Abstract
Since the first determination of structure of the HLA-A2 complex, >200 MHC/peptide structures have been recorded, whereas the available T cell receptor (TCR)/peptide/MHC complex structures now are <20. Among these structures, only six are TCR/peptide/MHC Class II (MHCII) structures. The most recent of these structures, obtained by using TCR-Ob.1A12 from a multiple sclerosis patient and the MBP85–99/HLA-DR2 complex, was very unusual in that the TCR was located near the N-terminal end of the peptide-binding cleft of the MHCII protein and had an orthogonal angle on the peptide/MHC complex. The unusual structure suggested the possibility of a disturbance of its signaling capability that could be related to autoimmunity. Here, homology modeling and a new simulation method developed for TCR/peptide/MHC docking have been used to examine the positioning of the complex of two additional TCRs obtained from the same patient (TCR-Ob.2F3 or TCR-Ob.3D1 with MBP85–99/HLA-DR2). The structures obtained by this simulation are compatible with available data on peptide specificity of the TCR epitope. All three TCRs from patient Ob including that from the previously determined crystal structure show a counterclockwise rotation. Two of them are located near the N terminus of the peptide-binding cleft, whereas the third is near the center. These data are compatible with the hypothesis that the rotation of the TCRs may alter the downstream signaling.
Keywords: human leukocyte antigen, multiple sclerosis, myelin basic protein, structural docking, signaling
The elucidation of structures of protein complexes is an arduous procedure particularly when the complexes are very large. The production of protein by recombinant techniques and subsequent crystallization of the complexes followed by x-ray diffraction analysis is a standard method. Structures can also be determined by NMR but that technique is presently limited to only relatively small proteins or complexes not >40–50 kDa. The third method, simulation of structures by homology modeling, has improved greatly in recent years. However, this technique is usable only when an appropriate template structure is available.
Crystallization and structure determination by x-ray diffraction of a MHC-encoded Class I (MHCI) protein/peptide complex was first accomplished in 1987 and MHCII in 1993 (1, 2). Since then, >200 structures of such complexes have been recorded (3). The β chain of the TCRs that recognize these complexes were first cloned in 1984 (4, 5), and the first structure of a TCR chain was published in 1995 (6). Approximately 40 complete TCR structures including both α and β chains are available now (3, 7). Similarly, the first TCR/peptide/MHC complex structures were published in 1996 (8, 9) but the number of such complex structures available now is <20 (3). Among these, only six are TCR/peptide/MHCII structures. The most recent of these structures is very unusual in that the TCR was located near the N-terminal end of the peptide-binding cleft of the MHCII protein and its orthogonal angle on the MHCII/peptide was 84° as compared with a diagonal angle of 40–53° for the other five structures (10). Also, it was rotated counterclockwise on the MHC molecule relative to the other structures. This unusual structure suggested the possibility of a disturbance of its signaling capability that could be related to autoimmunity because this TCR, termed Ob.1A12 had been obtained from an autoreactive clone derived from a patient with multiple sclerosis (10). In fact, eight clones were obtained from this patient, two of which represent unique isolates (TCR-Ob.1A12 and TCR-Ob.3D1) and six of which have identical sequences, TCR-Ob.2F3 being an example (11).
In this article, we have used homology modeling of TCR structures on the appropriate templates and a new simulation method developed for TCR/peptide/MHC docking to examine the structures of the TCR/peptide/MHCII complexes of TCR-Ob.2F3 and TCR-Ob.3D1 in complex with the same MHC/peptide, namely HLA-DR2 (DRB1*1501/DRA) binding the myelin basic protein peptide epitope MBP85–99. MBP85–99 has previously been identified as the autoreactive peptide epitope in humans (12).
Results and Discussion
The AutoDock procedure was originally developed for docking studies of small chemicals to their receptors, for example the docking of a substrate to an enzyme (13). It makes use of charge and hydrophobicity calculations for both the receptor and the ligand (see Materials and Methods). By using this method in the present context, the peptide in question was first docked to the appropriate MHC molecule and separately to the TCR protein. The conformation of the peptide used in the docking in each case was taken from the crystal structure of the MBP85–99/HLA-DR2 (DRB1*1501, DRA) complex (10). After TCR/peptide and peptide/MHC structures were simulated, the two structures were merged by using the conformation of the peptide as the basis for merging. To validate the procedure, the technique was carried out by using two known TCR/peptide/MHC structures, that of the HLA-DR1 (DRB1*0101/DRA)/hemagglutinin (HA) 306–318 molecule in complex with the HLA-DR1-restricted HA306–318-specific TCR-HA1.7 (PDB ID code 1FYT) and then of the HLA-DR2/MBP85–99 molecule in complex with TCR-Ob.1A12 (PDB ID code 1YMM).
Simulated Structure of the TCR-HA1.7/HA306–318/HLA-DR1 Complex and the TCR-Ob.1A12/MBP85–99/HLA-DR2 Complex.
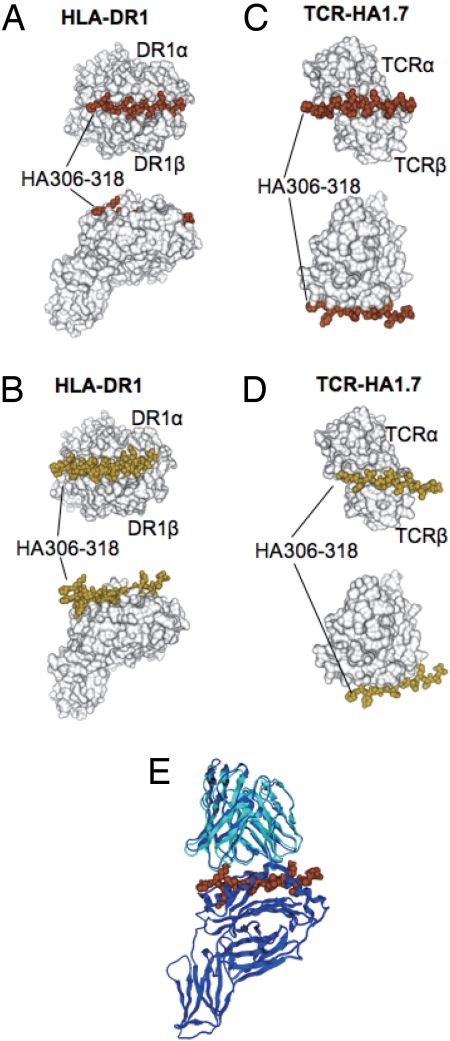
The docking of HA306–318 to HLA-DR1 was simulated 10 times. Six of the 10 simulations showed exactly the same conformation inside the peptide-binding groove with energy equal to −33.8 kcal/mol (Fig. 1A). Four of the 10 showed different conformations binding outside of the groove with higher energies of −5.59, −5.59, −5.68 and −8.73 kcal/mol (Fig. 1B). One of the clustered conformations inside the groove with the lowest energy was selected as representative.
Fig. 1.
Docking simulation of the HA306–318 peptide on HLA-DR1 and on TCR-HA1.7. (A) The six clustered docked peptides of HA306–318 on HLA-DR1 are indicated as space filling models in brown. (B) The four nonclustered docked peptides are indicated as space filling models in yellow. In A and B, HLA-DR1 (DRB1*0101/DRA) (20) is shown as a surface model in white. (C) The nine clustered docked peptides of HA306–318 on TCR-HA1.7 are indicated as space filling models in brown. (D) The one nonclustered docked peptide is shown as a space-filling model in yellow. In C and D, TCR-HA1.7 is shown as a surface model in white. (E) Merging of the docked TCR-HA1.7/HA306–318 and HA306–318/HLA-DR1 was carried out by using the conformation of the peptide as the basis for merging. Superposition was performed between the docked structure and the crystal structure of TCR-HA1.7/HA306–318/HLA-DR1 (20). Docked structure of TCR-HA1.7 in cyan, HA306–318 in brown, and crystal structures of both TCR-HA1.7 and HLA-DR1 in blue. The two TCR structures were superimposed by means of structures of HA306–318.
Similarly, HA306–318 docking to TCR-HA1.7 was simulated 10 times. Nine of the 10 simulations had almost the same conformation with energy equal to −5.59 kcal/mol five times and energy −5.67 four times (Fig. 1C). One of the 10 simulations yielded a different conformation with a higher energy of −5.02 kcal/mol (Fig. 1D). Again, one of the five clustered conformations with the lowest energy was selected as representative.
Next, the TCR-HA1.7/HA306–318 simulated complex was merged with the HA306–318/HLA-DR1 simulated complex to give the TCR-HA1.7/HA306–318/HLA-DR1 complex by using the structure of the peptide as the basis for merging (a related procedure was used in a docking study of the dimeric maltose-binding complex involving maltose-binding protein and aspartate receptor, although in this case the octapeptide used was from a functional region of the maltose-binding protein) (14).
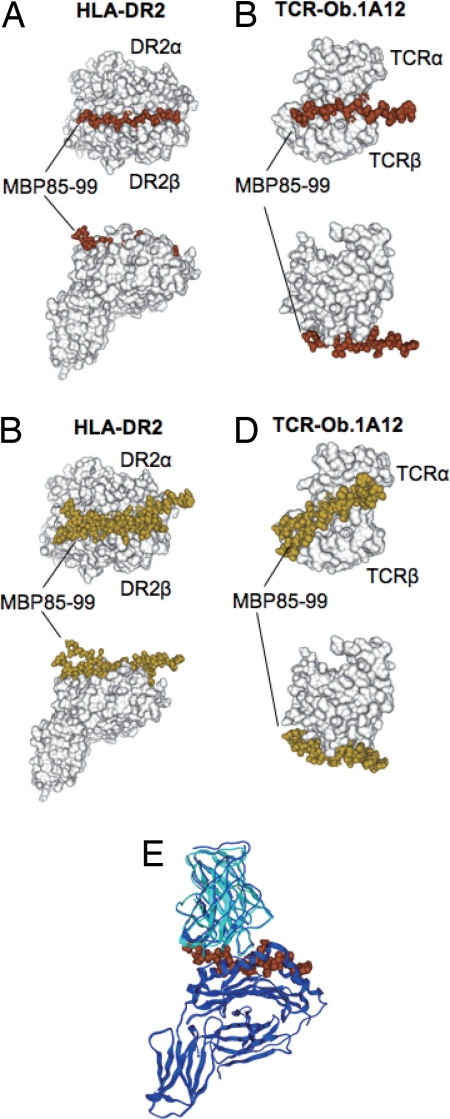
This simulated structure of the ternary complex was merged with the structure determined by crystallization and x-ray diffraction and gave excellent reproducibility with a rmsd of 1.64 Å (Fig. 1E). The same procedure was carried out to obtain the TCR-Ob.1A12/MBP85–99/HLA-DR2 structure. The docking of MBP85–99 to HLA-DR2 was simulated 10 times. Four of the 10 simulations showed exactly the same conformations inside the peptide-binding groove with energy equal to −26.8 kcal/mol (Fig. 2A). Six of the 10 showed binding outside of the groove each with a different conformation and with much higher energies of −3.02, −4.95, −4.95, −5.54, −5.64 and −5.91 kcal/mol (Fig. 2B). One of the clustered conformations inside the groove with the lowest energy was selected as representative.
Fig. 2.
Docking simulation of the MBP85–99 peptide on HLA-DR2 and on TCR-Ob.1A12. (A) The four clustered docked peptides of MBP85–99 on HLA-DR2 are indicated as space filling models in brown. (B) The six nonclustered docked peptides are indicated as space filling models in yellow. In A and B, HLA-DR2 (DRB1*1501/DRA) (10) is shown as a surface model in white. (C) The six clustered docked peptides of MBP85–99 on TCR-Ob.1A12 are indicated as space filling models in brown. (D) The four nonclustered docked peptides are indicated as space filling models in yellow. In C and D, TCR-Ob.1A12 is shown as a surface model in white. (E) Merging of the docked TCR-Ob.1A12/MBP85–99 and MBP85–99/HLA-DR2 was carried out by using the conformation of the peptide as the basis for merging. Superposition was performed between the docked structure and the crystal structure of TCR-Ob.1A12/MBP85–99/HLA-DR2 (10). Docked structure of TCR-Ob.1A12 in cyan, MBP85–99 in brown, and crystal structures of both TCR-Ob.1A12 and HLA-DR2 in blue. The two TCR structures were superimposed by means of structures of MBP85–99.
Similarly, MBP85–99 docking to TCR-Ob.1A12 was simulated 10 times. Six of the 10 simulations had almost the same conformation with energy equal to −4.78 five times and energy −4.65 once (Fig. 2C). Four of the 10 simulations each yielded a different conformation with a similar energy of −4.50, −5.08, −5.32, and −5.32 kcal/mol (Fig. 2D). Again, one of the six clustered conformations with the lowest energy was selected as representative.
The TCR-Ob.1A12/MBP85–99 simulated complex was also merged with the MBP85–99/HLA-DR2 simulated complex as described above to give the TCR-Ob1A12/MBP85–99/HLA-DR2 complex. Merging of the simulated structure with the structure determined by x-ray crystallography gave excellent reproducibility with a rmsd of 1.54 Å (Fig. 2E). Thus, by using two known TCR/peptide/MHC complexes, these data provided a validation for the docking procedure used.
Notably, the energies of docking of the two peptides to their respective MHC proteins (−34 and −27 kcal/mol) were much lower than the energies of their docking to their respective TCR (−5.6 and −4.8 kcal/mol). Thus, the binding of the peptide to the MHC proteins is much stronger than the binding to the TCR (13).
Simulated Structure of the TCR-Ob.2F3/MBP85–99/HLA-DR2 Complex and the TCR-Ob.3D1/MBP85–99/HLA-DR2 Complex.
Next, structures of two TCR complexes from patient Ob whose complexes with HLA-DR2/MBP85–99 had not been determined were modeled. The sequence alignment with the template TCR-Ob.1A12 of these two TCRs, TCR-Ob.2F3 and TCR-Ob.3D1, are shown in Fig. 3. Notably, TCR-Ob.2F3 has 99% sequence identify with TCR-Ob.1A12 differing at only three amino acid positions. One of these is near the CDR3 loop of the TCR α chain and the other two are in the CDR3 loop of the TCR β chain. By contrast, TCR-Ob.3D1 had only 89% identify in the TCR α chain differing from TCR-Ob.1A12 by 10 positions including two gaps. It had only 37% identify (but 59% similarity) in the β chain differing from the template structure by 40 aa including two insertions in the CDR3 loop.
Fig. 3.
Sequence alignment of TCR-Ob.2F3 and TCR-Ob.3D1 with TCR-Ob.1A12. (A) Sequence alignment between TCR-Ob.2F3 and TCR-Ob.1A12. (B) Sequence alignment between TCR-Ob.3D1 and TCR-Ob.1A12. Differences in amino acids between the two clones are indicated in yellow. *, identical amino acids; +, similar amino acids.
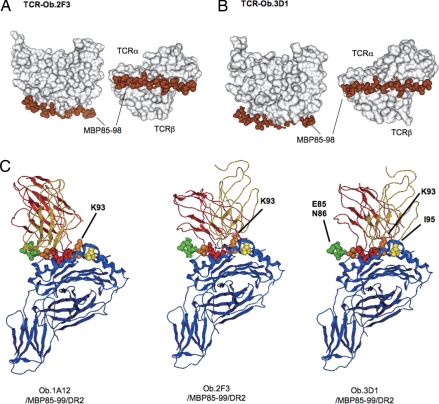
When MBP85–99 was docked to the modeled TCR-Ob.2F3, the 10 simulations showed almost the same conformation with energies of −5.77 kcal/mol five times, −5.76 four times, and −5.75 once (Fig. 4A). One of the clustered confirmations with the lowest energy was again selected as representative. The TCR-Ob.2F3/MBP85–99 docked structure was merged with the crystal structure of MBP85–99/HLA-DR2 again by using the conformation of the MBP85–99 peptide as the basis for merging.
Fig. 4.
Positioning of TCR-Ob.2F3 and TCR-Ob.3D1 on MBP85–99/HLA-DR2. (A) Docking simulation of the MBP85–99 peptide on TCR-Ob.2F3. The 10 clustered docked peptides are indicated as space filling models in brown. TCR-Ob.2F3 is shown as a surface model in white. (B) Docking simulation of the MBP85–99 peptide on TCR-Ob.3D1. The 10 clustered docked peptides are indicated as space filling models in brown. TCR-Ob.3D1 is shown as a surface model in white. (C) Comparison of positioning of the TCRs obtained from patient Ob. on the MBP85–99/HLA-DR2 structure. The structures of TCRs, MBP85–99 and HLA-DR2 (DRB1*1501/DRA) are shown as ribbon models. MBP85–99 and HLA-DR2 in blue, the α chains of the three TCRs in yellow, and the β chains of the three TCRs in red. Functionally important residues of MBP85–99 are shown as space filling models, E85 and N86 in green, V88 and K93 in orange, H90 and F91 in red, and I95 in yellow (see Results and Discussion).
Similarly, MBP85–99 was docked to the model TCR-Ob.3D1 10 times. All 10 simulations showed exactly the same conformation with energy −8.2 kcal/mol (Fig. 4B). One of these clustered conformations was again selected as representative and as before docked with the MBP85–99/HLA-DR2 structure.
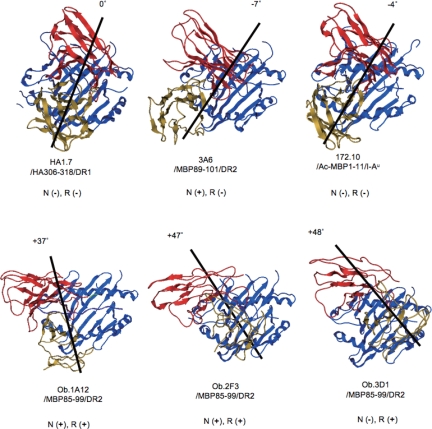
The structural models that resulted form these simulations are shown in Fig. 4C. Several features are obvious. First, like TCR-Ob.1A12 (10), TCR-Ob.2F3 is positioned toward the N- terminus of the peptide-binding groove. However, it is located very slightly more toward the C-terminal end of the peptide. With regard to rotation, the counterclockwise rotation of Ob.1A12 (+37°) as compared with HA1.7 (taken as 0°) is apparent (Fig. 5). The TCR-Ob.2F3 had an even greater counterclockwise rotation (+47°).
Fig. 5.
Comparison of positioning between autoimmune TCRs and nonautoimmune TCR. HA1.7/HA306–318/DR1 indicates the positioning of a nonautoimmune TCR. All of the others are autoimmune TCRs. Black lines are drawn between the S–S bonds of TCRα and TCRβ. Coloring is the same as in Fig. 4C. N, N-terminal shift; R, counterclockwise rotation. The degrees of rotation taking TCR-HA1.7 on HA306–318/HLA-DR1 as 0° were as follows: TCR-3A6, −7°; TCR-172.10, −4°; TCR-Ob.1A12, +37°; Ob.2F3, +47°; and TCR-Ob.3D1, +48°.
TCR-Ob.3D1 by contrast was located near the middle of the peptide-binding cleft at the apex of the helixes in a similar position to TCR-HA1.7 (Figs. 4C and 5). However, again its counterclockwise rotation with regard to TCR-HA1.7 was large (+48°) and similar to that of TCR-Ob.2F3. When viewed from the top, the orthogonal angle of all three TCRs was distinct from that of HA1.7 and the other TCR/peptide/MHC complexes whose structures had been determined (5, 3, 15, 16). All three autoimmune TCRs show a counterclockwise rotation and two of them are located near the N-terminus of the peptide-binding cleft. These data are compatible with the hypothesis (10) that the rotation of the TCRs may alter the downstream signaling.
Available data on peptide specificity of the TCR epitope are compatible with these structures (Table 1, compiled from refs. 11, 12, 17, 18). With regard to the peptide, both TCR-Ob.1A12 and TCR-Ob.2F3 are sensitive to deletion of the N-terminal residue 85 of MBP85–99, whereas TCR-Ob.3D1 can recognize even a truncation of two residues at the N-terminus of MBP85–99 (Figs. 4C and 5). Alanine scanning of MBP85–99 revealed that TCR recognition of His-90 and Phe-91 was essential for all three clones (Table 1). Only TCR-Ob.3D1 was sensitive to mutation of Asn-94 (P6) or partially Ile-95 (P7), however, K93 was not essential for TCR-Ob.1A12, and K93A revealed reduced sensitivity at this residue for TCR-Ob.2F3 (Table 1). However, both K93A and N94A eliminated reactivity with TCR-Ob.3D1 and I95A reduced reactivity. These data are compatible with the positions of the three TCR as shown in Figs. 4C and 5. Thus, good structure-function correlations are observed in the three autoimmune T cell clones.
Table 1.
Characters of the T cell clones from patient Ob
| Clone | Ob.1A12 | Ob.2F3 | Ob.3D1 |
|---|---|---|---|
| Ob.1C3 | |||
| Ob.1E10 | |||
| Ob.1E12 | |||
| Ob.1H8 | |||
| Ob.2G9 | |||
| Minimal peptide | MBP 85–98 | MBP 85–98 | MBP 87–98 |
| MBP 85–99 | ++++ | ++++ | ++++ |
| MBP 85–98 | ++++ | ++++ | ++++ |
| MBP 86–98 | − | − | ++++ |
| MBP 87–98 | − | − | ++++ |
| MBP 87–97 | − | − | − |
| TCR epitope | |||
| Major | HF—- | HF-K– | HF-K– |
| Minor | —K– | —–I | |
| MBP 85–99 | ++++ | ++++ | ++++ |
| Val88Ala | ++++ | ++++ | ND |
| His90Ala | + | − | − |
| Phe91Ala | − | − | − |
| Lys93Ala | ++++ | ++ | − |
| Asn94Ala | ++++ | +++ | + |
| Ile95Ala | ++++ | +++ | ++ |
| Val96Ala | ++++ | ++++ | ++++ |
The CD4 coreceptor binds to the membrane-proximal MHCII domains and is essential for T cell development and T cell function by recruiting the tyrosine kinase Lck. The alignment of the TCRs observed here showed that the geometry of the interaction with the CD4 coreceptor is altered for the TCRs as previously suggested (10). These findings raise the possibility that CD4 function is affected in immature T cells by an altered geometry of TCR binding to peptide-MHC during the formation of immunological synapses (10). These structures may add to our understanding of the molecular mechanism that could relate to autoimmunity.
Materials and Methods
Structure Modeling of TCR.
Structural modeling of TCR-Ob.2F3 and TCR-Ob.3D1 was performed by using MOE software (Chemical Computing Group, www.chemcomp.com) combined with the segment-matching procedure (19, 20). Briefly, the structure of TCR-Ob.1A12 complexed with MBP85–99/HLA-DR2 (10) (PDB ID code 1YMM) was used as a template for homology modeling of TCR-Ob.2F3 and TCR-Ob.3D1. The modeled structure was further energy minimized (MOE software).
Docking Studies of the TCR with Peptide/MHC.
Docking was performed by using the AutoDock software package running on Intel-based Xeon, ppcDarwin platform (13). The structure of HLA-DR1 (DRB1*0101/DRA), HLA-DR2 (DRB1*1501/DRA), TCR-HA1.7, TCR-Ob.1A12, TCR-Ob.2F3, or TCR-Ob.3D1 was used as the target structure. HA306–318 or MBP85–99 in the conformation found in their crystal structures (10, 21) was used as the ligand structure. AutoDock with a Lamarckian genetic search algorithm (LGA) was chosen for all dockings (13).
The optimized AutoDocking run parameters were similar to those described in ref. 13 with minor modification in grid size, a maximum number of energy evaluations, and a maximum number of generations. The proteins and ligands in the dockings were treated by using the united-atom approximation. Only polar hydrogens were added to the protein, and Kollman united-atom partial charges were assigned. All waters were removed. Atomic solvation parameters and fragmental volumes were assigned to the protein atoms by using an AUTODOCK utility, ADDSOL and the grid maps were calculated by using AUTOGRID (13).
The dimensions of the grids for docking were thus 180 × 80 × 90 points (67.5 Å × 30.0 Å × 33.7 Å) and a grid-point spacing of 0.375 Å, and the center of the grids were placed to cover the surface of the HLA or TCR structure. The ligand was treated initially as all atom entities, i.e., all hydrogens were added, then partial atomic charges were calculated by using the Gasteiger-Marsili method (13). AUTOTORS, an AUTODOCK utility, was used to define the rotable bonds in the ligand to unite the nonpolar hydrogens added by SYBYL for the partial atomic charge calculation. The partial charges on the nonpolar hydrogens were added to that of the hydrogen-bearing carbon also in AUTOTORs.
In the analyses, 10 dockings were performed; in the analysis of the docked conformations, the clustering tolerance as different conformations for the rmsd was 1.0 Å. The step sizes were 0.2 Å for translations and 5° for orientations and torsions. The α and β parameters determined the size of the mutation in the genetic algorithms, LGA. The Cauchy distribution parameters were: α = 0 and β = 1. Note that random changes were generated in the genetic algorithm by a Cauchy distribution.
In the LGA dockings, an initial population of random individuals with a population size of 50 individuals was used; a maximum number of 2.5 × 108 energy evaluations; a maximum number of generations of 2.7 × 104; an elitism value of 1, which was the number of top individuals that automatically survived into the next generation; a mutation rate of 0.02, which was the probability that a gene would undergo a random change; and a cross-over rate of 0.80, which was the probability that two individuals would undergo cross-over. Proportional selection was used, where the average of the worst energy was calculated over a window of the previous 10 generations. In the LGA dockings, the pseudoSolis and Wets local search method was used, having a maximum of 300 iterations per local search; the probability of performing local search on an individual in the population was 0.06; the maximum number of consecutive successes or failures before doubling or halving the local search step size, r, was 4, in both cases; and the lower bound on r, the termination criterion for the local search, was 0.01.
TCR/Peptide/MHC Complex Structure.
To make the whole TCR/peptide/MHC complex structure, superposition was done between the docked structure of the TCR/peptide and the docked structure of peptide/HLA or between the docked structure of the TCR/peptide and the crystal structure of the peptide/HLA complex. Then, the peptide structure was removed from the system. The structures obtained were further energy-minimized (MOE software).
Acknowledgments.
We thank M. Karplus for technical advice and D. Keskin for helpful comments. This work was supported by National Institutes of Health Research Grant 5RO1 AI049524 and National Multiple Sclerosis Society Award RG3796A3.
Footnotes
The authors declare no conflict of interest.
References
- 1.Bjorkman PJ, et al. The foreign antigen binding site and T cell recognition regions of class I histocompatibility antigens. Nature. 1987;329:512–518. doi: 10.1038/329512a0. [DOI] [PubMed] [Google Scholar]
- 2.Brown JH, Jardetzky TS, Gorga JC, Stern LJ, Urban RG. Three-dimensional structure of the human class II histocompatibility antigen HLA-DR1. Nature. 1993;364:33–39. doi: 10.1038/364033a0. [DOI] [PubMed] [Google Scholar]
- 3.Rudolph MG, Stanfield RL, Wilson IA. How TCRs bind MHCs, peptides, and coreceptors. Annu Rev Immunol. 2006;24:419–466. doi: 10.1146/annurev.immunol.23.021704.115658. [DOI] [PubMed] [Google Scholar]
- 4.Hedrick SM, Cohen DI, Nielsen EA, Davis MM. Isolation of cDNA clones encoding T cell-specific membrane-associated proteins. Nature. 1984;308:149–153. doi: 10.1038/308149a0. [DOI] [PubMed] [Google Scholar]
- 5.Yanagi Y, et al. A human T cell-specific cDNA clone encodes a protein having extensive homology to immunoglobulin chains. Nature. 1984;308:145–149. doi: 10.1038/308145a0. [DOI] [PubMed] [Google Scholar]
- 6.Bentley GA, Boulot G, Karjalainen K, Mariuzza RA. Crystal structure of the beta chain of a T cell antigen receptor. Science. 1995;267:1984–1987. doi: 10.1126/science.7701320. [DOI] [PubMed] [Google Scholar]
- 7.Mak TW. The T cell antigen receptor: “The Hunting of the Snark.”. Eur J Immunol. 2007;37(Suppl 1):S83–S93. doi: 10.1002/eji.200737443. [DOI] [PubMed] [Google Scholar]
- 8.Garboczi DN, et al. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature. 1996;384:134–141. [PubMed] [Google Scholar]
- 9.Garcia KC, et al. An alphabeta T cell receptor structure at 2.5 A and its orientation in the TCR-MHC complex. Science. 1996;274:209–219. [PubMed] [Google Scholar]
- 10.Hahn M, Nicholson MJ, Pyrdol J, Wucherpfennig KW. Unconventional topology of self peptide–major histocompatibility complex binding by a human autoimmune Tcell receptor. Nat Immunol. 2005;6:490–496. doi: 10.1038/ni1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wucherpfennig KW, et al. Clonal expansion and persistence of human T cells specific for an immunodominant myelin basic protein peptide. J Immunol. 1994;152:5581–5592. [PubMed] [Google Scholar]
- 12.Wucherpfennig KW, et al. Structural requirements for binding of an immunodominant myelin basic protein peptide to DR2 isotypes and for its recognition by human T cell clones. J Exp Med. 1994;179:279–290. doi: 10.1084/jem.179.1.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morris GM, Goodsell DS, Huey R, Olson AJ. Distributed automated docking of flexible ligands to proteins: Parallel applications of AutoDock 2.4. J Comput Aided Mol Des. 1996;10:293–304. doi: 10.1007/BF00124499. [DOI] [PubMed] [Google Scholar]
- 14.Stoddard BL, Koshland DE., Jr Prediction of the structure of a receptor-protein complex using a binary docking method. Nature. 1992;358:774–776. doi: 10.1038/358774a0. [DOI] [PubMed] [Google Scholar]
- 15.Maynard J, et al. Structure of an autoimmune T cell receptor complexed with class II peptide- MHC: Insights into MHC bias and antigen specificity. Immunity. 2005;22:81–92. doi: 10.1016/j.immuni.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 16.Li Y, et al. Structure of a human autoimmune TCR bound to a myelin basic protein self-peptide and a multiple sclerosis-associated MHC class II molecule. EMBO J. 2005;24:2968–2979. doi: 10.1038/sj.emboj.7600771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hausmann S, Martin M, Gauthier L, Wucherpfennig KW. Structural features of autoreactive TCR that determine the degree of degeneracy in peptide recognition. J Immunol. 1999;162:338–344. [PubMed] [Google Scholar]
- 18.Wucherpfennig KW, Hafler DA, Strominger JL. Structure of human T-cell receptors specific for an immunodominant myelin basic protein peptide: Positioning of T-cell receptors on HLA-DR2/peptide complexes. Proc Natl Acad Sci USA. 1995;92:8896–8900. doi: 10.1073/pnas.92.19.8896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fechteler T, Dengler U, Schomberg D. Prediction of Protein Three-Dimensional Structures in Insertion and Deletion Regions: A Procedure for Searching Databases of Representative Protein Fragments Using Geometric Scoring Criteria. J Mol Biol. 1995;253:114–131. doi: 10.1006/jmbi.1995.0540. [DOI] [PubMed] [Google Scholar]
- 20.Levitt M. Accurate Modeling of Protein Conformation by Automatic Segment Matching. J Mol Biol. 1992;226:507–533. doi: 10.1016/0022-2836(92)90964-l. [DOI] [PubMed] [Google Scholar]
- 21.Hennecke J, Carfi A, Wiley DC. Structure of a covalently stabilized complex of a human αβ T-cell receptor, influenza HA peptide and MHCII molecule, HLA-DR1. EMBO J. 2000;19:5611–5624. doi: 10.1093/emboj/19.21.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]