Abstract
Proper regulation of the intracellular ion concentration is essential to maintain life and is achieved by ion transporters that transport their substrates across the membrane in a strictly regulated manner. MgtE is a Mg2+ transporter that may function in the homeostasis of the intracellular Mg2+ concentration. A recent crystallographic study revealed that its cytosolic domain undergoes a Mg2+-dependent structural change, which is proposed to gate the ion-conducting pore passing through the transmembrane domain. However, the dynamics of Mg2+ sensing, i.e., how MgtE responds to the change in the intracellular Mg2+ concentration, remained elusive. Here we performed molecular dynamics simulations of the MgtE cytosolic domain. The simulations successfully reproduced the structural changes of the cytosolic domain upon binding or releasing Mg2+, as well as the ion selectivity. These results suggested the roles of the N and CBS domains in the cytosolic domain and their respective Mg2+ binding sites. Combined with the current crystal structures, we propose an atomically detailed model of Mg2+ sensing by MgtE.
Keywords: magnesium homeostasis, molecular dynamics simulation
Proper regulation of the intracellular ion concentration is crucial for organisms. Ion transporters are responsible for transporting ions across the cellular membrane in a strictly regulated manner. This regulation is accomplished by converting the physical stimuli or the chemical environment into the structural changes of the transporter protein. Although these regulatory mechanisms have been investigated from a structural point of view for several membrane proteins, including Mg2+ transporters (1–4), the Ca2+-activated K+ channel MthK (5) AmtB-GlnK (6, 7), and an acid-sensing ion channel (8), the molecular mechanism of the regulation, i.e., how these proteins provide a response to the change in the chemical environment, is still poorly understood.
MgtE is a Mg2+ transporter conserved in all three kingdoms of life. Its Mg2+-transporting activity was demonstrated by a complementation assay and the uptake of a Co2+ isotope in vivo (9). In bacteria, translation of its gene is regulated by a Mg2+-specific riboswitch (10). Its human homologues have been functionally characterized and may be involved in the Mg2+ homeostasis in the cell (11–13). Recently we solved the crystal structures of full-length MgtE and of its cytosolic domains in the presence and absence of Mg2+ (4). A comparison of these crystal structures revealed that a large structural change occurs in the cytosolic domain upon binding or releasing Mg2+. The cytosolic domain is therefore proposed to act as a sensor for the intracellular Mg2+ concentration, and its structural change may induce a rearrangement of the transmembrane (TM) helices, which control the gating of the ion-transduction pore.
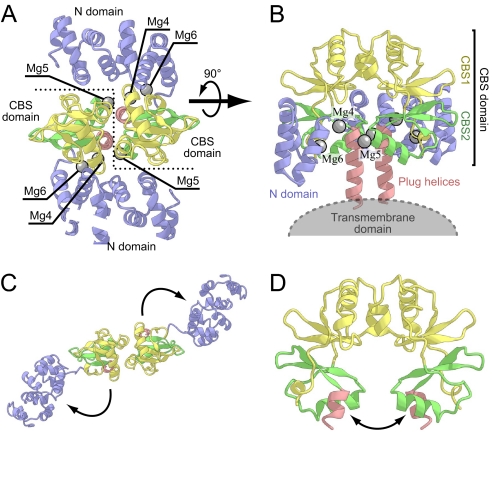
The cytosolic domain of MgtE is composed of the NH3+-terminal (N) domain and the cystathionine β-synthase (CBS) domain (Fig. 1). The crystal structure obtained under Mg2+-excess conditions revealed that MgtE forms a dimer, in which the CBS domains from both subunits face each other and several Mg2+ are bound at the CBS-CBS and N-CBS domain interfaces (“closed” form; Fig. 1 A and B). In contrast, in the crystal structure under the Mg2+-free conditions, the orientations of these domains were drastically changed (“open” form; Fig. 1 C and D); the two CBS domains are separated from each other by a 40° rotation, and the N domain moves away from the following CBS domain by a 120° rotation. These structural changes may control the gating of the ion-conducting pore through the “plug helices” (formerly referred to as the “connecting helix”)‖, composed of a long α-helix connecting the CBS and TM domains. Especially, the structural change of the CBS domain may play a crucial role in regulating the transporter activity by sensing the intracellular Mg2+ concentration, because it directly affects the orientation of the plug helices (Fig. 1 B and D). In fact, CBS domains are present in other transporter proteins and may be involved in the regulation of their transporting activities (14–16). On the other hand, despite its large movement, the functional role of the N domain remained elusive. The domain forms a right-handed superhelix, consisting of 10 α-helices, which shares structural similarity with the C-terminal domain of the human FANCF protein (17). However, no functional relationship has been found between the FANCF protein and MgtE.
Fig. 1.
Crystal structure of the MgtE cytosolic domain (PDB ID code 2YVY) used as the initial structure for the simulations. Protein backbones are depicted by ribbon models. The N domain, CBS1, CBS2, and the plug helix are colored blue, yellow, green, and red, respectively. Mg2+ ions (Mg4, Mg5, and Mg6) investigated in the present simulations are shown as gray spheres. (A) View from the intracellular region. The boundary of the subunits is indicated by a dashed line. (B) View in the plane of the membrane. Part of the N domain (1–88) of molecule B is not shown. (C and D) The crystal structure of the MgtE cytosolic domain under the Mg2+-free conditions viewed from the same directions as in A and B, respectively. The directions of the observed structural change are designated by arrows. This figure and the other figures were generated with the programs CueMol (http://www.cuemol.org/) and Warabi (http://park11.wakwak.com/∼warabiya/).
Here we performed a series of molecular dynamics (MD) simulations of the MgtE cytosolic domain (see Table 1). When the bound Mg2+ are removed from the closed form of MgtE, the cytosolic domain spontaneously converts to the open form. The resulting open form is in good agreement with the crystal structure of the Mg2+-unbound form. By reducing the number of bound Mg2+, we also examined the role of each Mg2+ in the stability of the closed-form structure. Furthermore, we observed the structural changes occurring upon the transition from the open to closed form by adding Mg2+ at the proper binding sites in the open structure of MgtE. These reversible simulations provided insights into the roles of the N and CBS domains and allowed us to propose a detailed dynamic model of Mg2+ sensing and transport by MgtE.
Table 1.
Bound ions, lengths, and initial structures of the simulation runs investigated in this work
| Run | Initial structure* | Bound ions at |
Simulation length, ns | ||
|---|---|---|---|---|---|
| Site 4 | Site 6 | Site 5 | |||
| C465 | X-ray; 5–262 | Mg2+ | Mg2+ | Mg2+ | 10.5 |
| C0 | X-ray; 5–262 | — | — | — | 20 |
| C5 | X-ray; 5–262 | — | — | Mg2+ | 10.5 |
| C46 | X-ray; 5–251 | Mg2+ | Mg2+ | — | 10 |
| Na5 | X-ray; 5–262 | — | — | Na+ | 10 |
| Ca5 | X-ray; 5–262 | — | — | Ca2+ | 12 |
| O5 | From C0 run; 132–251 | — | — | Mg2+ | 30 × 2 |
| O465 | From C0 run; 5–251 | Mg2+ | Mg2+ | Mg2+ | 50 |
*The numbers indicate protein residues included in the simulations.
Results
The Closed Crystal Structure of MgtE Is Stable, with 6 Mg2+.
The MgtE cytosolic domain is composed of the α-helical N domain (1–131) and the CBS (132–245) domain (Fig. 1). The CBS domain comprises the tandemly repeated CBS motifs, which are hereafter denoted as the CBS1 (132–203) and CBS2 (204–245) domains, respectively. The “plug helix” (246–263) is a long α-helix connecting the CBS and TM domains, which may affect the gating of the ion-conducting pore in the TM domains. The MgtE cytosolic domain forms a dimer, mainly through the interactions between the CBS1 and CBS2 domains. In the crystal structures, three Mg2+ binding sites are observed per protomer of the MgtE cytosolic domain: one is site 5 between the CBS domains of both subunits, and the others are sites 4 and 6 between the N and CBS domains (hereafter we refer to the Mg2+ ions bound at sites 4, 5, and 6 as Mg4, Mg5, and Mg6, respectively) (Fig. 1A). Electron densities for Mg4 and Mg6 with coordinated water molecules are clearly visible in the crystal structure of the Mg2+-bound cytosolic domains [Protein Data Bank (PDB) ID code 2YVY]. On the other hand, whereas a high-electron-density peak corresponding to Mg5 was observed in the crystal structure (PDB ID code 2YVY), we could not identify electron density peaks for the coordinated water molecules. We therefore included only two Mg2+ (Mg4 and Mg6) in the deposited coordinates (PDB ID code 2YVY), considering the uncertainty about whether the peak corresponding to Mg5 can actually be ascribed to Mg2+. One of the purposes of this study was to determine the lowest number of Mg2+ needed to stabilize the closed form of MgtE.
We first performed an MD simulation of MgtE with the three Mg2+ at the respective binding sites, starting from the crystal structure of the Mg2+-bound MgtE cytosolic domain (closed form; PDB ID code 2YVY) (Fig. 2). In the simulation with the three Mg2+ (C465; Table 1), the crystal structure remained stable without any interdomain and intradomain motions throughout the 10-ns simulation [Fig. 2 C and G and supporting information (SI) Figs. S1 and S2]. In the simulation, the Mg2+ binding at site 5 as well as those at sites 4 and 6 remained stable (Fig. S3). In addition, their coordination manners, including the water molecules, were unchanged: Mg2+-binding at site 4 involves the two carboxyl oxygens of Asp-91 (N domain) and Asp 247 (plug helix) as well as four coordinating water molecules, whereas the carboxyl oxygen of Asp-95 (N domain), the carbonyl oxygen of Gly-136 (CBS1), and four water molecules form site 6. At site 5, four water molecules were bound to the Mg2+, in addition to the carboxyl oxygen of Asp-226 and the carbonyl oxygen of Ala-223 (CBS2 in subunit A). Two of the water molecules were strictly fixed by the carboxyl oxygens of Asp-250 in the plug helix of subunit B (Fig. S3B). These results strongly support our interpretation that the high-electron-density peaks at sites 4, 5, and 6 observed in the crystal structure stem from Mg2+.
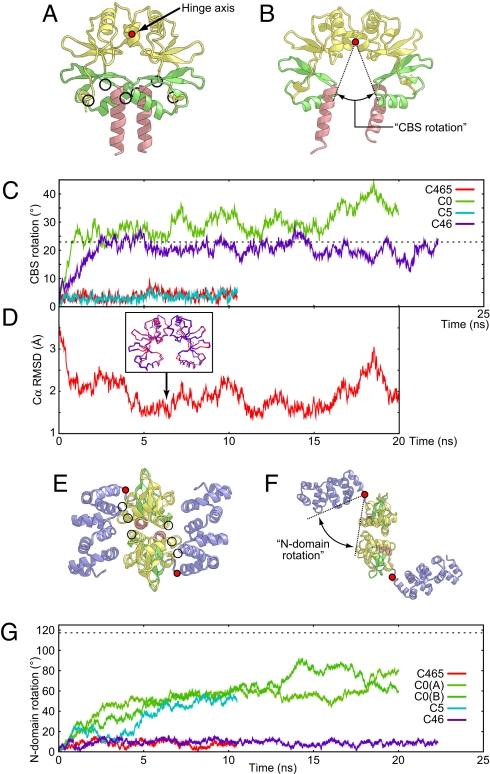
Fig. 2.
The closed-to-open form structural changes observed in the simulations. (A and B) The initial (A) and final (B) structures in the C0 simulation, viewed in the plane of the membrane. The protein backbones of the CBS domains and the plug helices (residues 132–262) are depicted by ribbon models, with the same coloring scheme as Fig. 1. The positions of the hinge axis (perpendicular to paper) of the CBS rotation are indicated as filled red circles. Mg2+ binding sites are indicated by circles in A. (C) CBS rotation as a function of time during the C465 (red), C0 (green), C5 (cyan), and C46 (blue) simulations. The value corresponding to the open-form crystal structure (23°) is indicated by the dashed horizontal line. (D) Structural deviation (Cα rmsd over residues 132–252) from the Mg2+-free crystal structure (PDB ID code 2YVZ) as a function of time during the C0 simulation. A superposition of the minimal rmsd snapshot from the C0 simulation (at 6.554 ns; blue) and the crystal structure (red) is shown in the Inset. (E and F) The initial (E) and final (F) structures in the C0 simulation viewed from the intracellular region. The protein backbones of the entire cytosolic domains (residues 5–262) are depicted by ribbon models, with the same coloring scheme as Fig. 1. The positions of the hinge axis (perpendicular to paper) of the N-domain rotation are indicated as filled red circles. Mg2+ binding sites are indicated by circles in E. (G) N-domain rotation as a function of time during the C465 (red), C0 (green and yellow green), C5 (cyan), and C46 (blue) simulations. For the C0 simulation, both of the rotations for the A and B subunits are shown. The value corresponding to the open-form crystal structure (117°) is indicated by the dashed horizontal line.
Structural Changes from the Closed to Open Form upon Mg2+ Removal.
In the simulation without any Mg2+ (C0; Table 1), the magnitude of the intradomain motions for the N and CBS domains was similar to that in the simulation with three Mg2+ (C465; Table 1), suggesting that the bound Mg2+ do not affect the internal conformations of the N and CBS domains (Fig. S2). By contrast, we observed large domain movements, which could be divided into the intersubunit movements of the CBS domains and the movements of the N domains from the following CBS domains. To study the former, we evaluated the rotational angle between the two CBS domains by fixing one subunit (“CBS rotation” in Fig. 2B). The latter movement was also evaluated by calculating the rotational angle of the N domain relative to the CBS domain in each protomer (“N-domain rotation” in Fig. 2F). Because the cytosolic domain of MgtE forms a homodimer, two N-domain rotations for the A and B subunits can be obtained.
Without Mg2+ in the C0 simulation, the CBS domains in the two subunits quickly opened, and the CBS rotation finally reached 45° after the 20-ns simulation (Fig. 2 A–C). The open conformation of the CBS domains that appeared in the C0 simulation is in good agreement with the crystal structure under the Mg2+-free conditions (PDB ID code 2YVZ), and the minimum rmsd against the crystal structure is 1.3 Å (Fig. 2D). At the same time, the N domain of each subunit also swung away from the CBS domain (Fig. 2 E and F). The N-domain rotations of the A and B subunits both underwent similar changes and attained about 90° after the 20-ns simulation (Fig. 2G). In the other simulations, the N-domain rotations of the A subunit also exhibited tendencies similar to those of the B subunit, and thus we selected those of the A subunit to clarify the following discussion. To characterize the domain rotation, we examined the hinge axes of the CBS and N domains. The hinge axis of the CBS rotation lies parallel to the membrane surface and passes between the Ile-190 side chains (in CBS1) of both subunits, which are far away from Mg5 (Fig. 2 A and B). The hinge axes of the N-domain rotation are perpendicular to the membrane and pass near the linker region between the N and CBS domains (residues 127–129; Fig. 2 E and F).
To investigate the structural role of each Mg2+ site, we also performed simulations after the removal of selected Mg2+ ions (C5 and C46; Table 1). In the C5 simulation, in which both Mg4 and Mg6 were removed, the N-domain rotation increased to the same degree as observed in the C0 simulation (Fig. 2G). By contrast, the orientation of the CBS domains did not change from the initial structure, as in the C465 simulation (Fig. 2C). In the C46 simulation, in which Mg5 was removed (Table 1), the N domain orientation was mostly locked in the closed form, as observed in the C465 simulation (Fig. 2G). On the other hand, the CBS rotation approaches the value in the open conformation, as observed in the C0 simulation and the Mg2+-free crystal structure (Fig. 2C). These simulations demonstrated that the CBS rotation exclusively depends on Mg5, supporting our interpretation that the electron density peak observed at site 5 in the crystal structure stems from Mg2+. They also showed that the conformational change of the N domains exclusively depends on Mg2+ binding at sites 4 and 6, which are located at the interface between the N and CBS domains.
Simulations with Cations Other than Mg2+.
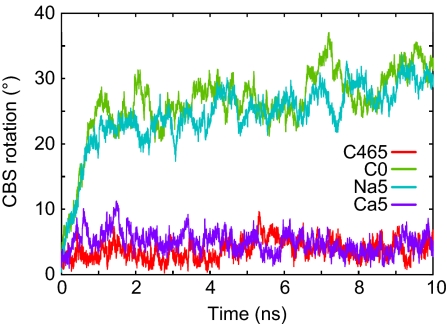
To investigate the ion selectivity at site 5, we performed simulations with the replacement of Mg5 with Na+ and Ca2+ (Na5 and Ca5; Table 1). In the Na5 simulation, a Na+ ion, which was initially bound at site 5 in one of the subunits, was dissociated immediately after the equilibration (Fig. S4 A and B), with the CBS rotation increasing quickly, as in the C46 and C0 simulations (Fig. 3). Finally, the Na+ at site 5 of the other subunit also diffused into the solvent after the 10-ns simulation (Fig. S4A). On the other hand, in the Ca5 simulation, the orientation of the CBS domains did not change from that in the initial structure, as in the C465 simulation (Fig. 3). Both of the Ca2+ ions were stably bound throughout the simulation. However, the Ca2+ was bound to site 5 with a different coordination manner from that of Mg2+; i.e., the Asp-250 of one subunit and the Asp-226 of the other subunit made bidentate interactions with Ca2+, with both subunits directly bridged by the Ca2+ (Fig. S3E).
Fig. 3.
CBS rotation as a function of time during the Na5 (cyan) and Ca5 (purple) simulations. Profiles for the C465 (red) and C0 (green) simulations are also shown for comparison.
Structural Changes from the Open to Closed Form upon Mg2+ Addition.
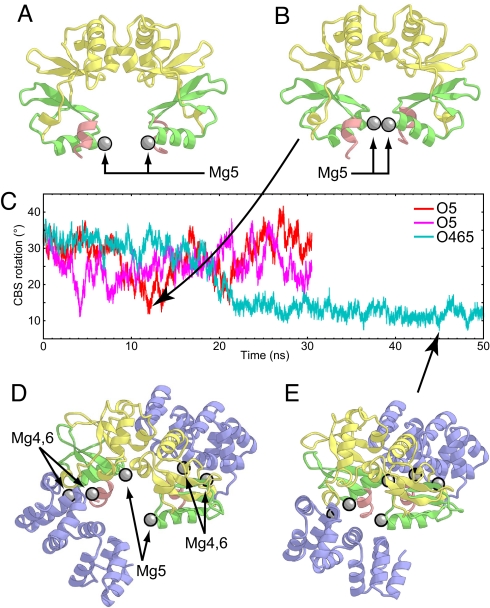
To investigate the open-to-closed transition of the CBS rotation, we performed MD simulations starting from the two different open-form structures (O5 and O465; Table 1). At first, we performed the 30-ns simulations starting from the open-form structure without the N domains (Fig. 4A) (O5; Table 1). In the course of the simulations, Mg5 stably bound to Asp-224 and Ala-223 despite the absence of a water-mediated interaction with Asp-250 (Fig. S5A). Unexpectedly, the CBS rotation fluctuated between the open and closed forms several times (Fig. 4C). This fluctuation was reproducibly observed in two independent simulations.
Fig. 4.
The open-to-closed form structural changes observed in the simulations. (A and B) The initial structure (A) and the most closed structure at 11.574 ns (B) in the O5 simulation viewed in the plane of the membrane. The protein backbones are depicted by ribbon models, with the same coloring scheme as in Fig. 1. Mg2+ ions are shown as gray spheres. (C) CBS rotation as a function of time during the two O5 (red and magenta) and O465 (cyan) simulations. (D and E) The initial structure (D) and the most closed structure at 44.912 ns (E) in the O465 simulation viewed from the intracellular region. The protein backbones are depicted by ribbon models, with the same coloring scheme as in Fig. 1. Mg2+ ions are shown as gray spheres.
Next we performed the 50-ns MD simulation starting from the initial structure including the N domain (O465; Table 1). The aim of this O465 simulation was to see how the closed-form N domain affects the CBS rotation. Therefore, we modeled the N domains to adopt the closed form, as in the crystal structure, whereas the orientation between the CBS domains of both subunits was modeled to adopt the same open form as in the initial structure of the O5 simulation (Fig. 4D). The six Mg2+ ions were placed at the proper Mg2+ binding sites (see Methods for the detailed modeling procedure). Throughout the simulation, Mg4, Mg5, and Mg6 remained stably bound to the protein without changing their coordination manners (Fig. S5B). As compared with the O5 simulation, the CBS domains underwent a gradual structural change from the open to closed form (Fig. 4C) through the 50-ns simulation, suggesting that the N domain in the closed form has considerable influence on the formation of the closed form of the CBS domains.
Surprisingly, in the O465 simulation, the water molecules between the CBS domains were gradually excluded, and several contacts between the CBS2 domains observed in the closed-form crystal structure, especially the van der Waals contact between the Leu-249 side chains (Fig. S6 A and C), were recovered after 40 ns of the simulation. The minimum rmsd value against the closed-form crystal structure over the CBS domains (132–252) is 1.48 Å. However, several water molecules still remained at the interface between the two CBS2 domains of both subunits. As a result, the CBS rotation plateaued at ≈10° in the O465 simulation (Fig. 4C), which is still larger than that in the C456 simulation (Fig. 2C). It is possible that these water molecules will also be excluded from the interface, but this phenomenon may transcend the time scale currently achievable by MD simulation.
Discussion
The CBS Domain Structure Facilitates Its Conformation Change upon Mg2+ Binding.
The cytosolic domain of MgtE forms a dimer through the CBS1 and CBS2 interfaces (Figs. 1 A and B and 5). Characteristically, the CBS2 interface consists of hydrophobic and conserved acidic residues (especially Asp-226 and Asp-250) but lacks basic residues (Fig. 5F). The Mg2+ binding sites 5 reside among these acidic residues in this CBS2 interface (Fig. 1B). The C0 and C46 simulations revealed that the CBS domains immediately undergo a closed-to-open structural transition without Mg2+ in site 5 (Fig. 2C), whereas the O5 and O465 simulations showed that the CBS domains of both subunits approached each other after the addition of Mg2+ to site 5 (Fig. 4C). Therefore, the electrostatic repulsion between these acidic residues in the subunit interface may facilitate the open form of the CBS domains under low Mg2+ concentration conditions. On the other hand, in an environment with a high Mg2+ concentration, Mg2+ bound to site 5 (Mg5) may relax this electrostatic repulsion and thereby facilitate the closed form of the CBS domains. However, the O5 and O465 simulations suggested the considerable contributions of both Mg5 and the N domain in the closed form to the formation of the closed form of the CBS domains. The formation of the closed form is discussed further below.
Fig. 5.
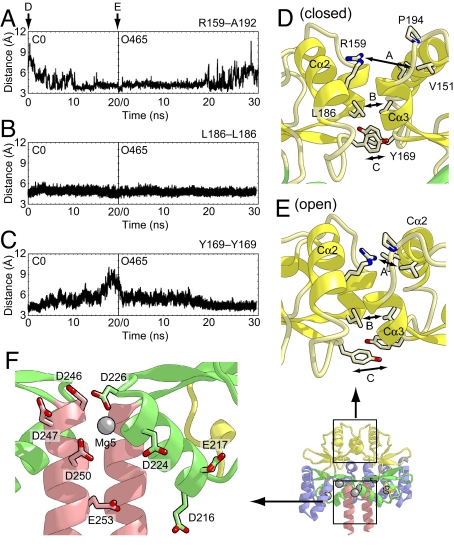
The interactions between the two CBS domains that enable the domain motion upon Mg2+ binding. (A) The distances between the Cζ atom of Arg-159 in one subunit and the main-chain carbonyl atom of Ala-192 in another subunit as a function of time during the O5 and O465 simulations. (B and C) The distances between the Cγ atoms of Leu-186 (B) and Tyr-169 (C) in both subunits during the simulations. (D and E) Close-up views of the subunit interface of CBS1 in the initial structures of the C0 (D) and O465 (E) simulations. The protein side chains discussed in the text are shown as stick models. The distances shown in A–C are indicated. (F) Close-up view of the subunit interface of CBS2 in the Mg2+-bound crystal structure. The protein side chains of acidic residues are shown as stick models.
In contrast, the interface between the CBS1 domains consists not only of hydrophobic and acidic residues, but also of basic residues, and lacks an ion-binding site. Throughout all of the simulations, the van der Waals interactions involving the side chains of Leu-155, Leu-158, Leu-186, Ile-190, Ile-168, and the aliphatic chain of Arg-187 (i.e., Cβ, Cγ, and Cδ) of both subunits were maintained (Fig. 5B), suggesting that these residues in CBS1 act as a hinge for the open-to-closed or closed-to-open structural transition. However, the hinge region is not completely rigid during the structural transition. The cavity between the Cα2 helices of both subunits shrinks in the closed-to-open transition, whereas it expands in the open-to-closed transition (Fig. 5 D and E). Consequently, the interactions between the side chains on these helices were formed and disrupted during these structural transitions. Especially, the guanidinium group of Arg-159 was accommodated in the negatively charged pocket formed by the carbonyl oxygens of Ile-190, Val-191, Ala-192 and the side chains of Pro-194, Ile-155, and Val-151 in the open form. On the other hand, the side chain of Arg-159 was withdrawn from this pocket in the open-to-closed transition (Fig. 5 A, D, and E). In addition, the stacking interaction between the side chains of Tyr-169, which are located beneath the hinge region, was disrupted in the closed-to-open form simulation and recovered in the open-to-closed form simulation (Fig. 5 C–E). These soft interactions between the two CBS1 domains may be crucial for converting the electrostatic force between the CBS2 domains into the domain rotating motion around the hinge axis.
Ion Selectivity of Mg2+ Binding Site 5.
The concentrations of Na+ and K+ in the bacterial cytosol are usually on the same order as and much higher than that of Mg2+, respectively (18). Therefore, to act as a Mg2+ sensor, the Mg2+ binding sites in the cytosolic domain of MgtE must discriminate between Mg2+ and monovalent cations. In the present study, we investigated the binding affinities for Na+ and Ca2+ and the structural changes of the MgtE cytosolic domain upon Na+ and Ca2+ binding at site 5. In the simulation with Na+ at site 5, the Na+ immediately dissociated from site 5, and then the CBS domains underwent the closed-to-open structural transition. After the structural change, the same or other Na+ ions iteratively bound to and dissociated from site 5 (Fig. S4B). Similarly, in the simulation C0, site 5 was also occasionally occupied by Na+ (Fig. S4C). However, the Na+ could not induce the open-to-closed structural transition, unlike the Mg2+ bound at site 5, suggesting that monovalent cations like Na+ cannot relax the electrostatic repulsion between the CBS2 domains and thus cannot induce the structural change (Fig. 2C). On the other hand, in the Ca5 simulation, the CBS domains formed the stable closed conformation with Ca2+ at site 5 (Fig. 3), but in a coordination manner different from that of Mg2+ (Fig. S3E). This result led to the hypothesis that site 5 cannot discriminate between Mg2+ and Ca2+, which is reasonable because the cytosolic concentration of Ca2+ is usually lower than that of Mg2+, by three orders of magnitude (18). To assess this hypothesis, we performed a protease protection assay of full-length MgtE (Fig. S7). The results showed that MgtE exhibits not only Mg2+ but also Ca2+-dependent protease resistance, which presumably reflects the closed conformation of the cytosolic domain upon binding to these divalent cations (see SI Text). For further discussions on Ca2+ binding by the MgtE cytosolic domain, including the significance of the difference in the coordination manner, the effects of divalent cations on the Mg2+ transport activity of MgtE should be explored by experimental methods.
Roles of the N Domain and Mg2+ Binding Sites 4 and 6.
Thus far, we have discussed the structural role of Mg5 observed in the computer simulations. However, we still need to address the roles of Mg4 and Mg6 in the Mg2+ sensing by MgtE. Is Mg2+ binding at sites 4 and site 6 essential for the ion sensing by MgtE? A comparison of the MD simulation starting from the open form bound to Mg5 (simulation O5) with that bound to Mg4, Mg6, and Mg5 (simulation O465) would help to answer this question.
In the O5 simulations without the N domain, the CBS rotation approached the closed form, but it immediately returned to the open form. This fluctuation of the CBS rotation was reproducibly observed in two independent simulations (Fig. 4C). As discussed in the previous section, Mg5 may facilitate the formation of the closed form of the CBS domain. However, the electrostatic attraction between Mg5 and the acidic residues may not be sufficient to stably maintain the closed form of the CBS domain. In contrast, the O465 simulation with the closed-form N domain showed that the CBS domains never opened again once they were converted to the closed form. In the initial structure of this simulation, residues 29–31 of the N domain are distant from CBS2 of another subunit (≈16 Å), and the interactions between them observed in the crystal structure were absent. After the 30-ns simulation, these interactions were strikingly recovered as the CBS2 domains of both subunits approached each other (Fig. S6 B and D). This recovery of the interactions between the N domain and the CBS2 domain of another subunit may lock the CBS domain in the closed form. Therefore, the N domain may function like a clamp, which locks the CBS domain into the closed form in conjunction with Mg5.
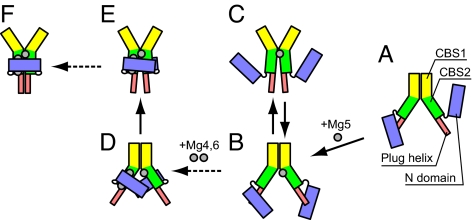
To summarize the findings from the present simulations, we propose a hypothetical pathway of the open-to-closed form structural transition, as shown in Fig. 6. In this pathway, the binding of the first Mg2+ to site 5 in the CBS domain initiates the structural transition (Fig. 6 A and B). After the binding of Mg5, the CBS domain may convert to the closed form, but it immediately returns to the open form, as observed in the O5 simulation (Fig. 6 B and C). Next, the binding of Mg4 and Mg6 fixes the N domain in the closed form (Fig. 6D), which may further lock the CBS domain in the stable closed form as a clump (Fig. 6 E and F). Although we hypothesized here that the binding of Mg5 precedes those of Mg4 and Mg6, it is possible that Mg4 and Mg6 binding occurs first. However, in either case, the role of the N domain is the same as that proposed above. Further discussion on the Mg2+ binding order may require a quantitative comparison of the binding free energies, calculated by computational methods, and an analysis of the binding kinetics by experimental methods.
Fig. 6.
The proposed pathway of the open-to-closed structural transition based on the present simulations. The structural changes indicated with the solid arrows were observed in the present simulations, and those with the dashed arrows were not directly observed, but were suggested from the crystal structures and simulations.
Conclusions
In this work, we successfully reproduced the Mg2+-dependent structural conversion of the MgtE cytosolic domain between the closed and open forms in silico using MD simulations. These results suggested that the MgtE cytosolic domain senses the Mg2+ concentration by Mg2+ binding site 5, wherein the electrostatic interactions among Mg5 and the CBS2 domains are converted to an intersubunit force. Subsequently, the interactions between CBS1 in each subunit enable the conversion of this force into the hinge motion of the overall CBS domains. Finally, this motion affects the orientation of the plug helices, which could result in the gating of the ion-conducting pore. Our simulations also showed that site 5 can discriminate between Mg2+ and Na+, mainly by their charge differences. The other abundant monovalent cation is K+, and its concentration in the bacterial cytosol is usually higher than that of Na+ (≈100 mM) (18). Site 5 presumably discriminates between Mg2+ and K+ in a similar manner. To understand how the structural changes observed in our simulations affect the structure of the TM domain, including the ion-conducting pore, the MD simulation of full-length MgtE, as well as the determination of the full-length MgtE structure under Mg2+-free conditions, is required.
Our simulations revealed that the stable form of the N domain depends exclusively on Mg2+ binding at sites 4 and 6. The closed form of the N domain allows the Mg2+-bound CBS domains to form the stable closed conformation by interacting with the CBS domain of another subunit. Furthermore, the full-length MgtE crystal structure suggested the existence of several Mg2+ binding sites between the TM and CBS domains. The presence of many Mg2+ binding sites, including Mg4 and 6, and the N domain may confer a cooperative, allosteric nature to Mg2+ sensing by MgtE.
Methods
MD Simulation.
The details of the modeling procedures for the initial structures of the simulations are described in SI Text. MD simulations were performed by using the NAMD 2.6 package (19) with the CHARMM27 set of force field parameters (20) with φ, ψ cross-term map (CMAP) correction (21). The TIP3P model (22) was used for water. The bond lengths between hydrogens and heavy atoms were restrained by the RATTLE method (23), allowing an integration step of 2 fs. The target pressure and temperature were set to 1.0 atm and 300 K, respectively. Constant pressure was maintained by using the Langevin piston Nose–Hoover method (24, 25). A Langevin damping coefficient γ of 5.0 ps−1 was used for temperature control. Periodic boundary conditions were implemented in all systems. The particle mesh Ewald method (26) was used to calculate the electrostatic forces without truncation. The initial structures were subjected to energy minimizations to remove unfavorable contacts. The systems were then relaxed by 500-ps MD simulations with gradually decreasing positional constraints from 10.0 to 0.0 kcal·mol−1·Å−2 on the protein heavy atoms. The O5 simulation was performed twice with different initial velocities. All simulations were conducted on the TSUBAME Grid Cluster at the Global Scientific Information and Computing Center (GSIC) of the Tokyo Institute of Technology.
MD Simulation of MgtE.
The hinge axes of the domain rotation were calculated by the program DynDom (27). The rotational angle between the two domains (e.g., domains A and B) of the simulation trajectories was calculated as follows. Each structure from the simulation trajectory was rotated and translated to fit over the Cα atoms in domain A of the crystal structure by the least-squares method. The Euler angles (φ, θ, ψ) for rotation and the translation vector to fit to the Cα atoms in domain B of the crystal structure were then calculated for each structure from the simulation trajectory. The Euler angles were then converted to a quaternion q [q0, q1, q2, q3]T by the following relation:
|
|
and then the quaternion q was decomposed to the axis e [ex, ey, ez] T and the rotational angle χ around that axis, by the following formula: q = [cos(χ/2), ex sin(χ/2), ey sin(χ/2), ez sin(χ/2)]T.
Supplementary Material
Acknowledgments.
We are grateful to Dr. T. Tsukazaki (Tokyo Institute of Technology) for helpful discussions. This work was supported by a Solution-Oriented Research for Science and Technology (SORST) program grant from the Japan Science and Technology Agency (to O.N.), by a grant for the National Project on Protein Structural and Functional Analyses from the Ministry of Education, Culture, Sports, Science and Technology (to O.N.), by grants from the Ministry of Education, Culture, Sports, Science and Technology (to R.I. and O.N.), and by Mitsubishi Foundation grants (to O.N.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
The regulation of the transporter activity by an α-helix called a “plug helix” has been proposed for several membrane transporter systems, such as the protein-conducting channel SecY (28) and the gap junction channel connexin26 (29). The gating by the long α-helix of MgtE is conceptually the same as that of a plug helix, and this may be a general mechanism among transporter systems. Therefore, we renamed this α-helix of MgtE as the “plug helix.”
This article contains supporting information online at www.pnas.org/cgi/content/full/0802991105/DCSupplemental.
References
- 1.Lunin VV, et al. Crystal structure of the CorA Mg2+ transporter. Nature. 2006;440:833–837. doi: 10.1038/nature04642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eshaghi S, et al. Crystal structure of a divalent metal ion transporter CorA at 2.9 angstrom resolution. Science. 2006;313:354–357. doi: 10.1126/science.1127121. [DOI] [PubMed] [Google Scholar]
- 3.Payandeh J, Pai EF. A structural basis for Mg2+ homeostasis and the CorA translocation cycle. EMBO J. 2006;25:3762–3773. doi: 10.1038/sj.emboj.7601269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hattori M, Tanaka Y, Fukai S, Ishitani R, Nureki O. Crystal structure of the MgtE Mg2+ transporter. Nature. 2007;448:1072–1075. doi: 10.1038/nature06093. [DOI] [PubMed] [Google Scholar]
- 5.Chakrapani S, Perozo E. How to gate an ion channel: Lessons from MthK. Nat Struct Mol Biol. 2007;14:180–182. doi: 10.1038/nsmb0307-180. [DOI] [PubMed] [Google Scholar]
- 6.Gruswitz F, O'Connell J, Stroud RM. Inhibitory complex of the transmembrane ammonia channel, AmtB, and the cytosolic regulatory protein, GlnK, at 1.96 Å. Proc Natl Acad Sci USA. 2007;104:42–47. doi: 10.1073/pnas.0609796104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conroy MJ, et al. The crystal structure of the Escherichia coli AmtB-GlnK complex reveals how GlnK regulates the ammonia channel. Proc Natl Acad Sci USA. 2007;104:1213–1218. doi: 10.1073/pnas.0610348104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jasti J, Furukawa H, Gonzales EB, Gouaux E. Structure of acid-sensing ion channel 1 at 1.9A resolution and low pH. Nature. 2007;449:316–323. doi: 10.1038/nature06163. [DOI] [PubMed] [Google Scholar]
- 9.Smith RL, Thompson LJ, Maguire ME. Cloning and characterization of MgtE, a putative new class of Mg2+ transporter from Bacillus firmus OF4. J Bacteriol. 1995;177:1233–1238. doi: 10.1128/jb.177.5.1233-1238.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dann CE, et al. Structure and mechanism of a metal-sensing regulatory RNA. Cell. 2007;130:878–892. doi: 10.1016/j.cell.2007.06.051. [DOI] [PubMed] [Google Scholar]
- 11.Goytain A, Quamme GA. Functional characterization of human SLC41A1, a Mg2+ transporter with similarity to prokaryotic MgtE Mg2+ transporters. Physiol Genomics. 2005;21:337–342. doi: 10.1152/physiolgenomics.00261.2004. [DOI] [PubMed] [Google Scholar]
- 12.Goytain A, Quamme GA. Functional characterization of the human solute carrier, SLC41A2. Biochem Biophys Res Commun. 2005;330:701–705. doi: 10.1016/j.bbrc.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 13.Sahni J, Nelson B, Scharenberg AM. SLC41A2 encodes a plasma-membrane Mg2+ transporter. Biochem J. 2007;401:505–513. doi: 10.1042/BJ20060673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ignoul S, Eggermont J. CBS domains: Structure, function, and pathology in human proteins. Am J Physiol. 2005;289:C1369–C1378. doi: 10.1152/ajpcell.00282.2005. [DOI] [PubMed] [Google Scholar]
- 15.Bennetts B, et al. Cytoplasmic ATP-sensing domains regulate gating of skeletal muscle ClC-1 chloride channels. J Biol Chem. 2005;280:32452–32458. doi: 10.1074/jbc.M502890200. [DOI] [PubMed] [Google Scholar]
- 16.Biemans-Oldehinkel E, Mahmood NA, Poolman B. A sensor for intracellular ionic strength. Proc Natl Acad Sci USA. 2006;103:10624–10629. doi: 10.1073/pnas.0603871103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kowal P, Gurtan AM, Stuckert P, D'Andrea AD, Ellenberger T. Structural determinants of human FANCF protein that function in the assembly of a DNA damage signaling complex. J Biol Chem. 2007;282:2047–2055. doi: 10.1074/jbc.M608356200. [DOI] [PubMed] [Google Scholar]
- 18.Maguire ME, Cowan JA. Magnesium chemistry and biochemistry. Biometals. 2002;15:203–210. doi: 10.1023/a:1016058229972. [DOI] [PubMed] [Google Scholar]
- 19.Phillips JC, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.MacKerell AD, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 21.Mackerell AD, Feig M, Brooks CL. Extending the treatment of backbone energetics in protein force fields: Limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J Comput Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 22.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 23.Andersen HC. Rattle—A velocity version of the shake algorithm for molecular-dynamics calculations. J Comput Phys. 1983;52:24–34. [Google Scholar]
- 24.Feller SE, Zhang YH, Pastor RW, Brooks BR. Constant-pressure molecular-dynamics simulation—The Langevin piston method. J Chem Phys. 1995;103:4613–4621. [Google Scholar]
- 25.Martyna GJ, Tobias DJ, Klein ML. Constant-pressure molecular-dynamics algorithms. J Chem Phys. 1994;101:4177–4189. [Google Scholar]
- 26.Darden T, York D, Pedersen L. Particle mesh Ewald—An N. log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 27.Hayward S, Berendsen HJC. Systematic analysis of domain motions in proteins from conformational change: New results on citrate synthase and T4 lysozyme. Proteins. 1998;30:144–154. [PubMed] [Google Scholar]
- 28.van den Berg B, et al. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- 29.Oshima A, Tani K, Hiroaki Y, Fujiyoshi Y, Sosinsky GE. Three-dimensional structure of a human connexin26 gap junction channel reveals a plug in the vestibule. Proc Natl Acad Sci USA. 2007;104:10034–10039. doi: 10.1073/pnas.0703704104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.