Abstract
Chromosome segregation in bacteria is rapid and directed, but the mechanisms responsible for this movement are still unclear. We show that Caulobacter crescentus makes use of and requires a dedicated mechanism to initiate chromosome segregation. Caulobacter has a single circular chromosome whose origin of replication is positioned at one cell pole. Upon initiation of replication, an 8-kb region of the chromosome containing both the origin and parS moves rapidly to the opposite pole. This movement requires the highly conserved ParABS locus that is essential in Caulobacter. We use chromosomal inversions and in vivo time-lapse imaging to show that parS is the Caulobacter site of force exertion, independent of its position in the chromosome. When parS is moved farther from the origin, the cell waits for parS to be replicated before segregation can begin. Also, a mutation in the ATPase domain of ParA halts segregation without affecting replication initiation. Chromosome segregation in Caulobacter cannot occur unless a dedicated parS guiding mechanism initiates movement.
Keywords: centromere, parS, ParA
Bacterial chromosomes are highly organized structures with predictable orientation and segregation patterns (1, 2). In vivo fluorescence microscopy showed that the speed of segregation of individual loci is too fast to be accounted for by attachment of sister chromosomes to a growing cell envelope (1, 3, 4), as had been proposed (5). These observations led to the suggestion that rapid segregation may be the consequence of nondedicated mechanisms, such as force exerted by the DNA or RNA polymerases (6, 7) or entropic exclusion of sister chromosomes (8), all of which predict that the order of segregation will follow the order of replication. Alternatively, segregation may be driven by a dedicated mechanism acting on a centromeric sequence (9, 10), in which case the first sequence to segregate would be the centromere regardless of when it is replicated.
The parABS locus is a large family of plasmid and chromosomal elements composed of a cis-acting sequence generally named parS and two transacting proteins: ParB, which binds to cognate parS sites, and ParA, a MinD-related Walker-type ATPase whose plasmid homologues polymerize in vitro and in vivo (11, 12). Although chromosomal parABS (chr-parABS) elements are phylogenetically distinct from those found in plasmids (13), inactivating or overexpressing chr-parABS components in several species leads to elevated numbers of anucleate cells (14–16) and introduction of chr-parABS stabilizes plasmids in heterologous hosts (17–19). In vivo observations of Vibrio cholerae's chromosome I dynamics suggested a mechanism by which ParAI (chromosome I's cognate ParA) pulls on the ParBI/parSI complex to effect chromosome segregation (20). However, although the absence of parAI in V. cholerae alters chromosome segregation, growth is not affected (19, 20). Indeed, despite widespread conservation of the parS sequence, except for V. cholerae chromosome II and Caulobacter crescentus (henceforth, Caulobacter), the absence of parABS elements only mildly impairs growth (18, 21), suggesting the presence of redundant chromosome segregation mechanisms (8, 22–24).
Caulobacter requires an active parABS system to live (21) and replicates its single chromosome only once per cell cycle (25), providing a good model organism to study chromosome segregation in its simplest form. Here, we show that the ancestral parS sequence is the site of force exertion during the initiation of Caulobacter chromosome segregation, and that ParA activity is required for this movement. We also find that in the absence of parS-directed movement, segregation of newly replicated loci does not begin. Finally, we demonstrate that parS is specifically targeted to the cell pole, and that the subcellular location of two other DNA loci depends on their chromosomal distance from parS.
Results
Extra Copies of parS Impair Viability of Caulobacter Cells.
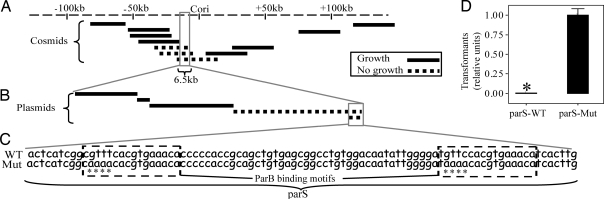
We hypothesized that the site of force exertion for chromosome segregation should show copy-number effects when present in trans. If many indistinguishable copies of this site are present, such as when Caulobacter is transformed with plasmid-borne extra copies, the cells will segregate a random subset of plasmids and chromosomes to each daughter. This will result in some daughters with zero or two chromosomes, thus causing significantly slower growth. Accordingly, we screened for growth impairment in the presence of extrachromosomal fragments of DNA. We first screened a library of cosmids (each ≈30 kb at five to eight copies per chromosome equivalent) that were tiled across the origin region of the chromosome (Fig. 1). We chose this region because previous experiments had shown that it segregates before the rest of the chromosome (1) and, therefore, should contain the centromeric site. Three cosmids that shared a 6.5-kb region prevented normal growth of Caulobacter colonies (Fig. 1A). We narrowed this region further by inserting individual subregions into a promoterless plasmid (≈10 copies per chromosome equivalent) and found a 100-bp stretch of DNA that could not be maintained in Caulobacter (Fig. 1B). This sequence, which lies upstream of the parAB genes, contains two ParB-binding boxes (Fig. 1C; ref. 26), which comparative genomics analysis identified as the Caulobacter parS site (27). Furthermore, previous work had shown that one of these predicted binding motifs is bound directly by Caulobacter ParB in vitro (28).
Fig. 1.
Extra copies of parS DNA impair cell viability. (A and B) DNA-copy-number screen. Cosmids (A) or plasmids (B), carrying an antibiotic resistance cassette and a region (lines) of the Caulobacter genome, were transformed into cells. Solid lines represent constructs that permitted growth of colonies on selective plates; dashed lines represent constructs that did not allow growth. (C) Sequence of the wild-type parS site (WT) and parS with mutated ParB-binding motifs (Mut). Asterisks denote the location of base-pair changes in parS. (D) Transformation efficiency (relative number of colonies at day 3) of plasmids carrying either the wild-type (parS-WT) or mutated (parS-Mut) versions of parS. * = 0.003. Bars represent the means of seven separate experiments. Here and elsewhere, error bars represent standard error of the mean (SEM).
To determine whether the ParB-binding sites in parS were responsible for the loss of viability seen in our screen (Fig. 1 A and B), we introduced four point mutations into each site (Fig. 1C). These mutations restored the ability of the plasmid to be maintained in cells (Fig. 1D), demonstrating that extrachromosomal copies of the ParB-binding boxes impair viability in Caulobacter.
Identification of a Chromosomal Region That Contains parS as the Site of Force Exertion During Chromosome Segregation.
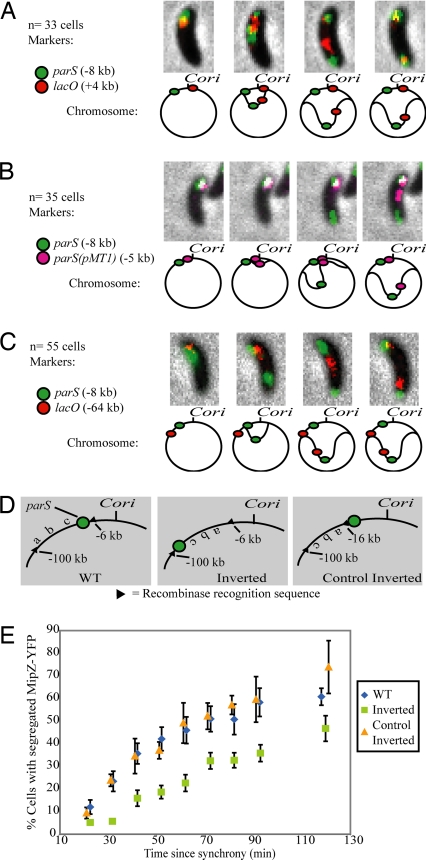
Having established a copy-number effect for the parS sequence, we asked whether parS is the first part of the chromosome to segregate, as is the case in V. cholerae (20). As is common in bacteria, chromosome replication in Caulobacter begins at a single origin of replication (Cori) [refs. 29 and 30; supporting information (SI) Fig. S1]. We used time-lapse fluorescence microscopy to track the order of segregation of the parS region relative to Cori and other nearby loci in vivo. We followed the cellular position of parS using a MipZ-YFP fusion, which binds ParB/parS directly (28), under the control of the native mipZ promoter. Concurrently, we followed the cellular position of an array of lacO operators located at +4 kb (all distances are relative to Cori; parS is located at −8 kb) using a LacI-CFP fusion driven by a xylose-inducible promoter. As shown in Fig. 2A and Movie S1, in 33 of 33 cells examined, parS segregated ahead of the lac operators.
Fig. 2.
A 10-kb region including parS contains the site of force exertion during segregation. (A–C) Segregation pattern of different loci and accompanying schematics of the position of markers used and order of segregation. Note that although micrographs show only one cell, the segregation order shown (parS segregating before the other tagged locus) was repeated in all cells observed. In all cases, parS visualized with MipZ-YFP. For clarity, schematics are not to scale. (A) lacO inserted at +4 kb and visualized with LacI-CFP. (B) parS(pMT1) inserted at −5 kb and visualized with CFP-pMT1Δ23ParB. (C) lacO inserted at −64 kb and visualized with LacI-CFP. (D) Schematic of the chromosomal configuration of inversion strains constructed by site-specific recombination. (E) Separating parS from Cori delays segregation. Plotted are the percentage of cells with two distinct MipZ-YFP foci as a function of time from synchrony. To avoid phototoxicity effects, a new field of cells was imaged for each time point. Chromosome configurations as in D. Symbols represent means of three experiments.
Next, we inserted a smaller DNA marker (the lacO arrays are ≈10 kb in length) between parS and Cori (Fig. 2B). The marker was the parS sequence from plasmid pMT1 (≈100 bp). CFP-pMT1Δ23ParB, which binds its cognate parS [denoted here as parS(pMT1)] was used to follow the cellular position of this locus (31). In agreement with previous reports (31), we did not observe any cross-talk between the Caulobacter and pMT1 parS systems (Fig. S2 A and B). Fig. 2B and Movie S2 show that in 35 of 35 cells observed, the Caulobacter parS sequence segregated ahead of parS(pMT1), despite having been replicated later.
Nondedicated chromosome segregation models predict that the order of segregation follows from the order of replication. When replication begins, entropic exclusion and/or the DNA/RNA polymerases would immediately force the nascent daughter strands apart. It follows then that when two nearby sequences are observed segregating, the one located closest to Cori would tend to move first. Our results contradict this prediction (Fig. 2B) and show that initial segregation in Caulobacter is driven by force exerted on the chromosome at the parS site.
To refine the location of the site of force exertion, we constructed the strain shown in Fig. 2C, which carries the lacO arrays at −64 kb. In this strain, parS moved ahead of the lacO arrays in all 55 cells observed (Fig. 2C), showing that the site of force exertion must be located between −5 and −64 kb.
Separating parS from Cori Delays Segregation.
As is commonly found in bacteria that have parABS (27), the Caulobacter parS sequence is located near Cori (8 kb away in the 4,000-kb genome). We asked whether parS would still be the first chromosomal site to segregate when it was moved farther from Cori. For this purpose, we used the phage PhiC31 site-specific recombinase (32) to create a strain carrying a chromosomal inversion that separates parS from Cori by ≈100 kb (“Inverted” in Figs. 2D and S3). We used this recombinase, because it has been shown to act unidirectionally (33), so that it will not reinvert the chromosome back to its wild-type configuration after it has catalyzed the inversion reaction (Fig. S3). We also created a similar inversion that excluded parS, to control for effects brought about by changes in the orientation of the inverted DNA (“Control Inverted” in Figs. 2D and S3).
We then followed the timing of parS segregation in synchronized populations of wild-type and the two inversion strains. Fig. 2E shows the percentage of cells which have begun segregation (measured as the appearance of two distinct MipZ-YFP foci) as a function of time from synchrony. We found that separating parS from Cori resulted in a delay in the timing of parS segregation (compare green squares and blue diamonds). Inverting a similar fragment of DNA lacking parS, in contrast, did not have a noticeable effect compared with wild type (yellow triangles and blue diamonds). The same behavior was observed when we followed the segregation of lacO arrays located at +4 kb (Fig. S4). These results show that the location of parS relative to Cori controls the timing of segregation. When parS is close to Cori, as in the WT and Control Inverted strains in Fig. 2 D and E, segregation happens quickly. When parS is located farther away, as in the Inverted strain, there is a delay before parS is replicated and segregation can begin. Furthermore, this result localized the site of force exertion to a 10-kb region that contains parS (−6 to −16 kb in Fig. 2D).
A ParAK20R Mutant Halts Segregation in Vivo and Induces the Production of Anucleate Minicells.
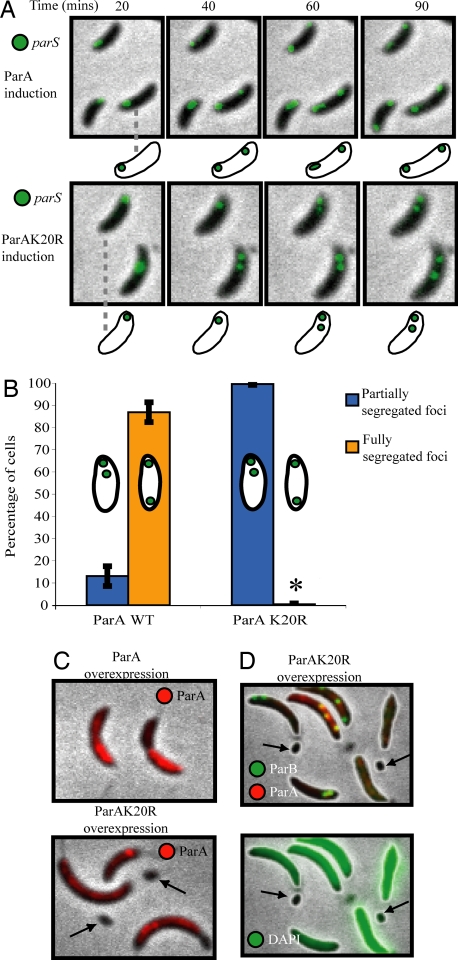
Plasmid parABS segregation systems rely on their cognate ParA ATPase to accurately move or position the parS/ParB complex (34–36). In Caulobacter, overexpressing ParA and/or ParB results in mislocalization of ParB foci and the appearance of 5–10% anucleate cells (16). We therefore asked whether Caulobacter ParA is involved in the directed movement of parS during segregation. Accordingly, we created a merodiploid strain with both the wild-type parA gene at its native position and one of two xylose-inducible genes: wild-type parA-mCherry or parAK20R-mCherry. Lysine 20 is a highly conserved amino acid in the Walker A ATP-binding motif of Caulobacter ParA. It has been shown that a homologous K to R mutation in phage P1 ParA greatly increases plasmid instability (37), so we predicted that it would have a dominant-negative phenotype in Caulobacter.
Fig. 3A and Movies S3 and S4 show synchronized cells in which we visualized parS using CFP-ParB, which binds to and colocalizes with parS (28). Under conditions of mild expression, ParA-mCherry did not perturb the segregation pattern of parS (Fig. 3A Upper). In cells with the ParAK20R-mCherry mutant, however, parS was replicated and moved a short distance, but segregation was not completed (Fig. 3A Lower, quantified in Fig. 3B).
Fig. 3.
A mutation in ParA abrogates chromosome segregation and produces anucleate minicells. (A) Time-lapse fluorescence micrographs of CFP-ParB in synchronized cells undergoing segregation after 60 min of ParA-mCherry (Upper) or ParAK20R-mCherry (Lower) induction with 0.03% xylose. (B) Percentage of cells with two foci that were either partially (blue bars) or fully (yellow bars) segregated 90 min after synchrony, in cultures treated as in A. * = 0.5%. Bars represent the average of two independent experiments, each with at least 150 cells counted for each strain. (C) ParA-mCherry (Upper) and ParAK20R-mCherry (Lower) localization in cells induced for 5 hours with 0.3% xylose. Arrows point to minicells produced only with the mutant ParA. (D) Localization of ParAK20R-mCherry and CFP-ParB (Upper) and DAPI (Lower) in cells carrying ParAK20R-mCherry driven by the xylose promoter and treated as in C. Arrows point to minicells that show no CFP-ParB foci and very low DAPI fluorescence. Levels of DAPI fluorescence have been increased to aid visualization of the very low fluorescence in minicells.
To determine the long term effects of the presence of ParAK20R-mCherry, we induced its expression with 10-fold higher concentration of xylose for 5 h. Under these conditions, the localization pattern of ParA-mCherry and ParAK20R-mCherry differed slightly (Figs. 3C and S5 and S6). Overexpression of both versions of ParA-mCherry caused the cells to grow somewhat filamentous in accord with previous observations (16), but only the mutant produced anucleate minicells (arrows in Fig. 3 C and D), likely as an indirect result of a parS segregation defect. MipZ, which colocalizes with parS, acts also as a negative regulator of the FtsZ division ring (28). Therefore, as the cell with ParAK20R-mCherry overexpression grows longer, MipZ remains sequestered at the end of the cell with two copies of parS, and the division ring is free to form at the opposite end, creating a DNA-free minicell. Taken together, these experiments show that a ParA mutant prevents chromosome segregation, causes a misplacement of the division ring, and leads to the creation of DNA-free minicells (Fig. 3).
Initiation of Chromosome Segregation Requires parS-Directed Movement.
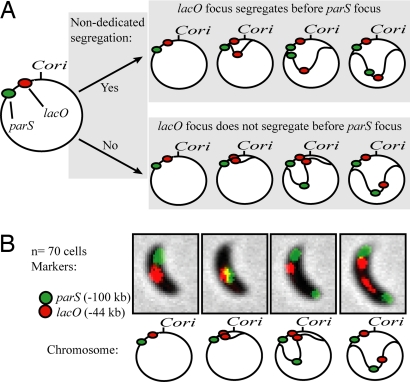
The ability to uncouple segregation from replication by moving parS away from Cori allowed us to determine whether nondedicated mechanisms are sufficient to effect chromosome segregation in Caulobacter, as has been suggested (8). The strain shown in Fig. 4A carries an insertion of the lacO arrays at −44 kb and the same 100-kb inversion described in Fig. 2D (“Inverted”), which separates parS from Cori by ≈100 kb. This inversion creates a significant time interval during which the lacO arrays have been replicated and parS has not (Fig. 4A). We can estimate the duration of this interval by referring to Fig. 2E. There, it can be seen that parS is replicated ≈20 min later than when it is at its wild-type position. Because the lacO arrays are placed roughly halfway between the “WT” and “Inverted” positions of parS, we estimate that the lacO arrays will be replicated ≈10 min before parS.
Fig. 4.
Replication is not sufficient to initiate chromosome segregation. (A) Schematic of the strain constructed to test for the presence of nondedicated segregation mechanisms, and possible outcomes. (Upper) Nondedicated segregation occurs. Replicated DNA is moved independently of the parABS mechanism, and movement of the lacO arrays takes place before parS duplication. (Lower) Nondedicated mechanisms are not able to initiate chromosome segregation, and lacO movement does not take place until after parS has begun segregating. (B) Results of the experiment outlined in panel A. parS was followed by using MipZ-YFP, lacO arrays were followed by using LacI-CFP. Seventy cells were observed segregating and in all cases parS segregated before the lacO arrays.
Nondedicated models predict that newly replicated lacO DNA will begin segregating immediately after being replicated, at speeds comparable to the 0.3 μm/min measured experimentally for Caulobacter (Fig. 4A; refs. 1, 6–8). Assuming the ≈10-min interval discussed above and a segregation speed of 0.3 μm/min, the lacO arrays should move across the entire ≈2.0 μm cell before parS is replicated (8). However, we never observed this behavior (Fig. 4B; Movie S5). In fact, no lacO array segregation was ever observed before the beginning of parS segregation (Movie S5).
Thus, our results strongly suggest that initial chromosome segregation in Caulobacter results from a mechanism that involves ParA exerting force at parS/ParB, and that entropy (8) or DNA/RNA polymerization reactions alone are not sufficient to initiate chromosome movement. Importantly, a change in the overall transcription orientation bias of the chromosome would modify the predictions made by the RNA polymerase model (7). This was not the case in our inversion strain. Of 110 genes inverted, 56 are transcribed away from Cori and 54 toward, rendering the change in transcription bias insignificant.
parS and not Cori Is Anchored at the Cell Pole.
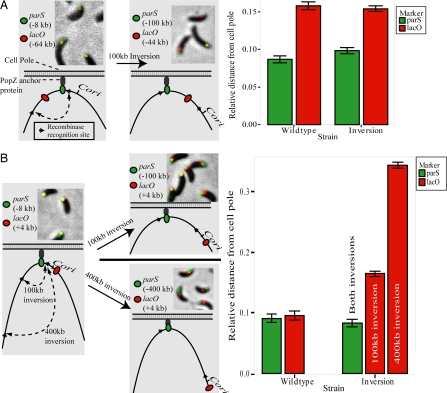
In the inverted strain shown in Figs. 4 and 5A, Cori was located closer to the lacO arrays than to parS (≈44 and ≈100 kb away, respectively). If Cori were the sequence that orients the Caulobacter chromosome with respect to the cell pole, we would expect the lacO arrays to be found nearer to the pole than parS. However, the opposite was true (Fig. 5A). This observation suggested that parS, and not Cori, is specifically anchored at the cell pole. To test this, we constructed the two additional inversion strains shown in Fig. 5B and measured the positions of parS and Cori. Before inversion, parS and the lacO arrays were located 8 and 4 kb away from Cori, respectively (left schematic in Fig. 5B), and both loci were found very close to the cell pole (Fig. 5B, “wildtype” bars on graph). In the inversion strains, parS remained close to the pole (Fig. 5B), even though it was now either 100 or 400 kb away from Cori. The position of the lacO arrays, however, shifted away from the pole proportionally to its distance from parS (Fig. 5B, red bars). Note that the distance from the lacO arrays to Cori was unchanged. These results demonstrate that the cellular positions of at least two DNA loci are determined by their relative distance to parS, rather than Cori.
Fig. 5.
parS is fixed at the cell pole, whereas Cori is not. (A and B) Fluorescent micrographs, schematic, and quantification of the relative positions of parS and lacO in cells before and after inversion of the chromosome region shown. (A) lacO originally positioned at −64 kb. (B) lacO positioned at +4 kb. Bars represent the average of three independent experiments, each with at least 100 cells counted.
Discussion
We have shown that Caulobacter employs the parS sequence as a centromere with which to segregate its chromosome through the action of the essential ParA ATPase. However, in most other bacteria studied, deleting ParA causes only mild segregation defects (14, 15, 17). A possible explanation for these differences is that when the parABS system is absent, nondedicated mechanisms of chromosome segregation take over. We have shown that in Caulobacter, this is not the case. When the Caulobacter parABS system cannot guide directional movement, segregation does not begin (Fig. 4). Earlier work had indicated that the initiation of chromosome segregation depends on the actin-like protein MreB, but our present results establish that the principal driving force for segregation is mediated by the ParABS system. Z, Gitai and coworkers have recently observed that chromosome segregation in Caulobacter depends on MreB only under certain conditions (Z. Gitai, personal communication).
It is important to note that the parS-driven initiation of chromosome segregation in Caulobacter does not preclude a role for forces exerted by the DNA/RNA polymerases (6, 7), compaction from condensins, or entropic exclusion (22), during bulk chromosome segregation. After parS has guided the newly replicated origin region to the opposite cell pole, these forces could contribute to the movement of the rest of the chromosome. Indeed, the documented rapid segregation of loci that are far away from parS (1) requires such contributions.
The Caulobacter chromosome is arranged in a highly organized manner within the cell, as is the case in other bacteria (1, 2, 38), but how this is achieved remains an open question. Recently, it was reported that the Caulobacter parS/ParB complex is anchored to the cell pole by the PopZ polymeric protein (Fig. 5A; refs. 39 and 40). This polar anchoring is required for effective chromosome segregation and cell division (39, 40). Here, we showed that the orientation of the Caulobacter chromosome within the cell appears to be achieved by “clocking” the DNA molecule relative not to Cori but rather to parS. It remains to be determined whether additional specific targeting sequences exist in the Caulobacter chromosome.
The mechanisms through which DNA is localized subcellularly vary considerably among species. During sporulation in Bacillus subtilis, for example, the RacA and DivIVA proteins combine to anchor the chromosomal origin region to the cell pole (41). RacA binds to 25 sites spread over a 612-kb region of origin-proximal DNA (42), and DivIVA (which does not share sequence similarity with PopZ) is required for RacA-mediated localization of this region to the pole (41). The Escherichia coli chromosome, in turn, does not contain a parABS locus, but it has been shown that the migS cis-acting sequence affects bipolar localization of the origin region in this species (43). In addition, the E. coli chromosome is organized with the left and right arms in separate cell halves (31), an organization that requires the SMC-like condensation protein MukB (44). Clearly, there is a strong selective pressure for bacterial chromosomes to remain organized inside the cell, perhaps to coordinate DNA segregation with cell division. Indeed, RacA-mediated anchoring in B. subtilis prevents the formation of DNA-free forespores (41), and loss of polar parS/ParB anchoring in Caulobacter leads to defects in cell division (39, 40).
In summary, the Caulobacter parABS system and its associated proteins initiate segregation, orient and anchor the chromosome, and signal the onset of segregation so that division may begin (28). They are thus emerging as the functional equivalent of a eukaryotic kinetochore.
Experimental Procedures
Tables S1–S3 list all strains and plasmids used in this study, along with a brief description of their construction. We grew all Caulobacter CB15N-derived strains in M2G minimal and PYE media at 28°C. Our procedures for ΦCR30 phage transductions and transformation of plasmids into Caulobacter (45) and DAPI staining (16) have been described. Detailed descriptions of our synchronization, microscopy, and image analysis methods and the induction of recombination are provided in SI Text.
Supplementary Material
Acknowledgments.
We thank M. Thanbichler (Max Planck Institute, Marburg, Germany), S. Austin (National Cancer Institute), M. P. Calos (Stanford University), and F. G. Hansen (Technical University of Denmark) for providing reagents; Stephanie Weber (Stanford University) for some Matlab code; and P. T. McGrath, A. A. Iniesta, B. Burkholder, and members of the Shapiro and McAdams laboratories for helpful discussions. This work was supported by National Institutes of Health Grants R01 GM51426 R24 and GM073011-04 (to L.S.) and Department of Energy Grant DE-FG02-05ER64136 (to H.H.M.). E.T. was funded by the Smith Stanford Graduate Fellowship.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0807448105/DCSupplemental.
References
- 1.Viollier PH, et al. Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. Proc Natl Acad Sci USA. 2004;101:9257–9262. doi: 10.1073/pnas.0402606101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Niki H, Yamaichi Y, Hiraga S. Dynamic organization of chromosomal DNA in Escherichia coli. Genes Dev. 2000;14:212–223. [PMC free article] [PubMed] [Google Scholar]
- 3.Fiebig A, Keren K, Theriot JA. Fine-scale time-lapse analysis of the biphasic, dynamic behaviour of the two Vibrio cholerae chromosomes. Mol Microbiol. 2006;60:1164–1178. doi: 10.1111/j.1365-2958.2006.05175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bates D, Kleckner N. Chromosome and replisome dynamics in. E coli: Loss of sister cohesion triggers global chromosome movement and mediates chromosome segregation. Cell. 2005;121:899–911. doi: 10.1016/j.cell.2005.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jacob F, Brenner S, Cuzin F. On the regulation of DNA replication in bacteria. Cold Spring Harbor Symp Quant Biol. 1963;28:329–348. [Google Scholar]
- 6.Lemon KP, Grossman AD. The extrusion-capture model for chromosome partitioning in bacteria. Genes Dev. 2001;15:2031–2041. doi: 10.1101/gad.913301. [DOI] [PubMed] [Google Scholar]
- 7.Dworkin J, Losick R. Does RNA polymerase help drive chromosome segregation in bacteria? Proc Natl Acad Sci USA. 2002;99:14089–14094. doi: 10.1073/pnas.182539899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jun S, Mulder B. Entropy-driven spatial organization of highly confined polymers: Lessons for the bacterial chromosome. Proc Natl Acad Sci USA. 2006;103:12388–12393. doi: 10.1073/pnas.0605305103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerdes K, Moller-Jensen J, Ebersbach G, Kruse T, Nordstrom K. Bacterial mitotic machineries. Cell. 2004;116:359–366. doi: 10.1016/s0092-8674(04)00116-3. [DOI] [PubMed] [Google Scholar]
- 10.Gitai Z, Dye NA, Reisenauer A, Wachi M, Shapiro L. MreB actin-mediated segregation of a specific region of a bacterial chromosome. Cell. 2005;120:329–341. doi: 10.1016/j.cell.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 11.Pogliano J. The bacterial cytoskeleton. Curr Opin Cell Biol. 2008;20:19–27. doi: 10.1016/j.ceb.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 12.Leonard TA, Butler PJ, Lowe J. Bacterial chromosome segregation: structure and DNA binding of the Soj dimer–a conserved biological switch. EMBO J. 2005;24:270–282. doi: 10.1038/sj.emboj.7600530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerdes K, Moller-Jensen J, Bugge Jensen R. Plasmid and chromosome partitioning: Surprises from phylogeny. Mol Microbiol. 2000;37:455–466. doi: 10.1046/j.1365-2958.2000.01975.x. [DOI] [PubMed] [Google Scholar]
- 14.Ireton K, Gunther N W t, Grossman AD. spo0J is required for normal chromosome segregation as well as the initiation of sporulation in Bacillus subtilis. J Bacteriol. 1994;176:5320–5329. doi: 10.1128/jb.176.17.5320-5329.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lasocki K, Bartosik AA, Mierzejewska J, Thomas CM, Jagura-Burdzy G. Deletion of the parA (soj) homologue in. Pseudomonas aeruginosa causes ParB instability and affects growth rate, chromosome segregation, and motility. J Bacteriol. 2007;189:5762–5772. doi: 10.1128/JB.00371-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mohl DA, Gober JW. Cell cycle-dependent polar localization of chromosome partitioning proteins in Caulobacter crescentus. Cell. 1997;88:675–684. doi: 10.1016/s0092-8674(00)81910-8. [DOI] [PubMed] [Google Scholar]
- 17.Dubarry N, Pasta F, Lane D. ParABS systems of the four replicons of Burkholderia cenocepacia: New chromosome centromeres confer partition specificity. J Bacteriol. 2006;188:1489–1496. doi: 10.1128/JB.188.4.1489-1496.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamaichi Y, Fogel MA, Waldor MK. par genes and the pathology of chromosome loss in Vibrio cholerae. Proc Natl Acad Sci USA. 2007;104:630–635. doi: 10.1073/pnas.0608341104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamaichi Y, Fogel MA, McLeod SM, Hui MP, Waldor MK. Distinct centromere-like parS sites on the two chromosomes of Vibrio spp. J Bacteriol. 2007;189:5314–5324. doi: 10.1128/JB.00416-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fogel MA, Waldor MK. A dynamic, mitotic-like mechanism for bacterial chromosome segregation. Genes Dev. 2006;20:3269–3282. doi: 10.1101/gad.1496506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mohl DA, Easter J, Jr, Gober JW. The chromosome partitioning protein, ParB, is required for cytokinesis in Caulobacter crescentus. Mol Microbiol. 2001;42:741–755. doi: 10.1046/j.1365-2958.2001.02643.x. [DOI] [PubMed] [Google Scholar]
- 22.Arnold A, Jun S. Time scale of entropic segregation of flexible polymers in confinement: Implications for chromosome segregation in filamentous bacteria. Phys Rev. 2007;76 doi: 10.1103/PhysRevE.76.031901. 031901. [DOI] [PubMed] [Google Scholar]
- 23.Woldringh CL, Nanninga N. Structural and physical aspects of bacterial chromosome segregation. J Struct Biol. 2006;156:273–283. doi: 10.1016/j.jsb.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 24.Elmore S, Muller M, Vischer N, Odijk T, Woldringh CL. Single-particle tracking of oriC-GFP fluorescent spots during chromosome segregation in Escherichia coli. J Struct Biol. 2005;151:275–287. doi: 10.1016/j.jsb.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 25.Marczynski GT. Chromosome methylation and measurement of faithful, once and only once per cell cycle chromosome replication in Caulobacter crescentus. J Bacteriol. 1999;181:1984–1993. doi: 10.1128/jb.181.7.1984-1993.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin DC, Grossman AD. Identification and characterization of a bacterial chromosome partitioning site. Cell. 1998;92:675–685. doi: 10.1016/s0092-8674(00)81135-6. [DOI] [PubMed] [Google Scholar]
- 27.Livny J, Yamaichi Y, Waldor MK. Distribution of centromere-like parS sites in bacteria: Insights from comparative genomics. J Bacteriol. 2007;189:8693–8703. doi: 10.1128/JB.01239-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thanbichler M, Shapiro L. MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell. 2006;126:147–162. doi: 10.1016/j.cell.2006.05.038. [DOI] [PubMed] [Google Scholar]
- 29.Dingwall A, Shapiro L. Rate, origin, and bidirectionality of Caulobacter chromosome replication as determined by pulsed-field gel electrophoresis. Proc Natl Acad Sci USA. 1989;86:119–123. doi: 10.1073/pnas.86.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marczynski GT, Shapiro L. Cell-cycle control of a cloned chromosomal origin of replication from Caulobacter crescentus. J Mol Biol. 1992;226:959–977. doi: 10.1016/0022-2836(92)91045-q. [DOI] [PubMed] [Google Scholar]
- 31.Nielsen HJ, Ottesen JR, Youngren B, Austin SJ, Hansen FG. The. Escherichia coli chromosome is organized with the left and right chromosome arms in separate cell halves. Mol Microbiol. 2006;62:331–338. doi: 10.1111/j.1365-2958.2006.05346.x. [DOI] [PubMed] [Google Scholar]
- 32.Groth AC, Olivares EC, Thyagarajan B, Calos MP. A phage integrase directs efficient site-specific integration in human cells. Proc Natl Acad Sci USA. 2000;97:5995–6000. doi: 10.1073/pnas.090527097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thorpe HM, Smith MC. In vitro site-specific integration of bacteriophage DNA catalyzed by a recombinase of the resolvase/invertase family. Proc Natl Acad Sci USA. 1998;95:5505–5510. doi: 10.1073/pnas.95.10.5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garner EC, Campbell CS, Weibel DB, Mullins RD. Reconstitution of DNA segregation driven by assembly of a prokaryotic actin homolog. Science. 2007;315:1270–1274. doi: 10.1126/science.1138527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Derman AI, Lim-Fong G, Pogliano J. Intracellular mobility of plasmid DNA is limited by the ParA family of partitioning systems. Mol Microbiol. 2008;67:935–946. doi: 10.1111/j.1365-2958.2007.06066.x. [DOI] [PubMed] [Google Scholar]
- 36.Saint-Dic D, Frushour BP, Kehrl JH, Kahng LS. A parA homolog selectively influences positioning of the large chromosome origin in Vibrio cholerae. J Bacteriol. 2006;188:5626–5631. doi: 10.1128/JB.00250-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fung E, Bouet JY, Funnell BE. Probing the ATP-binding site of P1 ParA: Partition and repression have different requirements for ATP binding and hydrolysis. EMBO J. 2001;20:4901–4911. doi: 10.1093/emboj/20.17.4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boccard F, Esnault E, Valens M. Spatial arrangement and macrodomain organization of bacterial chromosomes. Mol Microbiol. 2005;57:9–16. doi: 10.1111/j.1365-2958.2005.04651.x. [DOI] [PubMed] [Google Scholar]
- 39.Bowman G, et al. A polymeric protein anchors the chromosomal origin/ParB complex at a bacterial cell pole. Cell. 2008 doi: 10.1016/j.cell.2008.07.015. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ebersbach G, Briegel A, Jensen G J, Jacobs-Wagner C. A multimeric pole-organizing protein critical for chromosome attachment, division and protein localization in Caulobacter. Cell. 2008 doi: 10.1016/j.cell.2008.07.016. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ben-Yehuda S, Rudner DZ, Losick R. RacA, a bacterial protein that anchors chromosomes to the cell poles. Science. 2003;299:532–536. doi: 10.1126/science.1079914. [DOI] [PubMed] [Google Scholar]
- 42.Ben-Yehuda S, et al. Defining a centromere-like element in Bacillus subtilis by Identifying the binding sites for the chromosome-anchoring protein RacA. Mol Cell. 2005;17:773–782. doi: 10.1016/j.molcel.2005.02.023. [DOI] [PubMed] [Google Scholar]
- 43.Yamaichi Y, Niki H. migS, a cis-acting site that affects bipolar positioning of oriC on the Escherichia coli chromosome. EMBO J. 2004;23:221–233. doi: 10.1038/sj.emboj.7600028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Danilova O, Reyes-Lamothe R, Pinskaya M, Sherratt D, Possoz C. MukB colocalizes with the oriC region and is required for organization of the two Escherichia coli chromosome arms into separate cell halves. Mol Microbiol. 2007;65:1485–1492. doi: 10.1111/j.1365-2958.2007.05881.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ely B. Genetics of Caulobacter crescentus. Methods Enzymol. 1991;204:372–384. doi: 10.1016/0076-6879(91)04019-k. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.