla S, Thorpe C. Acyl-CoA dehydrogenases. A mechanistic overview. Eur J Biochem. 2004;271:494–508. doi: 10.1046/j.1432-1033.2003.03946.x. [DOI] [PubMed] [Google Scholar]
- 11.Almiron M, Link AJ, Furlong D, Kolter R. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 1992;6:2646–2654. doi: 10.1101/gad.6.12b.2646. [DOI] [PubMed] [Google Scholar]
- 12.Martinez A, Kolter R. Protection of DNA during oxidative stress by the nonspecific DNA-binding protein Dps. J Bacteriol. 1997;179:5188–5194. doi: 10.1128/jb.179.16.5188-5194.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Volkert MR, Hajec LI, Matijasevic Z, Fang FC, Prince R. Induction of the Escherichia coli aidB gene under oxygen-limiting conditions requires a functional rpoS (katF) gene. J Bacteriol. 1994;176:7638–7645. doi: 10.1128/jb.176.24.7638-7645.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Altuvia S, Almiron M, Huisman G, Kolter R, Storz G. The dps promoter is activated by OxyR during growth and by IHF and sigma S in stationary phase. Mol Microbiol. 1994;13:265–272. doi: 10.1111/j.1365-2958.1994.tb00421.x. [DOI] [PubMed] [Google Scholar]
- 15.Taverna P, Sedgwick B. Generation of an endogenous DNA-methylating agent by nitrosation in Escherichia coli. J Bacteriol. 1996;178:5105–5111. doi: 10.1128/jb.178.17.5105-5111.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rebeck GW, Samson L. Increased spontaneous mutation and alkylation sensitivity of Escherichia coli strains lacking the ogt O6-methylguanine DNA repair methyltransferase. J Bacteriol. 1991;173:2068–2076. doi: 10.1128/jb.173.6.2068-2076.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grant RA, Filman DJ, Finkel SE, Kolter R, Hogle JM. The crystal structure of Dps, a ferritin homolog that binds and protects DNA. Nat Struct Biol. 1998;5:294–303. doi: 10.1038/nsb0498-294. [DOI] [PubMed] [Google Scholar]
- 18.Tiffany KA, et al. Structure of human isovaleryl-CoA dehydrogenase at 2.6 A resolution: Structural basis for substrate specificity. Biochemistry. 1997;36:8455–8464. doi: 10.1021/bi970422u. [DOI] [PubMed] [Google Scholar]
- 19.Kim JJ, Wang M, Paschke R. Crystal structures of medium-chain acyl-CoA dehydrogenase from pig liver mitochondria with and without substrate. Proc Natl Acad Sci USA. 1993;90:7523–7527. doi: 10.1073/pnas.90.16.7523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Battaile KP, Nguyen TV, Vockley J, Kim JJ. Structures of isobutyryl-CoA dehydrogenase and enzyme-product complex: Comparison with isovaleryl- and short-chain acyl-CoA dehydrogenases. J Biol Chem. 2004;279:16526–16534. doi: 10.1074/jbc.M400034200. [DOI] [PubMed] [Google Scholar]
- 21.Lawley PD. Some chemical aspects of dose-response relationships in alkylation mutagenesis. Mutat Res. 1974;23:283–295. doi: 10.1016/0027-5107(74)90102-x. [DOI] [PubMed] [Google Scholar]
- 22.Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, Clardy J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J Mol Biol. 1993;229:105–124. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- 23.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 24.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 25.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 26.Nagpal A, Valley MP, Fitzpatrick PF, Orville AM. Crystal structures of nitroalkane oxidase: Insights into the reaction mechanism from a covalent complex of the flavoenzyme trapped during turnover. Biochemistry. 2006;45:1138–1150. doi: 10.1021/bi051966w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rocchia W, et al. Rapid grid-based construction of the molecular surface and the use of induced surface charge to calculate reaction field energies: Applications to the molecular systems and geometric objects. J Comput Chem. 2002;23:128–137. doi: 10.1002/jcc.1161. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.