Abstract
The twin-arginine translocation (Tat) system transports folded proteins across the bacterial cytoplasmic membrane and the thylakoid membrane of plant chloroplasts. The essential components of the Tat pathway are the membrane proteins TatA, TatB, and TatC. TatA is thought to form the protein translocating element of the Tat system. Current models for Tat transport make predictions about the oligomeric state of TatA and whether, and how, this state changes during the transport cycle. We determined the oligomeric state of TatA directly at native levels of expression in living cells by photophysical analysis of individual yellow fluorescent protein-labeled TatA complexes. TatA forms complexes exhibiting a broad range of stoichiometries with an average of ≈25 TatA subunits per complex. Fourier analysis of the stoichiometry distribution suggests the complexes are assembled from tetramer units. Modeling the diffusion behavior of the complexes suggests that TatA protomers associate as a ring and not a bundle. Each cell contains ≈15 mobile TatA complexes and a pool of ≈100 TatA molecules in a more disperse state in the membrane. Dissipation of the protonmotive force that drives Tat transport has no affect on TatA complex stoichiometry. TatA complexes do not form in cells lacking TatBC, suggesting that TatBC controls the oligomeric state of TatA. Our data support the TatA polymerization model for the mechanism of Tat transport.
Keywords: fluorescence, Tat protein transport
The bacterial cytoplasmic membrane and the chloroplast thylakoid membrane have a common evolutionary origin and contain equivalent protein transport pathways. The Sec pathway translocates proteins as unstructured chains (1), whereas the twin-arginine translocation (Tat) system is dedicated to transporting folded proteins (2–4). In both cases substrates must be translocated without compromising the membrane permeability barrier. This translocation is particularly challenging for the Tat apparatus because folded proteins are larger and more variable in size than the linear peptides transported by the Sec system. The mechanism of Tat transport has not been established but appears unrelated to that of other membrane transporters.
The essential Tat components in the bacterium Escherichia coli are the integral membrane proteins TatA, TatB, and TatC (5–8). TatB and TatC form a large membrane receptor complex (9) that recognizes an N-terminal signal peptide present on substrates (10, 11). Detergent-solubilized TatA is found as large, homooligomeric complexes of variable size (12–15). Low-resolution structures of purified TatA show doughnut-shaped particles with an internal cavity large enough to accommodate substrates (14). This and other evidence (10, 16) suggest that TatA forms the translocating element of the Tat system.
Tat transport is driven by the transmembrane protonmotive force (pmf) (17). Cross-linking studies show a pmf-dependent association between TatA and TatBC during transport (11, 16). In addition, the number of chemical cross-links that can be formed between TatA molecules increases under transport conditions (18). These observations have led to the suggestion that substrate interaction with TatBC triggers TatA polymerization (4, 18). We term this hypothesis the polymerization model. The observed heterogeneity in the size of detergent-solubilized TatA has led to the proposal that dynamic variation of TatA oligomeric state could maintain a seal around substrates of different sizes during transport (a bespoke channel model; ref. 14). Alternatively, it has been suggested that concentration of TatA by polymerization may alter local membrane bilayer structure to allow substrate movement (bilayer perturbation models; refs. 4 and 18–21).
Many questions about these models and their experimental basis remain. Most obviously, if TatA undergoes substrate-induced polymerization, why is TatA present as high-order oligomers in detergent solution in the absence of substrate or of other Tat components (12, 14)? This question, in turn, raises the issue of whether it is valid to study the oligomeric state of TatA in detergent solution when the phospholipids (and pmf) that might be expected to differentially stabilize the polymerized and dispersed states are absent. Although the TatA cross-linking studies have, by contrast, been carried out in a membrane environment it is difficult to confirm that the observed changes in cross-linking patterns reflect differences in oligomer size and not conformational change. In addition, cross-linking is unable to define the stoichiometry and stoichiometry distribution of the TatA complexes that are present.
Testing the proposed mechanistic models of TatA function requires a method that can directly determine the stoichiometry of individual TatA complexes in their native membrane environment. To this end we have used fluorescence microscopy to visualize individual yellow fluorescent protein-labeled TatA (TatA-YFP) complexes expressed at native levels in living E. coli cells. We show that it is possible to use the fluorescence intensity curves of tracked complexes to determine TatA stoichiometry. We find that TatA complexes have a heterogeneous size distribution controlled by other components of the Tat pathway. The methodology we describe to characterize the stoichiometries of individual mobile protein complexes should be widely applicable to other multicomponent systems.
Results
Our aim was to determine the organization and behavior of TatA in its native environment at physiological expression levels. We constructed a gene coding for a fusion of enhanced YFP to the carboxyl terminus of E. coli TatA. This gene was under the control of the native tatA promoter and was integrated into the phage lambda attachment site (att) on the E. coli chromosome in a range of tat backgrounds [supporting information (SI) Text]. The resulting strains are named according to the proteins they produce, with the TatA-YFP fusion designated “Ay.” For example, AyBC expresses the TatA-YFP fusion, TatB, and TatC, but not TatE or native TatA. YFP was used in preference to other GFP variants because it is relatively photostable and because the red-shifted wavelength minimizes phototoxicity and interference from cellular autofluorescence (22, 23).
The TatA-YFP fusion is stable, is expressed at similar levels in the different background strains, and localizes exclusively to the membrane fraction of broken cells (Fig. 1 A and B). Protease accessibility experiments (Fig. 1D) show that the YFP domain is at the cytoplasmic side of the membrane in agreement with the topology determined for native TatA by the same method (24, 25); note, however, that it has been suggested that the C tail of TatA can adopt the opposite orientation in the membrane (26, 27). The functionality of the fusion protein was investigated by assessing the ability to rescue a tatA mutation. The tatA-yfp allele corrected the cell-chaining and detergent-sensitivity phenotypes of the tatA mutant that arise from a failure to export the amidases AmiA and AmiC (28). It restores growth of the tatA mutant with trimethylamine N-oxide (TMAO) as electron acceptor, a metabolic mode that depends on the Tat substrate TMAO reductase. It also restores export of the Tat substrate CueO (Fig. 1E) to the periplasm. The amount of CueO transported is lower than that supported by the parental tatA allele, and the quantity of TMAO reductase activity in the periplasm was not detectably higher than that of a tatA strain (Table S1). Thus, although the TatA-YFP fusion is functional, it has a lower activity than WT TatA.
Fig. 1.
Characterization of strains expressing a TatA-YFP fusion. (A) Expression and stability of the TatA-YFP fusion in different tat backgrounds. TatA or YFP were detected by immunoblotting after SDS/PAGE of whole cells of the ΔtatABCDE strain DADE (lanes 1), the parental strain ABC (lanes 2), and TatA-YFP fusion-expressing strains AAyBC (lanes 3), AyBC (lanes 4), AyC (lanes 5), and Ay (lanes 6). Bands corresponding to native TatA and the TatA-YFP fusion are indicated as are nonspecific bands seen even in strains without TatA protein (*), and the molecular masses (kDa) of standard proteins (right). (B) Membrane localization of the TatA-YFP fusion. Cells were broken by passage through a French press, separated into pellet (P) and soluble (S) fractions by ultracentrifugation, and analyzed by immunoblotting with anti-YFP serum. (C) Assessment of the folding state of the TatA-YFP fusions. Cells of strains AyBC (lanes 1 and 3), AAyBC (lane 2), AyC (lane 4), and Ay (lane 5) were suspended in SDS/PAGE loading buffer and either heated to 100°C for 3 min (lane 1) or left at room temperature (lanes 2–5). Samples were then analyzed by SDS/PAGE and immunoblotting with anti-YFP serum. Bands corresponding to TatA-YFP with a folded (fYFP) or unfolded (uYFP) YFP domain are marked. (D) Topology of TatA-YFP. Sphaeroplasts produced from strain AyBC were subjected to treatment with proteinase K or Triton X-100 as indicated and analyzed by immunoblotting with anti-YFP serum. (E) Transport of the Tat substrate CueO. Strains containing plasmid pQE80-CueO expressing a hexahistidine-tagged version of E. coli CueO were fractionated. Equal portions of cytoplasmic (C) and periplasmic (P) fractions were subjected to SDS/PAGE and analyzed by immunoblotting with an antihexahistidine tag serum.
Epifluorescence images (SI Text) of strain AyBC (Fig. 2A) show bright spots of width ≈300 nm (similar in size to the point spread function of our microscope; ref. 29). These spots are independently mobile, suggesting that each corresponds to a distinct TatA complex (Movie S1). The spots move around the perimeter of the cell consistent with diffusion in the cell membrane. There is no obvious localization to subregions of the cell such as the poles. The number of TatA complexes per cell is 15 ± 9 (mean ± SD) based on an estimate of the number of spots detected per image multiplied by the proportion of the membrane being imaged relative to the whole cell.
Fig. 2.
Stoichiometry of TatA-YFP complexes in strain AyBC. (A) Overlaid bright-field (gray) and epifluorescence (yellow) images (single 40-ms frame) of AyBC cells. (B) Intensity trace of a tracked spot (black) with output from an edge-preserving filter (red) (41). (Inset) An expansion of the end of the trace. The mean autofluorescence of the parental strain is also plotted (magenta). (C) Intensity histogram for all spots of the same cell as B. (Inset) The power spectrum of the pairwise difference distribution for this cell (black with peak ± SD shown; arrow) and the mean power spectrum for all AyBC cells using 10 different cultures, 121 cells (solid red line; SD error-bounds, red dotted line). (D) Stoichiometry of TatA-YFP complexes in strain AyBC taken from the same data set as C Inset (dark gray) and 20 min after addition of 5 μM FCCP (light gray; three different cultures, 41 cells), together with lognormal fits (black and white, respectively). (Inset) The mean power spectrum generated from 10 different cultures is shown (solid line; SD error-bounds, dotted lines) with peak ± SD (arrow).
The subunit stoichiometries of individual TatA-YFP complexes were estimated from the stepwise changes in their photobleaching curves, modifying the protocol of ref. 30 for use with mobile complexes. An image mask was created for each cell based on its bright-field appearance. This masking technique generated measurements of cellular dimensions for use in coordinate transformations. Bright spots were identified and tracked automatically throughout a stack of consecutive images by using custom-written software (SI Text), which determined the intensity of each spot together with its position to a precision between ≈2 and ≈20 nm depending on spot intensity. Analysis of stepwise decreases in the spot intensity attributable to photobleaching of single YFP molecules gave an estimate of the fluorescence intensity IYFP of YFP molecules in each cell (ref. 30 and Fig. 2 B and C). Upward steps, indicative of YFP blinking as reported at this time scale (31, 32), were also seen. We fitted the intensity trace for each spot with an exponential decay function to estimate the initial intensity I0 before bleaching. We estimated the number of TatA-YFP molecules in each spot as I0 divided by IYFP (SI Text). SDS/PAGE (Fig. 1C) showed that all YFP in freshly harvested cells is folded, because folded YFP is resistant to SDS denaturation. In addition, cellular fluorescence increased by <10% after 2-h incubation with chloramphenicol to block protein synthesis (30). Thus the error in our estimate of the number of TatA-YFP molecules caused by a dark population of unfolded or immature YFP molecules is small.
The TatA-YFP complex stoichiometry in strain AyBC is highly heterogeneous (Fig. 2D). It is distributed in an approximately lognormal fashion with a median of ≈25 TatA-YFP molecules per complex. The variation in median complex size between different cultures grown under identical conditions was ≈15% (SI Text). Simulations indicate that our detection error, and thus detection sensitivity, is approximiately two YFP per spot (SI Text). Therefore the observed maximum in the TatA-YFP stoichiometry distribution is not an artifact arising from an inability to detect small complexes. We estimated the probability of two or more spots being separated by less than a point spread function width, and thus mistaken as a single spot, to be <8% (SI Text). Fourier analysis of the stoichiometry from individual cultures revealed a characteristic spacing of 4.3 ± 0.6 molecules (Fig. 2D Inset and SI Text). This periodicity suggests that the TatA-YFP complexes are made up of tetrameric units.
Cellular autofluorescence under our experimental conditions was quantified by using the parental strain. Subtraction of this autofluorescence from the membrane fluorescence of strain AyBC enabled us to determine the number of TatA-YFP molecules present in the membrane but not associated with identified spots (30). The size of this pool was ≈100 molecules per cell compared with ≈460 molecules per cell associated with complexes.
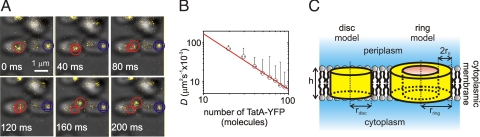
The mobility of individual TatA complexes was quantified. To allow an analysis of TatA complex movement solely in the plane of the membrane we transformed the spot positions into a coordinate system appropriate for diffusion on the membrane surface of a cell (SI Text). We then calculated the mean-squared displacement (MSD) versus time interval τ for each track (33). The coordinate-transformed MSD takes into account the curvature of the membrane surface to compute the real distances traveled by diffusing spots and resulted in values ≈20% greater than those obtained with the nontransformed version, consistent with previous membrane diffusion simulations on cells with similar surfaces (23). The diffusion coefficient D was based on a linear fit of the MSD versus τ trace (ref. 34 and SI Text). We found that most tracks had a goodness-of-fit for a straight line to the MSD relation of correlation coefficient (R2) >0.8, indicating normal Brownian diffusion. Visual inspection of consecutive fluorescence images of strain AyBC showed some variation in spot mobility. For example, some spots diffuse freely over a distance of >500 nm over the cell membrane surface in ≈1 s, whereas others of similar brightness diffuse <100 nm over a similar time scale (Fig. 3A). However, the tracks in our present study were too short to distinguish whether the less mobile spots showed restricted diffusion as opposed to being within the range of statistical variation expected from normal diffusion.
Fig. 3.
TatA-YFP complexes exhibit variation in diffusion. (A) Overlaid bright-field (gray) and epifluorescence (yellow) images (individual 40-ms frames) for two AyBC cells showing two different tracked TatA-YFP complexes (red and blue) of similar brightness. The initial position of each complex is marked with a square and the current position is indicated by a circle. (B) Variation of microscopic diffusion coefficient D (SD error bars) with TatA-YFP complex stoichiometry on a log-log plot with straight-line fit (red) of gradient −1.2 ± 0.4. (C) Schematic illustrating the disk and ring models.
Estimates for D were found to decrease with increasing complex size S, which could be fitted well (R2 = 0.96) by a straight line on a log-log plot (Fig. 3B). The range of D values was broadly consistent with previous estimates from other membrane proteins complexes (23, 30, 35, 36). We modeled complex movement as translational diffusion of a cylinder constrained two-dimensionally in a lipid bilayer bounded by periplasm and cytoplasm, using a heuristic treatment that assumes that D varies as the reciprocal of the complex radius r (ref. 35 and SI Text). We considered two different spatial distributions of protein within the cylinder (Fig. 3C), one in which subunits were uniformly spread across the circular cross-section of the cylinder (disk model) and another in which the subunits were located around its circumference (ring model). The disk model assumes r α S1/2, whereas in the ring model r α S, giving respective gradients of −0.5 and −1 on the log-log plot of Fig. 3B. The theoretical predictions from these two models compare with the measured gradient of −1.2 ± 0.4 and, within experimental error, indicates agreement with the ring model.
TatA interaction with substrate-bound TatBC complexes requires the transmembrane pmf (11, 16) as does substrate-induced structural reorganization of TatA (18). To determine whether pmf maintains the stoichiometry of TatA-YFP complexes we treated AyBC cells with the protonophore carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP) to abolish the transmembrane pmf. We found no significant change in the median number of molecules per complex (at a confidence level of P = 0.98) nor in the shape of the distribution (Fig. 2D). Control experiments showed that flagellar rotation, which depends on the pmf, was blocked at the FCCP concentration used.
We investigated how the oligomeric state of TatA-YFP is affected by removing other Tat components. In strains lacking TatB (AyC), or both TatB and TatC (Ay), fluorescence is spread diffusely around the cell periphery (Fig. 4 A and B and Movies S2 and S3). Granularity in fluorescence around the perimeter is seen on individual images, suggestive of dim spots moving rapidly against a bright background, but these cannot be distinguished reliably with automated tracking. Although the spot detection sensitivity for the AyBC strain is approxiately two molecules of YFP per spot, the effective background signal observed in strains Ay and AyC was about seven times higher, resulting in an equivalent reduction in detection sensitivity. Measuring image intensity profiles we estimate that at least 95% of the TatA-YFP molecules are associated with the cell membrane (Fig. 4C and SI Text) in agreement with biochemical data. We estimated the total TatA-YFP content in this membrane pool to be ≈680 and ≈650 molecules per cell for strains AyC and Ay, respectively. Addition of FCCP to Ay to disrupt the pmf results in no observable change in the behavior of TatA-YFP. Fluorescence recovery after photobleaching (FRAP) experiments were carried out by bleaching half of an Ay cell with a focused laser (ref. 30 and Fig. 4D), enabling us to track very dim spots diffusing into the bleached area. These spots contain a median of about four molecules (Fig. 4E), indicating that TatA-YFP complexes are significantly smaller when cells lack TatB and TatC. Our FRAP data indicate that TatA diffuses at 0.12 ± 0.05 μm2·s−1. This estimate for diffusion coefficient can be compared with an earlier FRAP study on E. coli cells overproducing a TatA-GFP fusion in which it was reported that the mobile TatA component diffuses at ≈0.13 μm2·s−1 (36).
Fig. 4.
Large TatA-YFP complexes are absent from strains lacking TatB and TatC. (A and B) Epifluorescence images of Ay (A) and AyC (B) cells (frame-average 0.4 s). (C) Mean intensity profile (solid black line) for the region covered by a red rectangle in A. SD error-bounds (dotted line) and a multiGaussian fit (red) are indicated. The intensity profile (green) and corresponding single Gaussian fit (blue) for the parental strain are shown (SI Text). (D) FRAP of Ay cells. Images, from left to right show the initial diffraction-limited bleach zone (red circle), fluorescence during the bleach (frame-average 0.4 s, bleach and imaging intensity ≈60 kW·cm−2 and ≈260 W·cm−2, respectively), and individual image frames acquired 1- and 3-s postbleach. (E) The TatA-YFP complex stoichiometry distribution obtained after FRAP. (F and G) Epifluorescence images (0.4-s time averages) of Ay containing plasmid pBAD-BC before induction (F) and after induction (G) of tatBC expression. (H) Immunoblot of the strain before induction (lane 1) and after induction (lane 2). (I) TatA-YFP complex stoichiometry distribution for postinduction cells (blue) compared with AyBC (orange).
To confirm that TatBC controls TatA-YFP complex size we transformed strain Ay with a plasmid expressing tatBC under the control of an arabinose-inducible promoter. Under noninduced conditions the cells have the same diffuse peripheral fluorescence as observed in the untransformed strain (Fig. 4F). After induction of TatBC mobile fluorescent spots similar to those found in AyBC appear (Fig. 4 G and H and Movie S4), confirming a causal link between TatBC and TatA-YFP complex size. The shape of the stoichiometry distribution of the induced complexes was similar to that of AyBC but with a smaller median of ≈15 molecules per complex (Fig. 4I).
Discussion
Current models of the mechanism of Tat protein transport make specific predictions about the oligomeric state of TatA and whether and how this changes during the transport cycle. The oligomeric state of TatA is, thus, key to understanding Tat translocation. Unfortunately, determining the molecular composition of large biological complexes under physiologically relevant conditions remains challenging. This determination is especially challenging for TatA because it can be anticipated that the organization of the Tat system may be perturbed by extraction from its native membrane environment. Variation in TatA protomer content between complexes is an important feature of some mechanistic models. Such a phenomenon can only be adequately explored by determining the stoichiometry of individual complexes. In the current work we have overcome the technical challenges involved in obtaining meaningful information on TatA oligomeric state by analyzing the in vivo fluorescence of individual, YFP-labeled TatA complexes.
Previous studies found that TatA exhibits heterogeneous oligomeric states when extracted from its membrane environment with detergents (13, 14). A key question has been whether this heterogeneity is an artifact of the detergent solubilization. In the current study we demonstrate that (YFP-labeled) TatA has a heterogeneous oligomer distribution in the membranes of living cells. Variability in oligomer size is, therefore, an inherent property of TatA. Importantly, our single-molecule analysis has allowed us to quantitatively define the oligomeric states of TatA that are present in the membrane. There is a range of at least 4 to 100, and an average of 25, TatA protomers per complex. The TatA stoichiometry distribution approximates to lognormal. However, this distribution can also be fit by models originally developed to describe the length variation of F-actin filaments in vitro (37), which may indicate some general similarity in the behavior of TatA to the kinetics of actin polymerization. The published electron microscopy structures of TatA in detergent solution were estimated to contain between 12 and 35 TatA protomers (14). These numbers are consistent with the peak of the TatA stoichiometry distribution determined here.
Previous analyses of detergent-solubilized TatA by blue native PAGE demonstrated that there is a distinct and constant step size between TatA complexes of increasing size and suggested that this step size corresponds to more than a single TatA protomer (13, 14). Data obtained in the current study indicate that the step size is most likely to be four. We observed a periodicity of approximately four TatA molecules in the power spectrum of the TatA stoichiometry distribution (Fig. 2D), which suggests that TatA complexes are assembled from TatA tetramers. This inference is consistent with the peak at four in the TatA-YFP stoichiometry profile of a strain that cannot assemble distinct TatA complexes (Fig. 4E).
We find that the relationship between the diffusion coefficients of TatA complexes and their subunit stoichiometry can be better modeled if TatA is assumed to form rings rather than being bundled as a filled disk (Fig. 3 B and C and SI Text). The difference in the modeled behavior of the two shapes arises because the diameter of a ring scales linearly with the number of TatA subunits, whereas the diameter of a disk scales as the square root. A ring structure would be consistent with the doughnut-shaped particles observed for detergent-solubilized TatA by negative-stain electron microscopy (14) and is a minimum requirement of channel, but not bilayer-perturbing patch, models of TatA mechanism (20).
The low transport activity of our TatA-YFP fusion is consistent with other studies that used fusions to the C terminus of TatA (refs 38 and 39 a nd T.P., unpublished observations) and occurs even though the last ≈50 aa of TatA have a random coil structure (12, 40). Steric hindrance to the movement of the substrate is a likely explanation for this phenomenon. However, the possibility that the tail of TatA undergoes topology inversion during transport (27), and that this is perturbed by the reporter domain, should also be considered.
Although we have observed that both native and detergent-extracted TatA form heterogeneous complexes, our experiments also reveal a striking difference in TatA behavior between the two environments. Although formation of TatA complexes in membranes requires the TatBC complex, detergent-solubilized TatA forms complexes even when expressed in the absence of other Tat components (12). A possible explanation for this difference is that detergent extraction may drive the TatA polymerization equilibrium toward the assembled state by removing phospholipid interactions that stabilize the dispersed state. This environment-dependent variation in TatA behavior suggests that observations on the structural organization of TatA obtained in detergent solution should be treated with caution.
One current model for the Tat mechanism postulates that TatA polymerizes in response to substrate binding to the TatBC complex. The direct analysis of TatA stoichiometry presented here provides substantial support for this model. First, the stoichiometry of TatA complexes is variable as expected if dynamic polymerization occurs. Second, cells contain a significant pool of TatA that is not part of distinct complexes. Such a pool would be necessary to provide the TatA units required for complex assembly. Finally, and most crucially, strains lacking a functional TatBC complex are unable to assemble defined TatA complexes. Thus, as predicted by the polymerization model, TatA oligomerization depends on TatBC. A further feature of the polymerization model is that TatA oligomerization is driven by the transmembrane pmf. However, we found that dissipation of the pmf did not affect the observed TatA stoichiometry distribution. This result might be reconciled with the polymerization model if disassembly, as well as assembly, of the TatA complexes requires the pmf. This situation could occur, for example, if substrate transport is driven by the pmf, with substrate release at the completion of transport being the trigger for TatA disassembly. Irrespective of model, the observation that the oligomeric state of TatA is unaffected by the removal of the pmf shows that a pmf is not required to maintain the polymerized state of TatA. It should, in principle, be possible to further test the polymerization model by analyzing how the composition of individual TatA complexes evolves with time. However, the development of methods to follow longer duration tracks over a wider range of illumination intensities than was possible in our present study will be required.
A specific prediction of the bespoke channel model for Tat transport is that TatA polymerizes around the substrate molecule to produce a pore that matches the size of the substrate. The observation of different-sized TatA oligomers is consistent with this model, and the number of TatA protomers we observe in the complexes should be sufficient to provide appropriately sized pores (14). However, we have been unable to show statistically significant alterations in TatA stoichiometry by saturating the Tat system with substrate proteins of different sizes.
Materials and Methods
Cell Strains and Preparation.
Strains expressing TatA-YFP were constructed, cultured, and prepared for microscopy as described in SI Text and Fig. S1.
Microscopy.
We used a home-built inverted epifluorescence microscope, excitation wavelength 532 nm (SI Text), intensity ≈260 W·cm−2, with a separate path allowing light to be focused to a width ≈1 μm, intensity ≈60 kW·cm−2 (30). For FRAP this microscope was used to bleach a cell pole for 0.5 s. The focal plane was set at the middle of a cell, ≈0.5 μm from the coverslip. Emission was imaged at 25 Hz by a 128 × 128-pixel (≈50 nm per pixel), cooled, back-thinned electron-multiplying CCD camera (iXon DV860-BI; Andor Technology), and sampled continuously for at least 8 s. Diffusing spots of fluorescence were tracked for at least five consecutive image frames (total duration 200 ms), with some tracks lasting for several seconds.
Data Analysis.
Automatic tracking using custom-written software allowed determination of fluorescence intensity, position, and number of spots in single cells. TatA-YFP stoichiometry was estimated from YFP stepwise changes in intensity caused by photobleaching or blinking (ref. 30 and SI Text). Data sets from different samples were compared statistically by using Wilcoxon testing, and stoichiometry variation with cell culture was measured (SI Text). Spot detection sensitivity and localization precision was assessed by simulations (SI Text), and the likelihood for erroneous multiple spot detection was estimated (SI Text). Position data were used to measure diffusion for each spot (SI Text). Diffusion was modeled as that of a cylinder in a lipid bilayer with fitting assessed by χ2 statistics (SI Text). A nonlinear model was applied to cellular intensity profiles to assess cytoplasmic YFP content (SI Text). Estimations for cellular TatA-YFP content and diffusion coefficients are in Table S2, and Tat phenotypes of strains used in this study are in Table S1.
Supplementary Material
Acknowledgments.
This work was supported by the Biotechnology and Biological Sciences Research Council through grant D004578 and a studentship to S.C., by a Leverhulme Trust Fellowship (to M.C.L.), a Royal Society University Research Fellowship (to M.C.L.), a Royal Society Dorothy Hodgkin Fellowship (to R.M.G.), by the Research Councils U.K. via the Interdisciplinary Research Collaboration in Bionanotechnology, by Wellcome Trust Grant 061780, and by a Medical Research Council Senior Nonclinical Fellowship Award (to T.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.W. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0806338105/DCSupplemental.
References
- 1.Rapoport TA. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. 2007;450:663–669. doi: 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- 2.Berks BC, Palmer T, Sargent F. Protein targeting by the bacterial twin-arginine translocation (Tat) pathway. Curr Opin Microbiol. 2005;8:174–181. doi: 10.1016/j.mib.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 3.Berks BC, Palmer T, Sargent F. The Tat protein translocation pathway and its role in microbial physiology. Adv Microb Physiol. 2003;46:187–254. doi: 10.1016/s0065-2911(03)47004-5. [DOI] [PubMed] [Google Scholar]
- 4.Cline K, Theg SM. The Sec and Tat protein translocation pathways in chloroplasts. In: Dalbey RE, Koehler C, Tamanoi F, editors. The Enzymes, Molecular Machines Involved in Protein Transport Across Cellular Membranes. Vol XXV. San Diego: Elsevier; 2007. pp. 455–485. [Google Scholar]
- 5.Weiner JH, et al. A novel and ubiquitous system for membrane targeting and secretion of cofactor-containing proteins. Cell. 1998;93:93–101. doi: 10.1016/s0092-8674(00)81149-6. [DOI] [PubMed] [Google Scholar]
- 6.Sargent F, et al. Overlapping functions of components of a bacterial Sec-independent protein export pathway. EMBO J. 1998;17:3640–3650. doi: 10.1093/emboj/17.13.3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sargent F, Stanley NR, Berks BC, Palmer T. Sec-independent protein translocation in Escherichia coli: A distinct and pivotal role for the TatB protein. J Biol Chem. 1999;274:36073–36083. doi: 10.1074/jbc.274.51.36073. [DOI] [PubMed] [Google Scholar]
- 8.Bogsch E, et al. An essential component of a novel bacterial protein export system with homologues in plastids and mitochondria. J Biol Chem. 1998;273:18003–18006. doi: 10.1074/jbc.273.29.18003. [DOI] [PubMed] [Google Scholar]
- 9.Bolhuis A, Mathers JE, Thomas JD, Barrett C, Robinson C. TatB and TatC form a functional and structural unit of the twin-arginine translocase from Escherichia coli. J Biol Chem. 2001;276:20213–20219. doi: 10.1074/jbc.M100682200. [DOI] [PubMed] [Google Scholar]
- 10.Cline K, Mori H. Thylakoid ΔpH-dependent precursor proteins bind to a cpTatC-Hcf106 complex before Tha4-dependent transport. J Cell Biol. 2001;154:719–729. doi: 10.1083/jcb.200105149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alami M, et al. Differential interactions between a twin-arginine signal peptide and its translocase in Escherichia coli. Mol Cell. 2003;12:937–946. doi: 10.1016/s1097-2765(03)00398-8. [DOI] [PubMed] [Google Scholar]
- 12.Porcelli I, et al. Characterization and membrane assembly of the TatA component of the Escherichia coli twin-arginine protein transport system. Biochemistry. 2002;41:13690–13697. doi: 10.1021/bi026142i. [DOI] [PubMed] [Google Scholar]
- 13.Oates J, et al. The Escherichia coli twin-arginine translocation apparatus incorporates a distinct form of TatABC complex, spectrum of modular TatA complexes and minor TatAB complex. J Mol Biol. 2005;346:295–305. doi: 10.1016/j.jmb.2004.11.047. [DOI] [PubMed] [Google Scholar]
- 14.Gohlke U, et al. The TatA component of the twin-arginine protein transport system forms channel complexes of variable diameter. Proc Natl Acad Sci USA. 2005;102:10482–10486. doi: 10.1073/pnas.0503558102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McDevitt CA, Buchanan G, Sargent F, Palmer T, Berks BC. Subunit composition and in vivo substrate-binding characteristics of Escherichia coli Tat protein complexes expressed at native levels. FEBS J. 2006;273:5656–5668. doi: 10.1111/j.1742-4658.2006.05554.x. [DOI] [PubMed] [Google Scholar]
- 16.Mori J, Cline K. A twin arginine signal peptide and the pH gradient trigger reversible assembly of the thylakoid ΔpH/Tat translocase. J Cell Biol. 2002;157:205–210. doi: 10.1083/jcb.200202048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yahr TL, Wickner WT. Functional reconstitution of bacterial Tat translocation in vitro. EMBO J. 2001;20:2472–2479. doi: 10.1093/emboj/20.10.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dabney-Smith C, Mori H, Cline K. Oligomers of Tha4 organize at the thylakoid Tat translocase during protein transport. J Biol Chem. 2006;281:5476–5483. doi: 10.1074/jbc.M512453200. [DOI] [PubMed] [Google Scholar]
- 19.Brueser T, Sanders C. An alternative model of the twin arginine translocation system. Microbiol Res. 2003;58:7–17. doi: 10.1078/0944-5013-00176. [DOI] [PubMed] [Google Scholar]
- 20.Cline K, McCaffery M. Evidence for a dynamic and transient pathway through the TAT protein transport machinery. EMBO J. 2007;26:3039–3049. doi: 10.1038/sj.emboj.7601759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greene NP, et al. Cysteine scanning mutagenesis and disulfide mapping studies of the TatA component of the bacterial twin arginine translocase. J Biol Chem. 2007;282:23937–23945. doi: 10.1074/jbc.M702972200. [DOI] [PubMed] [Google Scholar]
- 22.Tsien RY. The green fluorescent protein. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 23.Deich J, Judd EM, McAdams HH, Moerner WE. Visualization of the movement of single histidine kinase molecules in live Caulobacter cells. Proc Natl Acad Sci USA. 2004;101:15921–15926. doi: 10.1073/pnas.0404200101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee PA, et al. Cysteine-scanning mutagenesis and disulfide mapping studies of the conserved domain of the twin-arginine translocase TatB component. J Biol Chem. 2006;281:34072–34085. doi: 10.1074/jbc.M607295200. [DOI] [PubMed] [Google Scholar]
- 25.Mori H, Summer EJ, Ma X, Cline K. Component specificity for the thylakoidal Sec and ΔpH-dependent protein transport pathways. J Cell Biol. 1999;146:45–56. doi: 10.1083/jcb.146.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gouffi K, Gérard F, Santini CL, Wu LF. Dual topology of the Escherichia coli TatA protein. J Biol Chem. 2004;279:11608–11615. doi: 10.1074/jbc.M313187200. [DOI] [PubMed] [Google Scholar]
- 27.Chan CS, Zlomislic MR, Tieleman DP, Turner RJ. The TatA subunit of Escherichia coli twin-arginine translocase has an N-in topology. Biochemistry. 2007;46:7396–7404. doi: 10.1021/bi7005288. [DOI] [PubMed] [Google Scholar]
- 28.Ize B, Stanley NR, Buchanan G, Palmer T. Role of the Escherichia coli Tat pathway in outer membrane integrity. Mol Microbiol. 2003;48:1183–1193. doi: 10.1046/j.1365-2958.2003.03504.x. [DOI] [PubMed] [Google Scholar]
- 29.Lo C-J, Leake MC, Pilizota T, Berry RM. Nonequivalence of membrane voltage and ion-gradient as driving forces for the bacterial flagellar motor at low load. Biophys J. 2007;93:294–302. doi: 10.1529/biophysj.106.095265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leake MC, et al. Stoichiometry and turnover in single, functioning membrane protein complexes. Nature. 2006;443:355–358. doi: 10.1038/nature05135. [DOI] [PubMed] [Google Scholar]
- 31.McAnaney TB, et al. Protonation, photobleaching, and photoactivation of yellow fluorescent protein (YFP 10C): A unifying mechanism. Biochemistry. 2005;44:5510–5524. doi: 10.1021/bi047581f. [DOI] [PubMed] [Google Scholar]
- 32.Bagshaw CR, Cherny D. Blinking fluorophores: What do they tell us about protein dynamics? Biochem Soc Trans. 2006;34:979–982. doi: 10.1042/BST0340979. [DOI] [PubMed] [Google Scholar]
- 33.Gross DJ, Webb WW. Cell surface clustering and mobility of the liganded LDL receptor measured by digital video fluorescence microscopy. In: Loew LM, editor. Spectroscopic Membrane Probes II. Boca Raton, FL: CRC; 1988. pp. 19–45. [Google Scholar]
- 34.Kusumi A, Sako Y, Yamamoto M. Confined lateral diffusion of membrane receptors as studied by single-particle tracking (nanovid microscopy). Effects of calcium-induced differentiation in cultured epithelial cells. Biophys J. 1993;65:2021–2040. doi: 10.1016/S0006-3495(93)81253-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gambin Y, et al. Lateral mobility of proteins in liquid membranes revisited. Proc Natl Acad Sci USA. 2006;103:2098–2102. doi: 10.1073/pnas.0511026103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mullineaux CW, Nenninger A, Ray N, Robinson C. Diffusion of green fluorescent protein in three cell environments in Escherichia coli. J Bacteriol. 2006;188:3442–3448. doi: 10.1128/JB.188.10.3442-3448.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Edelstein-Keshet L, Ermentrout GB. A model for actin-filament length distribution in a lamellipod. J Math Biol. 2001;43:325–355. doi: 10.1007/s002850100102. [DOI] [PubMed] [Google Scholar]
- 38.Berthelmann F, Brüser T. Localization of the Tat translocon components in Escherichia coli. FEBS Lett. 2004;569:82–88. doi: 10.1016/j.febslet.2004.05.054. [DOI] [PubMed] [Google Scholar]
- 39.Ray N, Nenninger A, Mullineaux CW, Robinson C. Location and mobility of twin arginine translocase subunits in the Escherichia coli plasma membrane. J Biol Chem. 2005;280:17961–17968. doi: 10.1074/jbc.M413521200. [DOI] [PubMed] [Google Scholar]
- 40.Lange C, Müller SD, Walther TH, Bürck J, Ulrich AS. Structure analysis of the protein translocating channel TatA in membranes using a multi-construct approach. Biochim Biophys Acta. 2007;1768:2627–2634. doi: 10.1016/j.bbamem.2007.06.021. [DOI] [PubMed] [Google Scholar]
- 41.Leake MC, Wilson D, Gautel M, Simmons RM. The elasticity of single titin molecules using a two-bead optical tweezers assay. Biophys J. 2004;87:1112–1135. doi: 10.1529/biophysj.103.033571. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.