Abstract
The AbrB protein of the spore-forming bacterium Bacillus subtilis is a repressor of numerous genes that are switched on during the transition from the exponential to the stationary phase of growth. The gene for AbrB is under the negative control of the master regulator for entry into sporulation, Spo0A∼P. It has generally been assumed that derepression of genes under the negative control of AbrB is achieved by Spo0A∼P-mediated repression of abrB followed by rapid degradation of the AbrB protein. Here, we report that AbrB levels do decrease during the transition to stationary phase, but that this decrease is not the entire basis by which AbrB-controlled genes are derepressed. Instead, AbrB is inactivated by the product of a uncharacterized gene, abbA (formerly ykzF), whose transcription is switched on by Spo0A∼P. The abbA gene encodes an antirepressor that binds to AbrB and prevents it from binding to DNA. Combining our results with previous findings, we conclude that Spo0A∼P sets in motion two parallel pathways of repression and antirepression to trigger the expression of diverse categories of genes during the transition to stationary phase.
Keywords: sporulation, transcription
Bacteria ordinarily spend a relatively brief period of their existence in the exponential phase of growth (1–3). Nutrients become limiting, or other adverse environmental changes take place as cells reach a high population density, causing growth to slow and the bacteria to enter stationary phase. Coping with the transition to stationary phase involves dramatic changes in gene expression in which suites of genes are switched on that enable the cells to adapt to unfavorable circumstances. These changes are governed by signal transduction pathways that sense the onset of adverse circumstances and respond by activating (or inactivating) global regulatory proteins. One such global regulator is the general stress response transcription factor σS, which helps govern the transition to stationary phase in Escherichia coli (4, 5). In the spore-forming bacterium Bacillus subtilis, the subject of this investigation, the transition to stationary phase is principally governed by five regulatory proteins, CodY (6), σB (7, 8), σH (9), Spo0A∼P (10), and AbrB (11).
How CodY helps to govern the transition to stationary phase is well understood. Its activity as a repressor depends on either of two cofactors, GTP or a branched chain amino acid (12, 13). In the absence of either ligand, CodY's ability to bind DNA is impaired. Thus, because GTP or branched chain amino acid levels drop during nutrient limitation, repression is relieved and genes under the control of CodY are derepressed. Our understanding of how σB-, σH-, and Spo0A-controlled gene expression is coupled to the exit from the exponential phase of growth is less complete. The σB factor, for example, is activated by convergent pathways that sense, in an as yet undefined way, the lack of certain nutrients and the presence of certain kinds of physical-chemical signals (14). Spo0A, a member of the response regulator family of phosphoproteins, is activated by phosphorylation in response to nutrient limitation via a multicomponent phosphorelay (15). The phosphorelay is initiated by several kinases that are thought to recognize intra- or extracellular signals. When phosphorylated, Spo0A∼P acts as an activator or repressor of ≈120 genes under its direct control, including genes required for sporulation (16). However, how phosphorelay-mediated phosphorylation of Spo0A is coupled to a drop in nutrient availability has not been elucidated.
The fifth transcriptional control protein, the “transition-state regulator” AbrB and the focus of this report, has been of interest for almost 40 years (17–19). Yet little is known about the mechanisms that govern the derepression of AbrB-controlled genes at the end of the exponential phase of growth. The gene for AbrB was discovered because of the observation that mutations at the abrB locus suppressed some of the phenotypes characteristic of spo0A and other mutants blocked in the initiation of sporulation. However, how AbrB acted was mysterious for many years (20). An important clue came from studies of two promoters that depended on Spo0A∼P for their activation (21, 22). In both cases, an abrB mutation was found to bypass the dependence on spo0A, and in one case, it resulted in constitutive transcription. These findings indicated that AbrB is likely a repressor that is present in vegetatively growing cells and is inactivated or eliminated by the action of Spo0A at the end of the exponential phase of growth. Indeed, subsequent biochemical work confirmed that AbrB is a DNA-binding protein that acts by repressing target genes (23, 24). The further demonstration that Spo0A∼P directly represses abrB (25) led to the view that derepression of genes under AbrB control is mediated by a Spo0A∼P-imposed block in abrB transcription combined with rapid depletion of AbrB protein by degradation (24, 26, 27).
As we report here, AbrB levels do decrease as cells transition from exponential growth to stationary phase, but this drop in AbrB levels is not the sole basis for the derepression of genes under its control. Instead, AbrB is inactivated by the product of a previously uncharacterized gene, ykzF (for which we introduce the name abbA for antirepressor of abrB A) that is directly switched on by Spo0A∼P (16, 28). We show that abbA encodes an AbrB-binding protein that forms a complex with the repressor and prevents it from adhering to DNA. Thus, the derepression of some or all genes under the negative control of AbrB involves the Spo0A∼P-induced synthesis of an antirepressor. A parallel thereby emerges between AbrB and the SinR repressor of B. subtilis, which is also inactivated by an antirepressor (SinI) whose synthesis is induced by Spo0A∼P (29–31).
Results
The Cannibalism Operons sdp and skf Are Under the Indirect Control of AbbA.
The starting point for this investigation was the phenomenon of cannibalism in which cells that have activated Spo0A in response to nutrient limitation produce a toxin and a killing factor that kill sibling cells that have not activated the response regulator (32, 33). Colonies of cells that exhibit cannibalism are delayed in sporulation. It is presumed that nutrients released by the dead cells delay sporulation by reversing or slowing the activation of Spo0A in the toxin- and killing factor-producing cells. The toxin and the killing factor are produced under the direction of operons called sdpABC (hereafter simply sdp) and skfABCDE (hereafter simply skf), respectively. Both operons are under the direct negative control of AbrB (34). Colonies of cells mutant for sdp or skf are mutant for cannibalism and exhibit an accelerated sporulation phenotype. A previous survey of members of the Spo0A regulon for genes involved in cannibalism revealed an uncharacterized open-reading frame abbA (ykzF), which when mutant caused accelerated sporulation (16).
We asked whether the cannibalism phenotype of an abbA mutant was due to impaired expression of the sdp and skf operons. To do this, we examined the effect of an abbA mutation (ΔabbA) on the expression of lacZ fused to the promoters for sdp (Psdp-lacZ) and skf (Pskf-lacZ) during sporulation in DS medium. The results show that deletion of abbA led to decreased transcription from both promoters (Fig. 1Upper), thereby providing an apparent explanation for the cannibalism mutant phenotype of an abbA mutant.
Fig. 1.
Two cannibalism operons are under the indirect control of AbbA. (A and B) AbbA is required for maximal expression of the sdpABC and skfABCDE operons. (A) Strains harbored amyE::Psdp-lacZ, and were either wild-type (♦; RL4264), or mutant for abbA (●; AB149). (B) Strains harbored amyE::Pskf-lacZ, and either were wild-type (♦; RL3554), or mutant for abbA (●; AB148). (C and D) An abrB mutation is epistatic to the effect of an abbA mutation on expression of the sdpABC and skfABCDE operons. (C) Strains harbored amyE::Psdp-lacZ, and were wild-type (♦; RL4264), mutant for abbA (●; AB149), harbored the overexpression construct Phyperspank-abbA (○; AB151), mutant for abrB (■; AB183), mutant for both abbA and abrB (▴; AB184), or mutant for abrB and harbored Phyperspank-abbA (Δ; AB182). (D) Strains harbored Pskf-lacZ, and were either wild-type (♦; RL3554), mutant for abbA (●; AB148), harbored Phyperspank-abbA (○; AB150), mutant for abrB (■; AB188), mutant for both abbA and abrB (▴; AB189), or mutant for abrB and harbored Phyperspank-abbA (Δ; AB190). Cells were grown in liquid DS medium; hour 0 of sporulation was the end of the exponential phase of growth. Expression of Phyperspank-abbA was induced by the addition of 1 mM (final concentration) IPTG to the medium.
Next, we determined the effect of overproducing AbbA on the expression of sdp and skf. To do this, we constructed a fusion of the abbA gene to the IPTG-inducible promoter Phyperspank and examined the effect of inducing this construct on the expression of Psdp-lacZ and Pskf-lacZ during sporulation. We observed that expression of both genes was markedly elevated when abbA was overexpressed. Strikingly, the patterns of expression we observed were similar to those seen in an abrB mutant (Fig. 1 Lower).
AbrB directly represses both sdp and skf, and deleting abrB results in a dramatic increase in expression from both operons (34, 35). Could AbbA act by relieving AbrB-mediated repression of these operons? To test this hypothesis, we compared Psdp-lacZ and Pskf-lacZ expression in an abrB mutant, an abrB abbA double mutant, and an abrB mutant that also contained Phyperspank-abbA. We observed that removing or overexpressing abbA had no effect on Psdp-lacZ and Pskf-lacZ expression in the absence of AbrB (Fig. 1, Lower). In other words, the effect of the abrB mutation was epistatic to that of the abbA mutation. These results suggest that AbbA and AbrB act in the same pathway to control sdp and skf, and that AbbA acts upstream of AbrB. This observation raised the possibility that AbbA might be involved in other AbrB-regulated pathways as well.
We note that the abbA mutation only partially impaired expression of the sdp-lacZ and skf-lacZ fusions. If AbbA acts upstream of AbrB to reverse the effects of the repressor, then why did the abbA mutation not block expression of the lacZ fusions completely? In other work, we have found that a second, independently acting pathway contributes to the transcription of sdp (unpublished results). Similarly, skf is controlled by two pathways, one involving Spo0A indirectly via its induction of abbA and one involving Spo0A directly in which Spo0A∼P binds to and activates the skf promoter (34). Henceforth, and for simplicity, we will simply consider the contribution of AbbA to the transcription of sdp, skf and other operons under the negative control of AbrB.
Many Genes Under the Positive Control of AbbA Are Repressed by AbrB.
To determine whether AbbA was dedicated to cannibalism or governed a wider set of genes, we performed transcriptional profiling experiments that compared an abbA mutant strain (ΔabbA) to a strain that overexpressed AbbA (Phyperspank-abbA). Our analysis was carried out with RNA from cells growing exponentially in LB medium.
As a first step, we confirmed that sdp and skf were both up-regulated when AbbA was overexpressed, which gave us confidence that our findings represented members of the AbbA regulon (Table 1). Further analysis of our data revealed that AbbA promotes the transcription of genes beyond those involved in cannibalism. Strikingly, genes known to be repressed by AbrB represented the vast majority of the AbbA regulon (Table 1). Eleven of the 23 genes that we found to be controlled by AbbA had been reported to be either directly or indirectly (via the gene for σW) repressed by AbrB (36–38). Recently, nine additional AbbA-controlled genes have been identified as being under the negative control of AbrB (M. A. Strauch, personal communication). Interestingly, the gene that was controlled most strongly by AbbA, lip, was not known to be a direct target of AbrB, but, as shown below, the promoter region for lip contains a binding site for AbrB. The remaining three genes whose expression was stimulated by overproduction of AbbA are not well characterized. It will be interesting to see whether they too are direct or indirect targets of AbrB.
Table 1.
Genome-wide identification of genes regulated by AbrB
| Gene | Ratio* | Function | AbrB-repressed? |
|---|---|---|---|
| lip | 11.3 | Extracellular lipase | Yes† |
| pnbA | 3.6 | Intracellular esterase | Yes† |
| pspA | 4.9 | Cold and alkaline shock protein | Yes‡ |
| sdpA | 3.6 | Cannibalism | Yes |
| sipW | 3.8 | Sporulation/biofilm formation | Yes |
| skfD | 10.2 | Cannibalism | Yes |
| tasA | 3.9 | Sporulation/biofilm formation | Yes |
| tlpA | 6.6 | Chemotaxis | Unknown |
| ybdN | 6.7 | Unknown | Yes† |
| ybdO | 7.3 | Unknown | Yes† |
| ybfO | 4.4 | Similar to erythromycin esterase | Yes† |
| ycbJ | 3.4 | Similar to macrolide 2′-phosphotransferase | Yes† |
| ydaF | 3.2 | Similar to acetyltransferase | Unknown |
| ydjG | 3.6 | Unknown | Yes‡ |
| ydjH | 5.2 | Unknown | Yes‡ |
| yfhS | 2.9 | Unknown | Unknown |
| yhaP | 3.2 | Unknown | Yes† |
| yknW | 2.6 | Unknown | Yes |
| yknX | 5.2 | Unknown | Yes |
| yknY | 7.4 | Unknown | Yes |
| ylqB | 5.2 | Unknown | Yes† |
| ywoF | 4.3 | Unknown | Yes† |
| yxaL | 3.6 | Unknown | Yes† |
*Ratio of RNA levels in thrC::Phyperspank-abbA erm to ΔabbA::tet strains. The cutoff was a ratio of at least 2.5 in at least three of the five arrays.
†M. A. Strauch, personal communication.
‡Indirectly via σW.
AbbA Blocks the Binding of AbrB to DNA.
The results so far are consistent with the idea that AbbA acts by inhibiting the ability of AbrB to bind to DNA. To investigate this possibility, we carried out EMSA using purified His6-AbbA, purified His6-AbrB, and radiolabeled DNAs that contained the promoters for comK, sdp, skf, or lip. The promoter regions of comK, sdp, and skf have been shown to be bound by AbrB in vitro (35, 39), and lip emerged as a candidate for an AbrB-controlled gene from our microarray analysis (see above).
We first established that AbbA itself did not bind to any of the DNAs (Fig. 2, lane 2). This result suggests that AbbA does not exert its antagonistic effect by competing with His6-AbrB for DNA binding. Fig. 2 (lane 3) shows that the addition of His6-AbrB alone retarded the mobility of all four DNAs, including Plip. However, the additional presence of increasing quantities of His6-AbbA (lane 4–6) reversed the effect of His6-AbrB and did so over a concentration range (1–4 μM) that was similar to the concentration of His6-AbrB (2 μM). Interestingly, the reversal occurred in a step-like manner, which is similar to that observed in EMSA experiments carried out in the absence of AbbA with increasing concentrations of AbrB (data not shown). The simplest interpretation of these results is that AbrB has multiple DNA binding sites of various affinities and that increasing concentrations of AbbA remove AbrB from lower-affinity sites ahead of high-affinity sites.
Fig. 2.
AbbA prevents AbrB from binding to the promoter regions for four AbrB-controlled genes and operons. Shown are electrophoretic mobility shift assays in which 2 μM AbrB was mixed with AbbA or BSA control at the following concentrations: 0 μM (lane 3), 1 μM (lane 4), 2 μM (lane 5), and 4 μM (lane 6) before the addition of radiolabeled DNA (100 nM) containing the promoter for either sdp, skf, comK or lip. Lane 1 had promoter DNA but no protein and lane 2 had promoter DNA and 10 μM AbbA, but no AbrB.
Additionally, we performed order-of-addition experiments and observed that AbbA was equally efficient at disrupting preformed AbrB-DNA complexes as it was at preventing AbrB from binding to DNA (data not shown). As a control, BSA failed to prevent the binding of His6-AbrB to comK DNA at concentrations up to 10 μM (Fig. 2 Lower).
AbbA Binds AbrB.
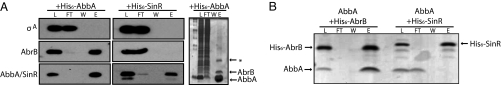
The simplest interpretation of our results so far is that AbbA binds to AbrB and thereby blocks AbrB's ability to bind to DNA. To determine whether AbbA and AbrB do indeed interact, we performed copurification experiments. We prepared a lysate from wild-type cells in the exponential phase of growth in LB medium, and incubated it with His6-AbbA. This mixture was applied to a Ni2+NTA-agarose affinity column and eluted with imidazole. Fig. 3A shows the results of an immunoblot (Fig. 3A Left) and Coomassie staining (Fig. 3A Right), which together indicate that AbrB was retained on the affinity column with impressive selectivity. Indeed, other than His6-AbbA, AbrB was the most abundant protein in the eluate (Fig. 3A Right), and only one other species (marked by an asterisk) could be detected. For comparison, the unrelated protein σA, was not detected in the eluate as judged by immunoblot analysis (Fig. 3A Left). As a control, when affinity chromatography was carried out with the unrelated protein His6-SinR (30), no AbrB (or σA) was detected in the eluate (Fig. 3A Center).
Fig. 3.
AbbA binds AbrB. A shows affinity chromatography of AbrB using immobilized His6-AbbA. Lysates prepared from exponentially growing wild-type cells (PY79) were incubated with purified His6-AbbA (Left and Right) or His6-SinR (Center) and applied to a Ni2+NTA-agarose column. The presence of AbrB, σA, His6-AbbA and His6-SinR was monitored in the load (L), flow-through (FT), wash (W) and eluate (E) by immunoblotting using specific antibodies (Left and Center) or Coomassie staining (Right). Benchmark prestained protein ladder (Invitrogen) is shown in the far left lane of the Coomassie-stained gel (Right). Elution fractions were concentrated 2.5-fold relative to the load. B shows affinity chromatography of AbbA using immobilized His6-AbrB. Purified AbbA was incubated with purified His6-AbrB or His6-SinR and applied to Ni2+NTA-agarose. The presence of AbbA, His6-AbrB, and His6-SinR was detected in the load (L), flow-through (FT), wash (W) and elution (E) fractions by Coomassie staining. Elution fractions were concentrated 10-fold relative to the load.
To determine whether AbbA and AbrB are in direct contact, we prepared highly purified unmodified AbbA using a Hitrap Q anion exchange column (see Materials and Methods). We then incubated AbbA with either His6-AbrB or His6-SinR. These mixtures were separately applied to Ni2+NTA-agarose and eluted with imidazole. Coomassie staining (Fig. 3B) shows that AbbA was retained quantitatively on the His6-AbrB-containing resin (little was seen in the flow through) and was eluted with imidazole. In contrast, when His6-SinR was used, AbbA was found in the flow through and not in the eluate.
AbrB Levels Decrease as Cells Exit the Exponential Phase of Growth.
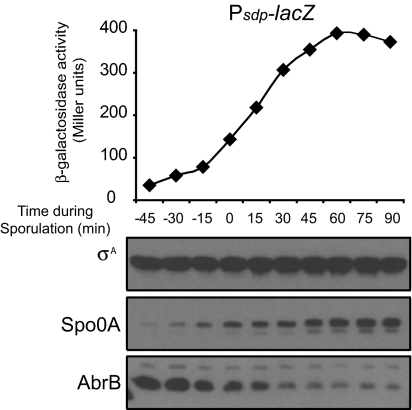
Our findings are consistent with a model for the derepression of AbrB-controlled genes in which Spo0A turns on the synthesis of the antirepressor AbbA, which in turn binds to AbrB and prevents it from repressing its target genes. Because Spo0A represses the transcription of abrB (25), an alternative model is that AbrB is unstable and is rapidly depleted or diluted because of continued cell growth when its synthesis is turned off or both (24, 26, 27). To investigate whether and how rapidly AbrB is depleted during sporulation, we monitored AbrB levels by immunoblot analysis over the course of growth and entry into stationary phase. The unrelated protein σA was used as a loading control, and Spo0A levels were also monitored and seen to increase after exponential phase as expected (Fig. 4). Our results reveal that AbrB levels decreased several fold during the period when derepression of sdp-lacZ was occurring (Fig. 4). Thus, decreasing AbrB could contribute to the derepression of sdp-lacZ, augmenting the effect of AbbA.
Fig. 4.
AbrB levels decrease as cells exit the exponential phase of growth. Upper shows the activation of sdp at the end of stationary phase. The strain harbored amyE::Psdp-lacZ, and was otherwise wild-type (♦; RL4264). Lower shows the kinetics of AbrB and Spo0A accumulation in cells taken from the same culture as followed by immunoblot analysis. σA was used as a loading control. Cells were grown in liquid DS medium; hour 0 of sporulation was the end of the exponential phase of growth.
We wondered whether AbbA was contributing to the drop in AbrB levels. To investigate this, we compared AbrB levels in an abbA mutant strain (ΔabbA) to those in a strain that overexpressed abbA (Phyperspank-abbA) during growth in LB medium. Overexpression of AbbA did not lead to a decrease in AbrB levels [supporting information (SI) Fig. S1].
Discussion
The principal contribution of this work is the elucidation of a previously unknown mechanism by which genes under the negative control of AbrB are derepressed during the transition to stationary phase. It has generally been assumed that derepression is achieved by Spo0A∼P-mediated repression of the abrB gene combined with depletion of AbrB protein by degradation. Indeed, earlier findings, which are confirmed here, do show a reduction in AbrB levels during the transition from growth to stationary phase. This reduction may contribute to the derepression of AbrB-controlled genes (see below), but it is not the exclusive basis for the derepression of genes and operons, such as skf and sdp, that are rapidly switched on as cells exit exponential phase. Instead, AbrB is inactivated by the action of an anti-repressor AbbA that binds to AbrB and thereby prevents the repressor from adhering to operator sites in target genes.
Does AbbA contribute to the derepression of all AbrB-controlled genes? Our evidence showed that AbbA reversed the binding of AbrB to four target genes, sdp, skf, comK, and lip. In addition, we identified 23 genes that were up-regulated by the overproduction of AbbA. A high proportion of these genes were identified as targets of AbrB. Nonetheless, AbrB is known to control many genes that did not appear in our microarray analysis (M. A. Strauch, personal communication). AbrB has various affinities for the operators of genes it controls (40), and AbbA may only be capable of removing AbrB from operators for which it has a relatively low affinity. If this is the case, then the intracellular drop in AbrB levels or other unknown mechanisms might account for the derepression of remaining targets that have a high affinity for the repressor and are not derepressed by the action of AbbA alone. Alternatively, the conditions we used for our transcriptional profiling analysis might not have allowed us to detect all of the members of the AbbA regulon. Our analysis was carried out by using cells in the exponential phase of growth, and AbbA is normally produced during sporulation. Thus, it is possible that AbbA contributes to the derepression of genes that are under the negative control of AbrB but that are not expressed during exponential phase growth because of a direct requirement for Spo0A or other postexponential phase transcription factors. In sum, the simplest interpretation of our results is that AbbA contributes to the derepression of all AbrB-controlled genes. Conceivably, however, AbbA governs the derepression of only a subset of AbrB-controlled genes, and depletion of AbrB or a yet-to-be-discovered mechanism or both is responsible for relieving repression of the remaining members of the AbrB regulon.
How might repression of abrB by Spo0A∼P contribute to the derepression of AbrB-controlled genes? We note from our immunoblot analysis that the level of AbrB decreased slowly and steadily from mid exponential phase growth into stationary phase. We therefore posit that Spo0A∼P-mediated repression of abrB and induction of abbA is a mechanism to adjust the ratio of AbbA to AbrB under different growth states. Thus, in exponential phase growth in rich medium when Spo0A∼P levels are expected to be at their lowest, the ratio of AbrB to AbbA would be at its highest, ensuring maximal repression of target genes. Conversely, during the transition to stationary phase, when Spo0A∼P levels begin to rise, the ratio of AbrB to AbbA would decrease, facilitating antirepression of AbrB and derepression of target genes.
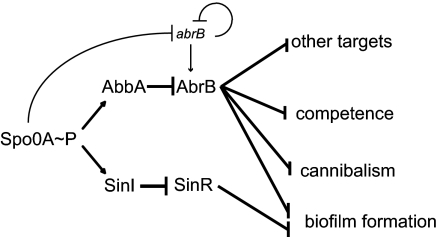
The discovery that the action of AbrB is reversed by an antirepressor reveals an interesting parallel to the circuitry that governs biofilm formation. Recent work has shown that genes involved in the production of the extracellular matrix are governed by two parallel pathways of repression mediated by the SinR and AbrB repressors (29, 30, 38). SinR is largely dedicated to the control of genes involved in biofilm formation whereas AbrB has a broad spectrum of targets and is not restricted to genes governing the production of extracellular matrix. Relief from SinR-mediated repression is known to be achieved by the action of an antirepressor, SinI, which binds to and blocks the action of SinR. SinI is produced under the direct control of Spo0A∼P. Likewise, relief from AbrB-mediated repression, as we have now shown, is mediated in part by AbbA, whose synthesis is also under the direct positive control of Spo0A∼P. Therefore, the regulatory logic governing biofilm formation is that of two parallel pathways of repression and antirepression, both of which are set in motion by the same master regulatory protein Spo0A∼P (Fig. 5). Of course, the AbrB circuit has the additional feature that Spo0A∼P also represses the gene for the AbrB repressor.
Fig. 5.
Spo0A∼P controls two parallel pathways of repression and antirepression during the transition to stationary phase. The SinR repressor is largely dedicated to genes involved in biofilm formation whereas AbrB controls a wide variety of genes, including genes for biofilm formation, competence and cannibalism. The AbrB and SinR repressors are each subject to antirepression by AbbA and SinI, respectively, whose synthesis is induced by Spo0A∼P. Also shown is that abrB is subject to repression by Spo0A∼P and autorepression.
The combination of Spo0A∼P inducing the synthesis of an antirepressor of AbrB and repressing the gene for AbrB conforms to a network motif known as a coherent feed-forward loop [specifically, a type 3 loop (41)]. Theoretical analysis indicates that coherent feed-forward loops display a delay in a response to input signal (increasing Spo0A∼P) and thus are insensitive to fluctuations in signal levels. In addition, the type 3 loop has the property that once a response has been achieved (AbrB inactivation) it persists after the input signal is removed (decreasing Spo0A∼P). Therefore, derepression of genes under AbrB control might be expected to require continuously rising levels of Spo0A∼P but once AbrB is inactivated, expression of target genes should persist even if Spo0A∼P levels subsequently drop.
In closing, we note that the abbA gene is found exclusively in the genomes of bacteria that have abrB, a finding in keeping with the idea that AbbA is a dedicated antirepressor for AbrB (Fig. S2). Interestingly, whereas AbrB is conserved in all Bacillus, Clostridium, Geobacillus and Listeria species examined, AbbA is only present in a subset of these species (those of Bacillus and Geobacillus). In AbrB-containing species that lack AbbA, an as-yet-unknown mechanism must be responsible for derepression of target genes. Especially interesting is the case of Listeria monocytogenes, which lacks both AbbA and Spo0A. Therefore, the mechanism(s) that governs relief from AbrB-mediated repression in L. monocytogenes must be unrelated to those that operate in B. subtilis. It may be that AbrB first arose as a general regulator of stationary phase and that AbbA appeared later as a sporulation-specific antagonist of AbrB in certain spore-forming species. Perhaps rapid inactivation of AbrB in AbbA-containing species is beneficial because it creates a variety of options in stationary phase, as exemplified by cannibalism, competence and biofilm formation in B. subtilis, before the cells commit to the costly and time-consuming process of sporulation.
Materials and Methods
Strain Construction.
All strains used are listed in Table S1.
General Methods.
Media, culture conditions, preparation of competent cells, and assays of β-galactosidase activity were as described (42, 43).
Transcriptional Profiling Assay.
RNA was isolated from midexponential phase cultures grown in LB medium with IPTG added to a final concentration of 1 mM. RNA was isolated by using the hot acid/phenol method (44). The strains used were AB141 (ΔabbA::tet) and AB147 (thrC::Phyperspank-abbA). RNA was prepared and hybridized as described (29). Oligoarrays were as described (9). Images were processed and analyzed with GenePix 4.0 software (Axon Instruments). We included only genes for which there was a 2.5-fold change in expression in at least three of our five replicates.
EMSA.
Radiolabeled probes were generated by PCR (with the primers listed in Table S2) by using PY79 chromosomal DNA and the following primer combinations: ECH343/ECH344 (PcomK) ECH337/ECH338 (Psdp), ECH339/ECH340 (Pskf) and AVB041/AVB042 (Plip). Each probe was 5′ end labeled with 10 mCi of [γ-32P]- ATP (NEG002A, New England Nuclear) by using polynucleotide kinase (New England Biolabs). Various concentrations of His6-AbrB, His6-AbbA or BSA were added to ≈100 nM radiolabeled probe. Binding reactions were carried out in 30-μl volumes including binding buffer [20 mM Tris·HCl (pH 8.0), 100 mM KCl, 5 mM MgCl2, 0.5 mM DTT, 10% glycerol, 0.05% Nonidet P-40] containing 25 mg ml−1 poly dI-dC, at 37°C for 20 min. A 6% polyacrylamide TAE gel was loaded with 10 μl of each binding reaction and resolved for 2.5 h at 50 V.
Protein Expression Constructs.
To generate plasmids for the expression of an N-terminal 6-histidine translation fusion to AbbA (pAB113) and unmodified AbbA (pAB111), PCR products of the abbA ORF inclusive of the stop codon were generated by using the primers AVB032/AVB026 (AVB032 contained a sequence to add 6 histidine residues to the N terminus of AbbA) and AVB015/AVB026, respectively. The PCR products were cloned into the NdeI and XhoI sites of plasmid pET21b (Novagen). The plasmids were then transformed into E. coli BL21(DE3) RIL-CodonPlus cells (Stratagene). Plasmids for the expression of His6-AbrB and His6-SinR (pEH213 and pDP90, respectively) have been described (30, 38).
Protein Purification.
His6-AbbA, His6-AbrB and His6-SinR were purified as described (38). To purify unmodified AbbA, BL21(DE3) pAB111 was grown at 30°C in 1 liter of LB supplemented with 100 μg ml−1 ampicillin until OD600 reached ≈0.5. IPTG was added to a final concentration of 1 mM and the culture grown for 2 h at 30°C. The cells were harvested, resuspended in 30 ml of binding buffer [50 mM Tris (pH 7.5), 25 mM NaCl] and lysed by sonication. The lysate was centrifuged at 32,000 × g and the supernatant was loaded onto a HiTrap Q Sepharose Fast Flow (GE Healthcare) anion exchange column that had been equilibrated with binding buffer. The column was washed with 50 ml of binding buffer, and then washed again with 10 ml of binding buffer containing 250 mM NaCl. A stepwise elution was performed with 4-ml aliquots of 50 mM Tris (pH 7.5) supplemented with 350, 400, 450, 500, and 550 mM NaCl. Purity was assessed by separating load, flow-through, wash and elutions by 15% PAGE.
Copurification of AbrB with His6-AbbA.
Overnight cultures of PY79 grown at 22°C were diluted into 50 ml of LB medium and grown until midexponential phase. Harvested cells were resuspended in 5 ml of sucrose buffer (500 mM sucrose, 20 mM MgCl2, 10 mM KPO4, and 0.1 mg ml−1 lysozyme) and incubated at 37°C for 20 min. The cells were pelleted, resuspended in 5 ml of binding buffer [25 mM Tris (pH 8.0), 100 mM KCl, 5 mM MgCl2, 0.2 mM DTT, and 10% (vol/vol) glycerol], and lysed by sonication. Lysate was cleared of debris as described above.
The cleared lysate was incubated with highly purified His6-AbbA or His6-SinR (20 μM final concentration of peptide) for 1 h at 4°C (final volume, 5 ml). Each binding reaction was placed onto 500 μl (bed volume) of Ni2+-NTA-agarose, washed with 50 ml of binding buffer containing 20 mM imidazole, and eluted with 2 ml of binding buffer containing 500 mM imidazole. Equal volumes of the various fractions were loaded onto a 15% polyacrylamide gel and AbrB, His6-AbbA, His6-SinR and σA were detected both by immunoblotting by using specific antisera and by Coomassie staining.
Copurification of AbbA with His6-AbrB.
Highly purified AbbA (2 μM) was added to binding buffer as above and incubated with highly purified His6-AbrB or His6-SinR (20 μM) (for a total volume of 1 ml) at 37°C for 10 min. One hundred microliters (bed volume) of Ni2+-NTA agarose was added to each binding reaction and incubated for 10 min at room temperature. The Ni2+-NTA agarose was pelleted, and the supernatant removed and set aside. Each reaction was then washed with 30 ml of binding buffer containing 20 mM imidazole, and eluted at 50°C with 100 μl of SDS sample buffer. AbbA, His6-AbrB, and His6-SinR were detected in the various fractions by Coomassie staining.
Immunoblot Analysis.
Cells were collected in 1-ml aliquots and resuspended in an equal volume of lysis buffer [20 mM Tris (pH 8.0), 100 mM NaCl]. Lysozyme was added to each sample to a final concentration of 0.1 mg ml−1 and incubated at 37°C for 15 min. Protein concentration was determined with Coomassie Plus Bradford kit (Pierce), and equal amounts of protein from each sample was loaded onto a 15% polyacrylamide gel. AbrB, Spo0A and σA levels were followed by immunoblot analysis by using specific antibodies. Anti-AbbA and anti-AbrB antibodies were raised in rabbits by using purified B. subtilis His6-AbbA and His6-AbrB proteins, respectively (Cocalico Biologicals). The anti-SinR antibodies were obtained from F. Chu (Harvard University, Cambridge, MA).
Acknowledgments.
We thank M. Strauch and A. L. Sonenshein for advice and comments on the manuscript. This work was supported by National Institutes of Health Grant GM 18568 (to R.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0805203105/DCSupplemental.
References
- 1.Sonenshein A. In: Bacterial Stress Responses. Storz G, Hengge-Aronis R, editors. Washington, DC: ASM Press; 2000. pp. 199–215. [Google Scholar]
- 2.Kolter R, Siegele DA, Tormo A. The stationary phase of the bacterial life cycle. Annu Rev Microbiol. 1993;47:855–874. doi: 10.1146/annurev.mi.47.100193.004231. [DOI] [PubMed] [Google Scholar]
- 3.Sonenshein AL. Control of sporulation initiation in Bacillus subtilis. Curr Opin Microbiol. 2000;3:561–566. doi: 10.1016/s1369-5274(00)00141-7. [DOI] [PubMed] [Google Scholar]
- 4.Hengge-Aronis R. Interplay of global regulators and cell physiology in the general stress response of Escherichia coli. Curr Opin Microbiol. 1999;2:148–152. doi: 10.1016/S1369-5274(99)80026-5. [DOI] [PubMed] [Google Scholar]
- 5.Lange R, Hengge-Aronis R. The cellular concentration of the sigma S subunit of RNA polymerase in Escherichia coli is controlled at the levels of transcription, translation, and protein stability. Genes Dev. 1994;8:1600–1612. doi: 10.1101/gad.8.13.1600. [DOI] [PubMed] [Google Scholar]
- 6.Sonenshein AL. CodY, a global regulator of stationary phase and virulence in Gram-positive bacteria. Curr Opin Microbiol. 2005;8:203–207. doi: 10.1016/j.mib.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 7.Boylan SA, Redfield AR, Price CW. Transcription factor sigma B of. Bacillus subtilis controls a large stationary-phase regulon. J Bacteriol. 1993;175:3957–3963. doi: 10.1128/jb.175.13.3957-3963.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boylan SA, Redfield AR, Brody MS, Price CW. Stress-induced activation of the sigma B transcription factor of Bacillus subtilis. J Bacteriol. 1993;175:7931–7937. doi: 10.1128/jb.175.24.7931-7937.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Britton RA, et al. Genome-wide analysis of the stationary-phase sigma factor (sigma-H) regulon of Bacillus subtilis. J Bacteriol. 2002;184:4881–4890. doi: 10.1128/JB.184.17.4881-4890.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoch JA. Regulation of the phosphorelay and the initiation of sporulation in Bacillus subtilis. Annu Rev Microbiol. 1993;47:441–465. doi: 10.1146/annurev.mi.47.100193.002301. [DOI] [PubMed] [Google Scholar]
- 11.Strauch MA, Hoch JA. Transition-state regulators: Sentinels of Bacillus subtilis post-exponential gene expression. Mol Microbiol. 1993;7:337–342. doi: 10.1111/j.1365-2958.1993.tb01125.x. [DOI] [PubMed] [Google Scholar]
- 12.Shivers RP, Sonenshein AL. Activation of the. Bacillus subtilis global regulator CodY by direct interaction with branched-chain amino acids. Mol Microbiol. 2004;53:599–611. doi: 10.1111/j.1365-2958.2004.04135.x. [DOI] [PubMed] [Google Scholar]
- 13.Ratnayake-Lecamwasam M, Serror P, Wong KW, Sonenshein AL. Bacillus subtilis CodY represses early stationary-phase genes by sensing GTP levels. Genes Dev. 2001;15:1093–1103. doi: 10.1101/gad.874201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hecker M, Pane-Farre J, Volker U. SigB-dependent general stress response in Bacillus subtilis and related gram-positive bacteria. Annu Rev Microbiol. 2007;61:215–236. doi: 10.1146/annurev.micro.61.080706.093445. [DOI] [PubMed] [Google Scholar]
- 15.Burbulys D, Trach KA, Hoch JA. Initiation of sporulation in B subtilis is controlled by a multicomponent phosphorelay. Cell. 1991;64:545–552. doi: 10.1016/0092-8674(91)90238-t. [DOI] [PubMed] [Google Scholar]
- 16.Molle V, et al. The Spo0A regulon of Bacillus subtilis. Mol Microbiol. 2003;50:1683–1701. doi: 10.1046/j.1365-2958.2003.03818.x. [DOI] [PubMed] [Google Scholar]
- 17.Guespin-Michel JE. Phenotypic reversion in some early blocked sporulation mutants of. Bacillus subtilis: Isolation and phenotype identification of partial revertants. J Bacteriol. 1971;108:241–247. doi: 10.1128/jb.108.1.241-247.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ito J, Mildner G, Spizizen J. Early blocked asporogenous mutants of. Bacillus subtilis 168 I Isolation and characterization of mutants resistant to antibiotic(s) produced by sporulating Bacillus subtilis 168. Mol Gen Genet. 1971;112:104–109. doi: 10.1007/BF00267488. [DOI] [PubMed] [Google Scholar]
- 19.Trowsdale J, Chen SM, Hoch JA. Genetic analysis of a class of polymyxin resistant partial revertants of stage O sporulation mutants of. Bacillus subtilis: Map of the chromosome region near the origin of replication. Mol Gen Genet. 1979;173:61–70. doi: 10.1007/BF00267691. [DOI] [PubMed] [Google Scholar]
- 20.Trowsdale J, Sheflett M, Hoch JA. New cluster of ribosomal genes in Bacillus subtilis with regulatory role in sporulation. Nature. 1978;272:179–181. doi: 10.1038/272179a0. [DOI] [PubMed] [Google Scholar]
- 21.Zuber P, Losick R. Role of AbrB in Spo0A- and Spo0B-dependent utilization of a sporulation promoter in Bacillus subtilis. J Bacteriol. 1987;169:2223–2230. doi: 10.1128/jb.169.5.2223-2230.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marahiel MA, Zuber P, Czekay G, Losick R. Identification of the promoter for a peptide antibiotic biosynthesis gene from. Bacillus brevis and its regulation in Bacillus subtilis. J Bacteriol. 1987;169:2215–2222. doi: 10.1128/jb.169.5.2215-2222.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strauch MA, et al. The transition state transcription regulator abrB of Bacillus subtilis is a DNA binding protein. EMBO J. 1989;8:1615–1621. doi: 10.1002/j.1460-2075.1989.tb03546.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Furbass R, Gocht M, Zuber P, Marahiel MA. Interaction of AbrB, a transcriptional regulator from Bacillus subtilis with the promoters of the transition state-activated genes tycA and spoVG. Mol Gen Genet. 1991;225:347–354. doi: 10.1007/BF00261673. [DOI] [PubMed] [Google Scholar]
- 25.Strauch M, Webb V, Spiegelman G, Hoch JA. The SpoOA protein of Bacillus subtilis is a repressor of the abrB gene. Proc Natl Acad Sci USA. 1990;87:1801–1805. doi: 10.1073/pnas.87.5.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Reilly M, Devine KM. Expression of AbrB, a transition state regulator from Bacillus subtilis, is growth phase dependent in a manner resembling that of Fis, the nucleoid binding protein from Escherichia coli. J Bacteriol. 1997;179:522–529. doi: 10.1128/jb.179.2.522-529.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strauch MA. Regulation of Bacillus subtilis gene expression during the transition from exponential growth to stationary phase. Prog Nucleic Acid Res Mol Biol. 1993;46:121–153. doi: 10.1016/s0079-6603(08)61020-x. [DOI] [PubMed] [Google Scholar]
- 28.Fujita M, Losick R. Evidence that entry into sporulation in Bacillus subtilis is governed by a gradual increase in the level and activity of the master regulator Spo0A. Genes Dev. 2005;19:2236–2244. doi: 10.1101/gad.1335705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chu F, Kearns DB, Branda SS, Kolter R, Losick R. Targets of the master regulator of biofilm formation in Bacillus subtilis. Mol Microbiol. 2006;59:1216–1228. doi: 10.1111/j.1365-2958.2005.05019.x. [DOI] [PubMed] [Google Scholar]
- 30.Kearns DB, Chu F, Branda SS, Kolter R, Losick RA. Master regulator for biofilm formation by Bacillus subtilis. Mol Microbiol. 2005;55:739–749. doi: 10.1111/j.1365-2958.2004.04440.x. [DOI] [PubMed] [Google Scholar]
- 31.Bai U, Mandic-Mulec I, Smith I. SinI modulates the activity of SinR, a developmental switch protein of Bacillus subtilis, by protein–protein interaction. Genes Dev. 1993;7:139–148. doi: 10.1101/gad.7.1.139. [DOI] [PubMed] [Google Scholar]
- 32.Ellermeier CD, Hobbs EC, Gonzalez-Pastor JE, Losick R. A three-protein signaling pathway governing immunity to a bacterial cannibalism toxin. Cell. 2006;124:549–559. doi: 10.1016/j.cell.2005.11.041. [DOI] [PubMed] [Google Scholar]
- 33.Gonzalez-Pastor JE, Hobbs EC, Losick R. Cannibalism by sporulating bacteria. Science. 2003;301:510–513. doi: 10.1126/science.1086462. [DOI] [PubMed] [Google Scholar]
- 34.Fujita M, Gonzalez-Pastor JE, Losick R. High- and low-threshold genes in the Spo0A regulon of Bacillus subtilis. J Bacteriol. 2005;187:1357–1368. doi: 10.1128/JB.187.4.1357-1368.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strauch MA, et al. Abh and AbrB control of. Bacillus subtilis antimicrobial gene expression. J Bacteriol. 2007;189:7720–7732. doi: 10.1128/JB.01081-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qian Q, Lee CY, Helmann JD, Strauch MA. AbrB is a regulator of the sigma(W) regulon in Bacillus subtilis. FEMS Microbiol Lett. 2002;211:219–223. doi: 10.1111/j.1574-6968.2002.tb11228.x. [DOI] [PubMed] [Google Scholar]
- 37.Hamon MA, Stanley NR, Britton RA, Grossman AD, Lazazzera BA. Identification of AbrB-regulated genes involved in biofilm formation by Bacillus subtilis. Mol Microbiol. 2004;52:847–860. doi: 10.1111/j.1365-2958.2004.04023.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chu F, et al. A novel regulatory protein governing biofilm formation in Bacillus subtilis. Mol Microbiol. 2008;68:1117–1127. doi: 10.1111/j.1365-2958.2008.06201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hamoen LW, et al. The Bacillus subtilis transition state regulator AbrB binds to the −35 promoter region of comK. FEMS Microbiol Lett. 2003;218:299–304. doi: 10.1111/j.1574-6968.2003.tb11532.x. [DOI] [PubMed] [Google Scholar]
- 40.Strauch MA. In vitro binding affinity of the Bacillus subtilis AbrB protein to six different DNA target regions. J Bacteriol. 1995;177:4532–4536. doi: 10.1128/jb.177.15.4532-4536.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mangan S, Alon U. Structure and function of the feed-forward loop network motif. Proc Natl Acad Sci USA. 2003;100:11980–11985. doi: 10.1073/pnas.2133841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilson GA, Bott KF. Nutritional factors influencing the development of competence in the Bacillus subtilis transformation system. J Bacteriol. 1968;95:1439–1449. doi: 10.1128/jb.95.4.1439-1449.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fujita M, Losick R. An investigation into the compartmentalization of the sporulation transcription factor sigmaE in Bacillus subtilis. Mol Microbiol. 2002;43:27–38. doi: 10.1046/j.1365-2958.2002.02732.x. [DOI] [PubMed] [Google Scholar]
- 44.Fawcett P, Eichenberger P, Losick R, Youngman P. The transcriptional profile of early to middle sporulation in Bacillus subtilis. Proc Natl Acad Sci USA. 2000;97:8063–8068. doi: 10.1073/pnas.140209597. [DOI] [PMC free article] [PubMed] [Google Scholar]