Abstract
The opioid peptides and receptors have prominent roles in pain transmission and reward mechanisms in mammals. The evolution of the opioid receptors has so far been little studied, with only a few reports on species other than tetrapods. We have investigated species representing a broader range of vertebrates and found that the four opioid receptor types (delta, kappa, mu, and NOP) are present in most of the species. The gene relationships were deduced by using both phylogenetic analyses and chromosomal location relative to 20 neighboring gene families in databases of assembled genomes. The combined results show that the vertebrate opioid receptor gene family arose by quadruplication of a large chromosomal block containing at least 14 other gene families. The quadruplication seems to coincide with, and, therefore, probably resulted from, the two proposed genome duplications in early vertebrate evolution. We conclude that the quartet of opioid receptors was already present at the origin of jawed vertebrates ≈450 million years ago. A few additional opioid receptor gene duplications have occurred in bony fishes. Interestingly, the ancestral receptor gene duplications coincide with the origin of the four opioid peptide precursor genes. Thus, the complete vertebrate opioid system was already established in the first jawed vertebrates.
Keywords: chromosome, G protein-coupled receptor, gene duplication
Several opioid peptides, including endorphin and enkephalins, are important regulators of nociceptive neurotransmission and reward mechanisms in mammals. Specific binding sites in the brain for opioid compounds were first reported in 1973 (1–3), and it was soon evident that more than one type of binding site existed (4). Subsequently three distinct opioid receptors were identified and designated delta, kappa, and mu. These receptors were cloned and found to be encoded by separate genes belonging to the superfamily of rhodopsin-like G protein-coupled receptors (GPCRs) (5–8). The genes for the opioid receptors (OPR) have been named OPRD1 (delta), OPRK1 (kappa), and OPRM1 (mu) by the HUGO Gene Nomenclature Committee (HGNC).
Homology searches resulted in the discovery of a fourth receptor in both rodents and humans initially named ORL1 for opioid receptor-like (9) or LC132 (10). This receptor shows 48–49% identity to the other three human receptors, which display 55–58% identity among one another. The receptor has been named NOP by the International Union of Basic and Clinical Pharmacology and its gene has been named OPRL1 by HGNC. An endogenous peptide ligand with some similarity to the other opioid peptides was discovered and named nociceptin (11) or orphanin FQ (12).
The evolution of the endogenous opioid peptide ligands has been studied extensively and the major peptide ligands are generated from four prepropeptides that are encoded by separate genes in tetrapods. The genes arose by duplications in the common ancestor of tetrapods and bony fishes (13). Opioid receptor sequences have been reported for a few nonmammalian tetrapods (14–18) and a few teleost fishes (19–24), and a partial sequence has been reported for a hagfish (19). Functional studies in amphibians and bony fishes have shown that the opioid system is involved in nociception also in these species (25, 26).
Many vertebrate gene families have been found to have expanded in the early stages of vertebrate evolution, before the radiation of jawed vertebrates. However, the high degree of sequence divergence over such large evolutionary distances often obscures orthology–paralogy relationships. Investigation of conserved synteny may facilitate identification of orthologs and gives important clues to the mechanisms by which the genes were duplicated. We used this approach to investigate the evolution of a few other gene families, namely the neuropeptide Y (NPY) family of peptides (27) and the large family of NPY receptors (28). These families were found to have expanded as a result of extensive chromosome duplications, most likely resulting from two tetraploidizations, i.e., genome duplications, that occurred early in vertebrate evolution (29). These genome duplications, often referred to as 1R and 2R, occurred after the divergence of tunicates and lancelets (30) from vertebrates but before the divergence of cartilaginous fishes and bony vertebrates (31). For the cyclostomes (lampreys and hagfishes) the picture is not completely clear but based on analyses of a limited number of gene families they seem to have undergone the first tetraploidization (1R) but not the second (2R) (32–36).
As the opioid receptor genes are located on four different chromosomes in human (1, 6, 8, and 20) we decided to investigate whether they arose by duplication of a single ancestral opioid receptor gene in the two tetraploidizations. Other investigators have also suggested that studies of chromosomal location may shed light on opioid receptor evolution (37). We describe here an investigation, using a combination of sequence-based phylogenies and gene locations for the opioid receptors and their neighboring families that shows that they expanded by gene duplications in conjunction with the proposed tetraploidizations in early vertebrate evolution.
Results
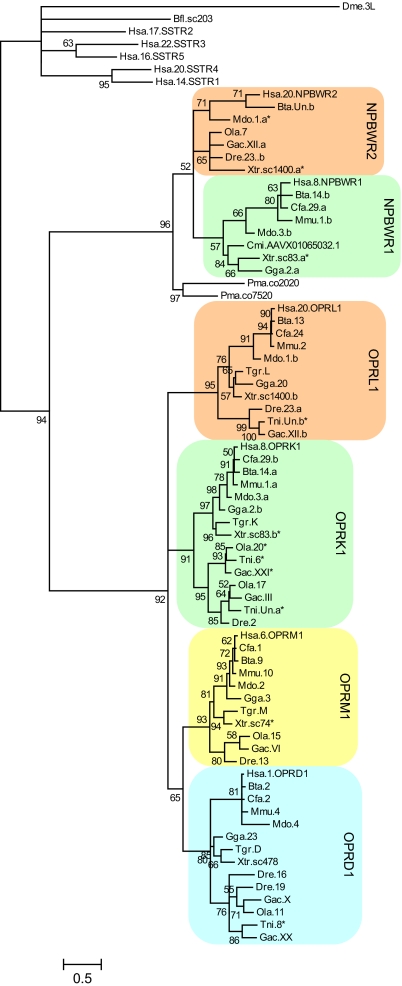
To investigate whether the opioid receptor genes arose by duplications of a single ancestral gene in the two basal vertebrate tetraploidizations we have analyzed the opioid receptor gene family and 20 of the neighboring gene families phylogenetically. Specifically, we wanted to find out whether these gene families were duplicated in the same time period, i.e., after the divergence of invertebrate chordates and vertebrates but before the divergence of bony fishes and tetrapods because this is the time span in which the two tetraploidizations took place. The gene families were analyzed phylogenetically by making both neighbor-joining (NJ) trees and quartet-puzzling maximum likelihood (QP) trees in which species that diverged before 2R were used as outgroups to provide relative dating of the gene duplications. The opioid receptors were analyzed in human (Homo sapiens), mouse (Mus musculus), dog (Canis familiaris), cow (Bos taurus), gray short-tailed opossum (Monodelphis domestica), chicken (Gallus gallus), western clawed frog (Xenopus tropicalis), rough-skinned newt (Taricha granulosa), zebrafish (Danio rerio), medaka (Oryzias latipes), stickleback (Gasterosteus aculeatus), and spotted green pufferfish (Tetraodon nigroviridis). We found at least four opioid receptor genes in the genome databases for most of these species. The zebrafish and medaka have duplicates of the OPRK1 and/or the OPRD1 genes, whereas the OPRL1 gene is missing in medaka and the OPRM1 gene is missing in spotted green pufferfish [Fig. 1 and supporting information (SI) Fig. S1]. However, this does not necessarily mean that these genes have been lost because their absence may simply be due to incomplete sequencing of the genomes or poor genome assembly in the databases. The human opioid receptor sequences were used for blastp searches of the Florida lancelet (Branchiostoma floridae) database and the elephant shark (Callorhinchus milii) database, but this produced no reasonable hits.
Fig. 1.
Quartet-puzzling maximum likelihood tree of the opioid and NPBW receptors with bootstrap values shown in percentage at each node. The tree is color-coded based on the opioid receptor bearing chicken chromosomes (chromosome 20, orange; chromosome 2, green; chromosome 3, yellow; and chromosome 23, blue). The first three letters of the sequences names are abbreviations of the species names and the number represents the chromosome on which the gene is located. In cases where two genes are located on the same chromosome, “a” or “b” has been added to the sequence names to separate them. For the human sequences, the approved HGNC symbol has also been added to the sequence name. An asterisk after the sequence name means that the sequence has been extended after inspection of the database entries as described in Methods. Hsa, Homo sapiens; Mmu, Mus musculus; Cfa, Canis familiaris; Bta, Bos taurus; Mdo, Monodelphis domestica; Gga, Gallus gallus; Xtr, Xenopus tropicalis; Tgr, Taricha granulosa; Dre, Danio rerio; Ola, Oryzias latipes; Gac, Gasterosteus aculeatus; Tni, Tetraodon nigroviridis; Cmi, Callorhinchus milii; Pma, Petromyzon marinus; Bfl, Branchiostoma floridae; Dme, Drosophila melanogaster.
The phylogenetic tree (Fig. 1) shows that the neuropeptide B/W (NPBW) receptors are closely related to the opioid receptors, i.e., closer than to any other GPCRs. This is supported by the chromosomal locations because NPBWR1 is located next to OPRK1 on human chromosome 8 (287 kb downstream) and NPBWR2 is situated on human chromosome 20 next to OPRL1 (only 26 kb downstream) (Fig. 2 and Fig. 3). Such close linkage can be seen in most of the species that have NPBW receptors. An earlier split of the somatostatin receptors from the opioid/NPBW receptors is suggested by the fact that the vertebrate somatostatin receptors cluster with a Florida lancelet sequence and a fruit fly (Drosophila melanogaster) allostatin C receptor sequence in the phylogenetic tree (Fig. 1). Both the opioid and the NPBW receptor families seem to have expanded in the time period coinciding with the tetraploidizations in early vertebrate evolution.
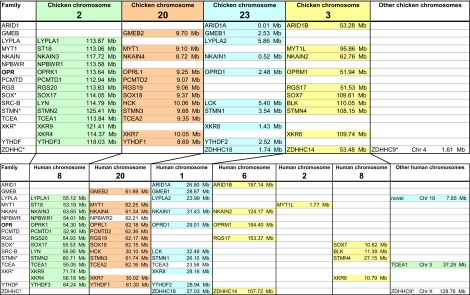
Fig. 2.
The figure shows the analyzed gene families, including the OPR family, and the chromosomal location of the genes they contain. All human gene names in the figure are approved HGNC symbols and the chicken genes have been given the name of their human orthologs. The tables are color-coded based on the chicken chromosomes (chromosome 2, green; chromosome 20, orange; chromosome 23, blue; and chromosome 3, yellow). An asterisk after the family name indicates that the NJ and QP trees display different topologies (see Figs. S1–S15). The gene family abbreviations are explained in Figs. S2–S15 along with brief descriptions of their properties.
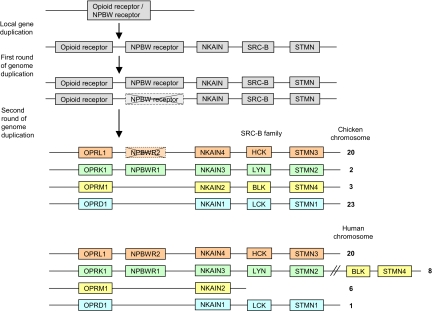
Fig. 3.
Proposed opioid receptor evolution. The opioid and NPBW receptors are shown together with three other gene families that have maintained all four copies after the two tetraploidizations. The BLK and STMN4 genes have been translocated to chromosome 8 in human and the NPBWR2 gene has been lost in chicken.
We have investigated the genomic regions extending 7 Mb on each side of the opioid receptor genes in human (see Methods for details). These regions contain 698 genes, of which 304 belong to families with at least two members anywhere in the genome. In total, 24 families in addition to the opioid receptors have members in at least two of the selected regions and were investigated further. Four gene families had to be left out because their multitude of members made phylogenetic analyses unreliable. For the remaining 20 families, NJ trees with protein sequences from human, mouse, dog, chicken, western clawed frog, spotted green pufferfish, tunicate (Ciona intestinalis), and fruit fly were constructed. After analysis of the initial NJ trees six additional families were excluded because their sequences were unalignable or because the families clearly did not expand in the two tetraploidizations (for description of the excluded families see SI Text). QP trees were constructed for the 14 remaining families and the topologies were compared with those of the NJ trees (Figs. S2–S15). A presentation of all of the neighboring families used in the study is also available in Figs. S2–S15.
For 10 of the 14 families both the NJ trees and the QP trees display topologies that support expansion in 2R. For three of the families (SOX, STMN, and XKR) the NJ trees support expansion in 2R but the QP trees have unresolved branches and are therefore inconclusive (Figs. S9, S11, and S13). For the ZDHHC family the QP tree supports an expansion in 2R but the NJ tree shows a topology inconsistent with expansion in 2R (Fig. S15).
The chromosomal locations for all members of the 15 families are shown for chicken and human in Fig. 2. The genes have been color-coded according to the chicken chromosome on which they are located to facilitate comparisons between different species. The chicken chromosomes were used as starting point for color-coding because they show more similarity to the ancestral vertebrate chromosomes than the human chromosomes do (38). This dataset demonstrates conserved synteny between the two species; genes that are located next to each other in human are linked in the chicken genome (Fig. 2). Our results also show that chromosomal rearrangements are frequent. As an example, genes on chicken chromosome 3 are represented on three different human chromosomes (see Fig. 2).
Discussion
Using a combination of positional and phylogenetic data, we have found that the OPR genes are located in four genomic regions that share a common evolutionary history. By using relative dating, we found that the opioid receptors, together with their neighboring families, seem to have expanded in the two tetraploidizations early in vertebrate evolution, indicating that the quartet of opioid receptors was already present at the origin of jawed vertebrates. Taking advantage of genome sequencing projects we found four opioid receptors in most of the species investigated (Fig. 1). Zebrafish has an extra copy of the delta receptor gene (20) and medaka has duplicates of both the delta and kappa receptor genes. The extra copies are likely to be a result of a third whole-genome duplication (3R) that took place early in ray-finned fish evolution. The kappa receptor duplicates in medaka are located on chromosome 20 and 17 and the delta receptor duplicates in zebrafish are located on chromosome 16 and 19. These chromosome pairs are considered to be 3R copies according to genome studies in medaka and zebrafish and ancestral karyotype reconstructions (39).
The repertoire of receptors in other species than tetrapods and bony fishes is still unknown. It was not possible to identify any opioid receptors by searches in the elephant shark genome database, probably because of the presence of introns in the receptor genes and because the genome is only available as relatively short scaffolds. An ortholog to one of the intronless NPBW receptors was identified in the elephant shark genome. Fragments from three putative opioid receptors have been cloned in thresher shark (Alopias vulpinus) (19, 40), and some fragments of opioid-like receptors from a jawless fish, the Pacific hagfish (Eptatretus stoutii), have also been cloned (19, 40). Our analyses are not able to determine which receptor type the cloned hagfish sequence corresponds to (data not shown). The genome of the sea lamprey (Petromyzon marinus), a species from another lineage of jawless fishes, has been sequenced but is only assembled into small unlinked contigs and our blast searches only gave partial sequences (data not shown). These sequences did not contain enough information to be assigned to specific opioid receptor types. However, two intronless NPBW receptor-like sequences were found in the sea lamprey genome (Fig. 1). We were not able to find any opioid receptor sequences in the Florida lancelet or the tunicate genome databases.
A report of a mu receptor sequence obtained from the blue mussel (Mytilus edulis) by RT-PCR that displayed 95% nucleotide sequence identity to the human mu receptor has been published (41, 42). The most likely explanation for this extraordinary degree of sequence identity between a mollusc and a mammal is either horizontal gene transfer or contamination. Until this has been resolved, we prefer not to include the claimed blue mussel mu receptor sequence in our phylogenetic trees.
Genomic regions with a common evolutionary history comprise a so called paralogon (43). Paralogons usually consist of quartets of chromosomes due to the two tetraploidizations early in vertebrate evolution. However, extensive gene loss is common after whole-genome duplications (29). As a result, many gene families that expanded in the two tetraploidizations only have two or three members rather than four. In the opioid receptor family, all four members have been kept in most of the vertebrate lineages, indicating important functions. The opioid receptor genes and adjacent gene families are primarily located on four different chromosomes in human (1, 6, 8, and 20), thus forming a paralogon. In chicken, the paralogon consists of the four chromosomes, 2, 3, 20, and 23. Chicken chromosome 3 corresponds largely to human chromosome 6, from which smaller pieces have ended up on chromosomes 2 and 8 (Fig. 2). Somewhat confusingly, chromosome 8 also contains the paralogon member that corresponds to chicken chromosome 2 (see Fig. 2). This is supported by comparisons with the orthologous genes in dog and opossum (Fig. S16). The dog has three chromosomes and the opossum has two chromosomes that correspond to chicken chromosome 3 indicating that the translocations in mammals occurred in two steps.
It has been suggested that the NPBW receptors are closely related to both the opioid and the somatostatin receptors (44, 45). The phylogenetic tree (Fig. 1) suggests that the ancestral somatostatin receptor diverged before the duplication resulting in the opioid and NPBW receptors. In Fig. 3, we present one possible scenario for the evolution of the opioid receptor family and four neighboring gene families. The opioid receptors and the NPBW receptors are the result of a local duplication before the tetraploidizations. In 2R several gene families, represented here by the NKAIN, SRC-B, and STMN families, expanded together with the opioid receptors, as shown by both their phylogenetic trees and their chromosomal positions (for descriptions of the families, see Figs. S2–S15). After the tetraploidizations the NPBWR2 gene has been lost in chicken, and, in human, the BLK and STMN4 genes have been translocated to chromosome 8 (together with some other genes; see above).
For the opioid receptors, it has been suggested, based on both sequence identity and chromosomal location, that the mu and delta receptors are more closely related to each other and that the kappa and NOP receptors are more closely related to each other (17). Our data support this pattern, but it should be noted that regarding sequence identity the kappa receptor is nevertheless more similar to the mu and delta receptors than to the NOP receptor. Our interpretation of this is that OPRL1 has had a higher evolutionary rate than the other opioid receptor genes. This is also supported by a longer branch length in the tree leading to the NOP receptors.
In most species, NPBWR1 is located in a tail to tail fashion with OPRK1, and NPBWR2 is located in a tail to tail fashion with OPRL1 with no other genes in-between, indicating a common origin by duplication before the tetraploidizations (Fig. 3). Although the mammalian NPBWR2 sequences do not cluster with the fish and western clawed frog NPBWR2 sequences in the tree (Fig. 1), the fish and frog sequences can still be identified as NPBWR2 because of their chromosomal location.
Studies of nonmammalian opioid receptors with regard to ligand-binding properties (in vitro) noted that some ligands did not display the same receptor-type preferences as in mammals (15, 17), and it was proposed that the nonmammalian receptors are less type-selective (17). However, these exogenous ligands were developed in mammals to distinguish the mammalian receptor types. The nonmammalian receptors may display other unique differences that might be useful for development of type-selective ligands in these species. Regarding receptor selectivity for endogenous ligands, each species must of course be studied using its own repertoire of peptides.
Our study shows that all four opioid receptor types are present in the major lineages of bony vertebrates and that the quartet was formed in the two tetraploidizations early in vertebrate evolution. This establishes that from an evolutionary perspective the NOP receptor is a member of the opioid receptor family on equal terms with the other three members. This also means that already the first jawed vertebrates had all four opioid receptor genes. An early origin of the opioid peptides has been suggested (13) and the time point for these duplications indicates that at least some of them could have arisen in 2R. Detailed studies of chromosomal location should help clarify this picture, although several chromosomal rearrangements make the situation more complicated for the opioid prepropeptide genes than for the receptor genes. In any event, these observations show that the vertebrate opioid system was already quite complex before the radiation of jawed vertebrates ≈450 million years ago, and at least the receptors quadrupled in the basal vertebrate tetraploidizations.
Methods
Database Searches.
The opioid receptor genes were identified in the Ensembl database (www.ensembl.org) release 38 for the 11 vertebrate species listed below. In this Ensembl version, the opioid receptors belonged to a protein family that also contained the somatostatin, NPBW, and galanin receptors and MCHR1, MCHR2, KISS1R, and UTS2R. All protein sequences included in this family were collected for human (Homo sapiens), mouse (Mus musculus), dog (Canis familiaris), cow (Bos taurus), gray short-tailed opossum (Monodelphis domestica), chicken (Gallus gallus), western clawed frog (Xenopus tropicalis), zebrafish (Danio rerio), medaka (Oryzias latipes), stickleback (Gasterosteus aculeatus), spotted green pufferfish (Tetraodon nigroviridis), fruit fly (Drosophila melanogaster), and tunicate (Ciona intestinalis). The human opioid receptors were used for blastp searches of the Florida lancelet (Branchiostoma floridae) database, Version 1.0 (http://genome.jgi-psf.org/Brafl1). In cases where one gene had more than one transcript, only the longest transcript was used in the phylogenetic analysis.
Phylogenetic Analysis of the Opioid Receptors.
The sequences were aligned using the Windows version of Clustal X 1.81 (46, 47), and an initial unrooted 1,000 times bootstrapped neighbor-joining (NJ) tree was constructed in Clustal X 1.81 with standard settings. Using the invertebrate sequences to relatively date gene duplications, the Ensembl protein family could be divided into subfamilies. Because we were interested in duplications that occurred in 2R i.e., after the split between vertebrates and invertebrate chordates like the tunicate and the Florida lancelet, we defined a subfamily as all of the sequences that share an invertebrate outgroup. The opioid receptors and NPBW receptors were defined as one subfamily because they clustered in the tree without any invertebrate sequences in between. The somatostatin receptors also seemed to be closely related to the opioid receptors, but they clustered with a Florida lancelet sequence and were therefore defined as a separate subfamily. A new alignment was made for the opioid and NPBW receptor subfamily and the human somatostatin receptors were included as an outgroup together with the Florida lancelet sequence and a fruit fly allostatin C receptor sequence. The sequences of the cloned opioid receptors from rough-skinned newt (Taricha granulosa) (14–16) were included in the alignment as well as a translated genomic NPBWR1-sequence from elephant shark (Callorhinchus milii) (http://esharkgenome.imcb.a-star.edu.sg) and two predicted genes from sea lamprey (Petromyzon marinus) (http://pre.ensembl.org). The alignment was edited manually to remove unalignable sequences. Short sequences were extended by a search for missing exons in the sequences flanking the genes or inside introns. Extended sequences are indicated with an asterisk after the sequence name in Fig. 1 and in the alignment of the opioid receptor sequences (Fig. S17). Phylogenetic trees were constructed using both the NJ and quartet-puzzling maximum likelihood (QP) methods (for details, see below). Gene IDs and accession numbers are available in Table S1.
Selection of Genomic Regions.
Genomic regions of 7 Mb on each side of the human opioid receptor genes were checked for gene content using the Ensembl database. The opioid receptor genes are located on human chromosomes 1 (OPRD1, 29.01 Mb), 6 (OPRM1, 154.40 Mb), 8 (OPRK1, 54.30 Mb), and 20 (OPRL1, 62.18 Mb); therefore, the regions selected for investigation were chromosomes 1 (22.01–36.01 Mb), 6 (147.40–161.40 Mb), 8 (47.30–61.30 Mb), and 20 (55.18–62.44 Mb). On human chromosome 20, the OPRL1 gene is positioned close to the end of the q arm; therefore, only ≈7.2 Mb was investigated on this chromosome. A list of all genes in these regions was compiled and gene families with representation in at least two of the selected regions were chosen for phylogenetic analysis. In total, 25 gene families, including the opioid receptors, fulfilled the selection criteria.
Phylogenetic Analysis of Neighboring Families.
For the 25 selected families, amino acid sequences from version 38 of the Ensembl database were downloaded for human, mouse, dog, chicken, western clawed frog, and spotted green pufferfish. In addition to the vertebrate sequences, family members from the tunicate and the fruit fly were included to relatively date the gene duplications. The sequences were aligned using the Windows version of Clustal X 1.81, producing an initial alignment. Tblastn (48) with standard settings in the Ensembl database and other searches in the National Center for Biotechnology Information database were used to find additional sequences not included in the Ensembl families. In cases where gene families were not represented in the genome databases of Ciona intestinalis and Drosophila melanogaster sequences from the tunicate Ciona savignyi, the Florida lancelet or the nematode Caenorhabditis elegans were included to relatively date the phylogenetic trees. (For sequence accession numbers, see Table S1).
The initial alignments were edited manually to remove poorly aligned or incomplete sequences. The edited alignments were used to produce an initial unrooted 1,000 times bootstrapped NJ tree with standard settings in Clustal X 1.81. This tree was used to find subfamilies defined as clusters that expanded after the split of invertebrate chordates and vertebrates. A refined NJ tree was made for each subfamily of interest. A QP tree was also constructed for each family using the Windows version of Treepuzzle 5.2 (49). The analysis was made using the JTT matrix, with the amino acid frequencies estimated from the dataset. The model of rate heterogeneity was set to gamma distributed rates with eight gamma rate categories and the alpha parameter estimated from the dataset. Parameters were estimated using the “exact” and “quartet sampling + NJ tree” options and the number of puzzling steps were automatically decided by Treepuzzle and varied between 1,000 and 25,000, depending on the dataset.
Supplementary Material
Acknowledgments.
We thank Ulrika Pettersson who participated in the earlier stages of this work. This work was supported by grants from the Swedish Research Council and Carl Trygger's Foundation, Sweden.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0805590105/DCSupplemental.
References
- 1.Pert CB, Snyder SH. Opiate receptor: Demonstration in nervous tissue. Science. 1973;179:1011–1014. doi: 10.1126/science.179.4077.1011. [DOI] [PubMed] [Google Scholar]
- 2.Simon EJ, Hiller JM, Edelman I. Stereospecific binding of the potent narcotic analgesic (3H) Etorphine to rat-brain homogenate. Proc Natl Acad Sci USA. 1973;70:1947–1949. doi: 10.1073/pnas.70.7.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Terenius L. Stereospecific interaction between narcotic analgesics and a synaptic plasma membrane fraction of rat cerebral cortex. Acta Pharmacol Toxicol (Copenhagen) 1973;32:317–320. doi: 10.1111/j.1600-0773.1973.tb01477.x. [DOI] [PubMed] [Google Scholar]
- 4.Martin WR, Eades CG, Thompson JA, Huppler RE, Gilbert PE. The effects of morphine- and nalorphine-like drugs in the nondependent and morphine-dependent chronic spinal dog. J Pharmacol Exp Ther. 1976;197:517–532. [PubMed] [Google Scholar]
- 5.Evans CJ, Keith DE, Jr, Morrison H, Magendzo K, Edwards RH. Cloning of a delta opioid receptor by functional expression. Science. 1992;258:1952–1955. doi: 10.1126/science.1335167. [DOI] [PubMed] [Google Scholar]
- 6.Kieffer BL, Befort K, Gaveriaux-Ruff C, Hirth CG. The delta-opioid receptor: Isolation of a cDNA by expression cloning and pharmacological characterization. Proc Natl Acad Sci USA. 1992;89:12048–12052. doi: 10.1073/pnas.89.24.12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Y, Mestek A, Liu J, Hurley JA, Yu L. Molecular cloning and functional expression of a mu-opioid receptor from rat brain. Mol Pharmacol. 1993;44:8–12. [PubMed] [Google Scholar]
- 8.Yasuda K, et al. Cloning and functional comparison of kappa and delta opioid receptors from mouse brain. Proc Natl Acad Sci USA. 1993;90:6736–6740. doi: 10.1073/pnas.90.14.6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mollereau C, et al. ORL1, a novel member of the opioid receptor family. Cloning, functional expression and localization. FEBS letters. 1994;341:33–38. doi: 10.1016/0014-5793(94)80235-1. [DOI] [PubMed] [Google Scholar]
- 10.Bunzow JR, et al. Molecular cloning and tissue distribution of a putative member of the rat opioid receptor gene family that is not a mu, delta or kappa opioid receptor type. FEBS Lett. 1994;347:284–288. doi: 10.1016/0014-5793(94)00561-3. [DOI] [PubMed] [Google Scholar]
- 11.Meunier JC, et al. Isolation and structure of the endogenous agonist of opioid receptor-like ORL1 receptor. Nature. 1995;377:532–535. doi: 10.1038/377532a0. [DOI] [PubMed] [Google Scholar]
- 12.Reinscheid RK, et al. Orphanin FQ: A neuropeptide that activates an opioidlike G protein-coupled receptor. Science. 1995;270:792–794. doi: 10.1126/science.270.5237.792. [DOI] [PubMed] [Google Scholar]
- 13.Dores RM, Lecaude S, Bauer D, Danielson PB. Analyzing the evolution of the opioid/orphanin gene family. Mass Spectrom Rev. 2002;21:220–243. doi: 10.1002/mas.10029. [DOI] [PubMed] [Google Scholar]
- 14.Bradford CS, Walthers EA, Searcy BT, Moore FL. Cloning, heterologous expression and pharmacological characterization of a kappa opioid receptor from the brain of the rough-skinned newt, Taricha granulosa. J Mol Endocrinol. 2005;34:809–823. doi: 10.1677/jme.1.01711. [DOI] [PubMed] [Google Scholar]
- 15.Bradford CS, Walthers EA, Stanley DJ, Baugh MM, Moore FL. Delta and mu opioid receptors from the brain of a urodele amphibian, the rough-skinned newt Taricha granulosa: Cloning, heterologous expression, and pharmacological characterization. Gen Comp Endocrinol. 2006;146:275–290. doi: 10.1016/j.ygcen.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 16.Walthers EA, Bradford CS, Moore FL. Cloning, pharmacological characterization and tissue distribution of an ORL1 opioid receptor from an amphibian, the rough-skinned newt Taricha granulosa. J Mol Endocrinol. 2005;34:247–256. doi: 10.1677/jme.1.01687. [DOI] [PubMed] [Google Scholar]
- 17.Stevens CW, Brasel CM, Mohan S. Cloning and bioinformatics of amphibian mu, delta, kappa, and nociceptin opioid receptors expressed in brain tissue: Evidence for opioid receptor divergence in mammals. Neurosci Lett. 2007;419:189–194. doi: 10.1016/j.neulet.2007.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deviche P, Cotter P, Gulledge CC. Identification, partial characterization, and hypothalamic distribution of kappa, mu, and delta opioid receptors in a passerine songbird (Junco hyemalis) Brain Res. 1993;614:220–226. doi: 10.1016/0006-8993(93)91038-t. [DOI] [PubMed] [Google Scholar]
- 19.Li X, Keith DE, Jr, Evans CJ. Mu opioid receptor-like sequences are present throughout vertebrate evolution. J Mol Evol. 1996;43:179–184. doi: 10.1007/BF02338825. [DOI] [PubMed] [Google Scholar]
- 20.Pinal-Seoane N, et al. Characterization of a new duplicate delta-opioid receptor from zebrafish. J Mol Endocrinol. 2006;37:391–403. doi: 10.1677/jme.1.02136. [DOI] [PubMed] [Google Scholar]
- 21.Darlison MG, et al. Opioid receptors from a lower vertebrate (Catostomus commersoni): Sequence, pharmacology, coupling to a G-protein-gated inward-rectifying potassium channel (GIRK1), and evolution. Proc Natl Acad Sci USA. 1997;94:8214–8219. doi: 10.1073/pnas.94.15.8214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barrallo A, Gonzalez-Sarmiento R, Alvar F, Rodriguez RE. ZFOR2, a new opioid receptor-like gene from the teleost zebrafish (Danio rerio) Brain Res Mol Brain Res. 2000;84:1–6. doi: 10.1016/s0169-328x(00)00152-2. [DOI] [PubMed] [Google Scholar]
- 23.Barrallo A, Gonzalez-Sarmiento R, Porteros A, Garcia-Isidoro M, Rodriguez RE. Cloning, molecular characterization, and distribution of a gene homologous to delta opioid receptor from zebrafish (Danio rerio) Biochem Biophys Res Commun. 1998;245:544–548. doi: 10.1006/bbrc.1998.8496. [DOI] [PubMed] [Google Scholar]
- 24.Rodriguez RE, et al. Characterization of ZFOR1, a putative delta-opioid receptor from the teleost zebrafish (Danio rerio) Neurosci Lett. 2000;288:207–210. doi: 10.1016/s0304-3940(00)01239-8. [DOI] [PubMed] [Google Scholar]
- 25.Stevens CW. Opioid research in amphibians: An alternative pain model yielding insights on the evolution of opioid receptors. Brain Res Brain Res Rev. 2004;46:204–215. doi: 10.1016/j.brainresrev.2004.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sneddon LU. Evolution of nociception in vertebrates: Comparative analysis of lower vertebrates. Brain Res Brain Res Rev. 2004;46:123–130. doi: 10.1016/j.brainresrev.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 27.Sundstrom G, Larsson TA, Brenner S, Venkatesh B, Larhammar D. Evolution of the neuropeptide Y family: New genes by chromosome duplications in early vertebrates and in teleost fishes. Gen Comp Endocrinol. 2008;155:705–716. doi: 10.1016/j.ygcen.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 28.Larsson TA, et al. Early vertebrate chromosome duplications and the evolution of the neuropeptide Y receptor gene regions. BMC Evol Biol. 2008;8:184. doi: 10.1186/1471-2148-8-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Panopoulou G, Poustka AJ. Timing and mechanism of ancient vertebrate genome duplications–the adventure of a hypothesis. Trends Genet. 2005;21:559–567. doi: 10.1016/j.tig.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 30.Putnam NH, et al. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;453:1064–1071. doi: 10.1038/nature06967. [DOI] [PubMed] [Google Scholar]
- 31.Venkatesh B, et al. Survey sequencing and comparative analysis of the elephant shark (Callorhinchus milii) genome. PLoS Biology. 2007;5:e101. doi: 10.1371/journal.pbio.0050101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Irvine SQ, et al. Genomic analysis of Hox clusters in the sea lamprey Petromyzon marinus. J Exp Zool. 2002;294:47–62. doi: 10.1002/jez.10090. [DOI] [PubMed] [Google Scholar]
- 33.Force A, Amores A, Postlethwait JH. Hox cluster organization in the jawless vertebrate Petromyzon marinus. J Exp Zool. 2002;294:30–46. doi: 10.1002/jez.10091. [DOI] [PubMed] [Google Scholar]
- 34.Escriva H, Manzon L, Youson J, Laudet V. Analysis of lamprey and hagfish genes reveals a complex history of gene duplications during early vertebrate evolution. Mol Biol Evol. 2002;19:1440–1450. doi: 10.1093/oxfordjournals.molbev.a004207. [DOI] [PubMed] [Google Scholar]
- 35.Stadler PF, et al. Evidence for independent Hox gene duplications in the hagfish lineage: A PCR-based gene inventory of Eptatretus stoutii. Mol Phylogenet Evol. 2004;32:686–694. doi: 10.1016/j.ympev.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 36.Fried C, Prohaska SJ, Stadler PF. Independent Hox-cluster duplications in lampreys. J Exp Zool B Mol Dev Evol. 2003;299:18–25. doi: 10.1002/jez.b.37. [DOI] [PubMed] [Google Scholar]
- 37.Stevens CW. Molecular evolution of vertebrate opioid receptor proteins: A preview. Recent Dev Pain Res. 2005:13–29. [Google Scholar]
- 38.Nakatani Y, Takeda H, Kohara Y, Morishita S. Reconstruction of the vertebrate ancestral genome reveals dynamic genome reorganization in early vertebrates. Genome Res. 2007;17:1254–1265. doi: 10.1101/gr.6316407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kasahara M, et al. The medaka draft genome and insights into vertebrate genome evolution. Nature. 2007;447:714–719. doi: 10.1038/nature05846. [DOI] [PubMed] [Google Scholar]
- 40.Li X, Keith DE, Jr, Evans CJ. Multiple opioid receptor-like genes are identified in diverse vertebrate phyla. FEBS Lett. 1996;397:25–29. doi: 10.1016/s0014-5793(96)01126-x. [DOI] [PubMed] [Google Scholar]
- 41.Cadet P, Stefano GB. Mytilus edulis pedal ganglia express mu opiate receptor transcripts exhibiting high sequence identity with human neuronal mu1. Brain Res Mol Brain Res. 999;74:242–246. doi: 10.1016/s0169-328x(99)00287-9. [DOI] [PubMed] [Google Scholar]
- 42.Cadet P, Zhu W, Mantione KJ, Baggerman G, Stefano GB. Cold stress alters Mytilus edulis pedal ganglia expression of mu opiate receptor transcripts determined by real-time RT-PCR and morphine levels. Brain Res Mol Brain Res. 2002;99:26–33. doi: 10.1016/s0169-328x(01)00342-4. [DOI] [PubMed] [Google Scholar]
- 43.Coulier F, Popovici C, Villet R, Birnbaum D. MetaHox gene clusters. J Exp Zool. 2000;288:345–351. doi: 10.1002/1097-010X(20001215)288:4<345::AID-JEZ7>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 44.Singh G, Davenport AP. Neuropeptide B and W: Neurotransmitters in an emerging G-protein-coupled receptor system. Br J Pharmacol. 2006;148:1033–1041. doi: 10.1038/sj.bjp.0706825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O'Dowd BF, et al. The cloning and chromosomal mapping of two novel human opioid-somatostatin-like receptor genes, GPR7 and GPR8, expressed in discrete areas of the brain. Genomics. 1995;28:84–91. doi: 10.1006/geno.1995.1109. [DOI] [PubMed] [Google Scholar]
- 46.Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ. Multiple sequence alignment with Clustal X. Trends Biochem Sci. 1998;23:403–405. doi: 10.1016/s0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
- 47.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 49.Schmidt HA, Strimmer K, Vingron M, von Haeseler A. TREE-PUZZLE: Maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002;18:502–504. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.