Abstract
Quantum dots (QDs) are being investigated as novel in vivo imaging agents. The leaching of toxic metals from these QDs in biological systems is of great concern. This study compared the cytotoxic mechanisms of two QD species made of different core materials (cadmium selenide [CdSe] vs. indium gallium phosphide [InGaP]) but similar core sizes (5.1 vs. 3.7 nm) and surface compositions (both ZnS capped, lipid-coated and pegylated). The CdSe QD was found to be 10-fold more toxic to porcine renal proximal tubule cells (LLC-PK1) than the InGaP QD on a molar basis, as determined by MTT assay (48 h IC50 10nM for CdSe vs. 100nM for InGaP). Neither of the QD species induced appreciable oxidative stress, as determined by lipid peroxide and reduced glutathione content, suggesting that toxicity was not metal associated. In agreement, treatment of cells with CdSe QDs was not associated with changes in metallothionein-IA (MT-IA) gene expression or Cd-associated caspase 3 enzyme activation. By contrast, incubation of the LLC-PK1 cells with the InGaP QD resulted in a dramatic increase in MT-IA expression by 21- and 43-fold, at 8 and 24 h, respectively. The most remarkable finding was evidence of extensive autophagy in QD-treated cells, as determined by Lysotracker Red dye uptake, TEM, and LC3 immunobloting. Autophagy induction has also been described for other nanomaterials and may represent a common cellular response. These data suggest that QD cytotoxicity is dependent upon properties of the particle as a whole, and not exclusively the metal core materials.
Keywords: quantum dots, autophagy, anomaterials
Quantum dots (QDs) are nanoscale (2–10 nm core), fluorescent colloids composed of semiconductor materials (i.e., cadmium selenide [CdSe], cadmium telluride [CdTe], indium arsenide, indium gallium phosphide [InGaP]) (Bruchez et al., 1998). These particles have unique properties from traditional small molecule fluorescent dyes, in that they have wide, highly efficient excitation ranges, and narrow, size-tunable emissions. QDs are also resistant to photobleaching. These properties make QDs ideal for use in biomedical applications, such as diagnostic imaging (Rhyner et al., 2006) or sentinel lymph node mapping (Ballou et al., 2007). QDs are also undergoing development as vectors for drug delivery (Derfus et al., 2007). These biomedical applications require that QDs have a high degree of biocompatibility.
As QDs represent a diverse group of materials, it is likely that their toxicities will depend on many factors, including core composition and physicochemical characteristics. QD biocompatability is thought to be at least partially dependent upon stability of the outer surface layers (Chan et al., 2006; Cho et al., 2007; Derfus et al., 2004; Kirchner et al., 2005). QDs often contain an outer zinc sulfide shell that enhances the quantum yield, as well as hydrophilic surface coatings to increase solubility and decrease nonspecific interactions with macromolecules, such as proteins (Bentzen et al., 2005; Uyeda et al., 2005). This outer coating also shields the inner metal core, which for some QDs is composed of toxic metal (i.e., Cd, As, Se, Te, Pb). These toxic metals have the potential to leach when the outer coating is deteriorated, by such conditions as photolysis, oxidation, or low pH (Aldana et al., 2001; Aldana et al., 2005; Derfus et al., 2004). In addition to leaching of toxic metals, exposure of the bare QD metal core can result in generation of reactive oxygen species (ROS) (Ipe et al., 2005; Lovrić et al., 2005b). ROS generation can result in cell death through lipid peroxidation and loss of membrane integrity, as well as oxidative damage to DNA and proteins (Valko et al., 2005).
The present study sought to determine if metal-associated mechanisms were responsible for the cytotoxicity of polyethylene glycol (PEG)–coated, zinc sulfide–capped CdSe and InGaP QDs in a porcine renal proximal tubule cell line (LLC-PK1). The LLC-PK1 cell line has been used extensively to examine metal-associated responses, including stress gene induction, apoptosis, and oxidative stress (Gennari et al., 2003; Liu et al., 2007; Wispriyono et al., 1998). In the absence of evidence for these metal-associated responses, observed QD-induced autophagy is proposed as a potential nanoparticle-mediated toxic mechanism.
MATERIALS AND METHODS
Materials
CdSe Maple Red Orange (Part# AWN16J2N) and InGaP Macoun Red (Part #AWN13J2N) QD were supplied by Evident Technologies (Troy, NY). Both these QD have a ZnS cap covering the metal core and pegylated neutral lipid surface coating.
Bovine serum albumin, 1-butanol, butylated hydroxytoluene, cispatin, Costar six-well, flat-bottomed, cell culture plates, dimethyl sulfoxide, diethyl maleate, 5-5′-dithiobis(2-nitrobenzoic acid), glycine, malondialdehyde tetraethylacetal (1,1,3,3 tetraethoxypropane), methanol, 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide, β-nicotinamide adenine dinucleotide 2′-phosphate reduced tetrasodium salt, ethylenediaminetetraacetic acid tetrasodium salt dihydrate, oxidized glutathione, protease inhibitor cocktail, phenyl methyl sulphonyl fluoride, 5-sulfosalicylic acid dihydrate, sodium phosphate, sodium carbonate, sodium chloride, thiobarbituric acid, trichloroacetic acid, Triton-X-100, and Tween-20 were purchased from Sigma, Inc. (St Louis, MO). L-glutamine, RPMI-1640 (phenol-free), and fetal bovine serum were purchased from Hyclone, Inc. (Logan, UT). Medium 199 media was purchased from Cambrex (East Rutherford, NJ). Ninety-six– and six-well, flat-bottomed, cell culture plates were purchased from Costar. Quick Start Bradford dye reagent, 1× was purchased from Bio-Rad Laboratories, Inc. (Hercules, CA). 1-Methyl-4-vinyl-pyridinium was purchased from OXIS International. Cell extraction buffer, Hank's balanced salt solution (with calcium and magnesium), NuPAGE LDS 4× sample buffer with reducing agent (10×), SeeBlue® Plus2 prestained standard, 4–20% tris-glycine gels, tris-glycine running buffer (10×), transfer buffer (25×), Lysotracker Red DND-99, and Celltracker Green CMFDA were purchased form Invitrogen, Inc. (Carlsbad, CA). Westran S polyvinylidene fluoride (PVDF) protein blotting membrane and blotting paper were purchased from Fisher Scientific, Inc. (Pittsburgh, PA). Tris-buffered saline (25×) was purchased from Amresco, Inc. (Solon, OH). Branched chain amino acid (BCA) protein assay, StartingBlock blocking buffer, and electrochemiluminescent (ECL) western blotting substrate reagent were purchased from Pierce (Rockford, IL). The mouse monoclonal anti-LC3 antibody was purchased from NanoTools (Teningen, Baden-Württemberg, DE). Peroxidase-conjugated AfiniPure donkey anti-mouse IgG was purchased from Jackson ImmunoResearch Labs, Inc. (West grove, PA). Hyperfilm ECL was purchased from Amersham Biosciences, Inc. (Piscataway, NJ). The Biovision Caspase-3 Fluorometric Assay Kit was purchased from Biovision, Inc. (Mountain View, CA).
Cell Line Maintenance
The porcine renal proximal cell line (LLC-PK1 cells, American Type Culture Collection, Rockville, MD) was maintained in 95% air/5% CO2 environment at 37°C in Medium 199 with 3% fetal bovine serum. The cells were split 1:5, and passage number was limited to 20 passages.
MTT Cell Viability Assay
Cytotoxicity was determined by the microtiter MTT assay (Alley et al., 1988). MTT is a yellow water-soluble tetrazolium dye that is reduced by live cells to a water-insoluble purple formazan. Cells were plated at a density of 2.5 × 105 cells/ml in 96-well microtiter format and allowed to reach approximately 80% confluence. Cells were then treated for 24 and 48 h with 4–1000nM of CdSe QD or InGaP QD, diluted directly in media, or media control. At each time point, plates were centrifuged at 700 × g for 3 min, media was removed, and 200 μl of fresh media and 50 μl of MTT solution (5 mg/ml in PBS, pH 7.4) were added to all wells. The plates were covered in aluminum foil and incubated at 37°C for 4 h. Following incubation, plates were again centrifuged at 700 × g for 3 min, MTT incubation media was removed, and 200 μl of dimethyl sulfoxide and 25 μl of glycine buffer (0.1M glycine, 0.1M NaCl, pH 10.5) were added to each well. The plates were vortexed, and the absorbance was read at 570 nm on a microplate spectrophotometer using a reference wavelength of 680 nm. Viability was expressed as a percentage of the media-treated control.
Caspase 3 Activity Assay
The Biovision Caspase 3 Fluorometric Assay Kit was used for caspase 3 activity measurement. LLC-PK1 cells were plated at a density of 2.5 × 105 cells/ml in a six-well plate (5 × 105 cells per well) format. Cells were treated for 24 and 48 h with 10nM CdSe QD, 100nM InGaP QD, 50μM cisplatin positive control, or media negative control. Following treatment, plates were washed with 1 ml of room temperature PBS, followed by addition of 200 μl ice-cold lysis buffer. The wells were then scraped, and the cell suspension transferred to 0.6 ml eppendorf tubes. The cell suspensions were incubated on ice for 10 min, centrifuged at 8000 × g for 5 min, and the resulting cell lysate supernatant was transferred to a new eppendorf tube. A 50-μl aliquot of the cell lysate was then transferred to a 96-well plate. To each sample well, 50 μl of 2× reaction buffer (with DTT) and 5 μl of DEVD-AFC substrate (50μM final concentration) was added. The plate was gently vortexed on an orbital shaker and incubated at 37°C for 1–2 h. Following incubation, fluorescence was measured at the excitation (400 nm) and emission (505 nm) wavelengths on a microtiter plate reader. The remaining cell lysate was used for protein determination by the BCA method. Data are presented as a percentage of the media-treated control caspase 3 activtiy normalized to total cellular protein.
Confocal Microscopy Images
LLC-PK1 cells were seeded in eight-well glass-bottomed chambers at a density of 125,000 cells/ml. Cells were preincubated for 24 h prior to addition of test sample, reaching an approximate confluence of 80%. Cells were incubated with treatment solutions, containing 10μM CdSe QD or 20μM InGaP QD, diluted directly in media, or media control for 30 min, followed by a 15-min incubation with 5 μg/ml Hoechst 33342 trihydrochloride trihydrate nuclear stain in media. Cells were washed two times with media to remove any cell-associated dye. Images were acquired using confocal laser scanning microscope LSM 510 Zeiss equipped with an Axiovert 200 inverted microscope (Carl Zeiss, Thornwood, NY) and a 63× 1.3 NA plan-Neofluar oil differential interference contrast objective lens. The QD and Hoechst nuclear stain were excited with Argon laser lines at 488 and 405 nm, respectively, by using a dichroic beam splitter (HFT 405/488). Emissions were collected with a bandpass filter of 420–480 nm for Hoechst staining and longpass filter of 615 nm for the QD species. The same microscope settings (laser power, filters, detector gain, amplification gain) were used for treated and control samples.
Reduced Glutathione Assay
The amount of reduced glutathione in the cell lysates was determined by the kinetic method of Shaik and Mehvar (2006), with adaptation for cell culture and protein normalization. This method is based on reduction of 5-5′-dithiobis(2-nitrobenzoic acid) (DTNB) by reduced glutathione (GSH) to form the colored product 2-nitro-5-thiobenzoic acid, which is measured at 415 nm. Oxidized glutathione (GSSG) is then reduced by glutathione reductase to form GSH, allowing another cycle of DTNB reduction. Preincubation of the samples with the thiol-masking agent 1-Methyl-4-vinyl-pyridinium (M4VP) prevents measurement of the GSH, so that only GSSG is measured. GSH in samples is quantified by comparing the rates of 2-nitro-5-thiobenzoic acid production in the presence or absence of M4VP to that of a glutathione standard curve.
LLC-PK1 cells were plated at a density of 1.5 × 106 cells per well in 35-mm2 six-well plates and grown to approximately 80% confluence prior to treatment. Cells were treated with 10nM CdSe QD, 100nM InGaP QD, positive control (0.1mM diethyl maleate) or media for 3, 6, or 24 h in the dark. Following treatment, cells were washed with 1 ml of room temperature PBS. Following washing, cells were scraped into 100 μl of ice-cold 5% 5-sulfosalicylic acid (SSA). Cells were then incubated for 10 min on ice in 0.6-ml eppendorf tubes and centrifuged at 8000 × g for 5 min. The supernatant was used for glutathione determinations, while the pellet was retained for determination of cellular protein by the Bradford assay. For total glutathione (GSH + GSSG), 5 μl of supernatant was diluted 1:2 with 5% SSA, then further diluted to 1:2 with 400mM sodium carbonate, and then finally diluted 1:8 with phosphate buffer (100mM Na3PO4–1mM EDTA buffer, pH 7.4), (total dilution 1:32). For GSSG determination, 5 μl of supernatant was diluted 1:2 with 5 μl of 0.5mM M4VP-masking reagent in 5% SSA, then further diluted to 1:2 with 400mM sodium carbonate (total dilution 1:4). Assay samples were then incubated for 5 min at room temperature and transferred to the 96-well microtiter plate. GSSG standards (20 μl) (5, 4, 3, 2, 1, and 0.5 ng/μl) and diluted samples (20 μl) were added to 170 μl of reaction mixture, containing 1.9 units/ml glutathione reductase and 0.4mM NADPH in phosphate buffer, and the plate was gently mixed on a rotary shaker. To this reaction mixture, 10 μl of 4.5mM DTNB substrate solution in phosphate buffer was added, the plate was gently mixed and incubated for 5 min at room temperature. The absorbance was read at 415 nm on a microplate spectrophotometer every 5 min for 25 min at room temperature. GSH concentration was determined by subtraction of GSSG from total glutathione concentrations and was normalized to total protein in the cell pellet, determined by the Bradford method.
Lipid Peroxidation Assay
Lipid peroxides in media were determined by the thiobarbituric reactive substances (TBARS) assay, adapted from Wey et al. (1993) for use with a microtiter plate. The TBARS assay measures lipid hydroperoxides and aldehydes, such as malondialdehyde (MDA), in the cell lysate. MDA combines with thiobarbituric acid (TBA) in a 1:2 ratio to form a fluorescent adduct that is measured at its excitation (521 nm) and emission (552 nm) wavelengths.
LLC-PK1 cells were plated at a density of 1.5 × 106 cells per well in 35-mm2 six-well plates and grown to approximately 80% confluence prior to treatment. Cells were treated with 10nM CdSe QD in media, 100nM InGaP QD in media, 5mM diethyl maleate in media (positive control), or blank media (negative control) for 3, 6, or 24 h in the dark. Following the incubation period, the cell culture media was removed and used for lipid peroxide determination. The wells were washed with ice-cold PBS, then the cells were scraped into 2 ml of 2.5% TCA and transferred to a collection tube. The collection tube was then centrifuged at 13,000 g for 2 min, and the resulting pellet was retained for determination of total cellular protein by Bradford Assay. A total of 500 μl of media or MDA standard (0.125–4 nmoles/ml in ddw) was added in duplicate to 400 μl 15% TCA and 800 μl of 0.67% TBA/0.01% BHT. The mixture was vortexed and then heated for 20 min in a 95°C water bath. The mixture was then allowed to cool to room temperature, 3 ml of butanol was added, the phases were gently mixed, and 200 μl of the butanol phase was transferred to a 96-well plate. The fluorescent TBA adducts were then measured at their excitation (521 nm) and emission (552 nm) wavelengths on a microplate spectrophotometer. The quantity of lipid peroxide in the sample wells was determined by comparison to the MDA standard curve and expressed as MDA equivalents normalized to total cellular protein.
Protein Determination
Bradford assay.
Cellular protein was determined using the Quick Start Bradford Dye Reagent, 1× kit from Bio-Rad Laboratories, Inc. Cellular protein pellets from the reduced glutathione and lipid peroxidation assays were resuspended in 0.5 ml of 0.05N NaOH. For protein quantitation, a BSA standard curve from 0.125 to 1.0 mg/ml was prepared in 0.05N NaOH. A 5-μl sample of the BSA standard, cellular protein sample, or 0.05N NaOH blank was added to wells of a 96-well microtiter plate in duplicate. Next, 250 μl of 1× Bradford dye reagent was added to each well, the plate was gently vortexed using an orbital shaker, then incubated at room temperature for 30 min. Following incubation, the plate was read at 595 nm on a microplate spectrophotometer.
BCA assay.
Cell lysate protein concentrations for LC3 western blot analysis and caspase 3 activity assay were determined using the Pierce BCA protein assay. The working reagent was prepared according to product instructions by mixing 25 parts of Micro BCA™ Reagent MA and 24 parts Reagent MB with one part of Reagent MC (25:24:1, Reagent MA:MB:MC). The standard curves for the cell lysates were prepared in their respective cell extraction buffers using BSA, from 0.5 to 200 μg/ml. A 150-μl sample of each standard, unknown, or extraction buffer blank was transferred to the microplate wells in duplicate. To these sample wells, 150 μl of the working reagent was added, and the plate was gently mixed on an orbital shaker for 30 s. The plate was then covered and incubated at 37°C for 2 h. Following incubation, the plate was allowed to cool to room temperature, and the absorbance was measured at 562 nm on microplate spectrophotometer.
Metallothionein Induction Study
LLC-PK1 cells were plated at a density of 200,000 cells/ml and grown to approximately 80% confluency in triplicate on six-well plates. LLC-PK1 cells were incubated for 8 and 24 h at 37°C with either CdSe QD at 2 or 10nM, InGaP QD at 20 or 100nM, media negative control, or 10μM zinc sulfate positive control. Media was removed at the end of the incubation period, and RNA protect cell reagent (Qiagen, Valencia, CA) was added for immediate stabilization of RNA. RNA was extracted from cell cultures using the RNeasy Mini Kit (Qiagen). The quality of total RNA was evaluated in representative samples subjected to agarose gel electrophoresis and stained with SYBR Green II RNA Gel Stain (Molecular Probes/Invitrogen Corporation) to visualize the RNA 28S and 18S subunits by band size discrimination under UV transillumination. RNA was quantified and characterized for purity by UV spectrophotometric analysis of the A260 and A260/280 ratio. Reagents for cDNA synthesis (TaqMan Reverse Transcription Reagents) were obtained from Applied Biosystems, Inc. (ABI). Reverse transcription was carried out in a final volume of 50 μl according to the manufacturers' instructions for a two-step RT-PCR of 500 ng of total RNA. Reagents for PCR (FastStart Taq Polymerase) were purchased from Roche Diagnostics. RT-PCR was performed on the Bio-Rad iCycler. Sequences were selected for optimum melting temperatures (Tm) and negligible secondary structure properties using PrimerQuest software from Integrated DNA Technologies (IDT). Basic Local Alignment Search Tool (BLAST) searches were conducted to confirm gene specificity of the primers and probes and absence of genomic DNA amplification. Synthesis of the metallothionein-IA (MT-IA) primers and probes were performed at IDT. Specific porcine primer and probe sequences: MT-IA forward primer, 5′-CACGTGCAAAGCCTGCAGAT-3′; MT-IA reverse primer, 5′-CCCTTTGCAGATGCAGCCC-3′. Expression of MT-IA was determined by multiplex real-time PCR utilizing 18S rRNA as internal control. Relative quantification of gene expression was calculated by the ΔΔCT method.
LC3 Immunoblot
LLC-PK1 cells were treated in T-75 flasks with positive control (Hank's balanced salt solution with calcium and magnesium), negative control (medium 199 with 3% fetal bovine serum), 10nM CdSe QD, or 100nM InGaP QD, for 6 h. Cells were then washed three times with ice-cold PBS, scraped into 1-ml ice-cold PBS, transferred to 15 ml conical tubes, and centrifuged at 700 × g for 3 min at 4°C. The supernatant was discarded, and cells were then lysed with 200 μl of Invitrogen Cell Extraction Buffer containing protease inhibitors. Lysed cells were placed on ice for 30 min, with vortexing every 10 min. The resulting lysate was centrifuged at 8000 × g for 5 min at 4°C, and the clear cell lysate was aliquoted into clean microcentrifuge tubes. The lysate samples were used immediately or stored at − 80°C until use. The protein content in the cell lysate samples was determined by the BCA protein assay. The cell lystate samples were diluted to 0.8 mg/ml, and 10 μl of 4× NuPAGE buffer was added to 30 μl of the diluted sample. The samples were then vortexed, heated at 95°C for 5 min, and centrifuged at 8000 × g for 30 min. The supernatants were transferred to clean tubes, and 20 μl of the duplicate test sample and control supernatants were loaded onto 4–20% tris-glycine gels. The gels were run at 125 V for approximately 2 h, rinsed with deionized water, and transferred to PVDF membranes overnight at 30 mA. The transfer membrane was washed three times with 50–100 ml of tris-buffered saline (TBS) (0.01% Tween-20) for approximately 15 min each and blocked with 50 ml StartingBlock blocking buffer (0.01% Tween-20) at room temperature for approximately 1 h. The membrane was then incubated with anti-LC3 primary antibody solution (1:200 dilution in 5 ml of the StartingBlock blocking buffer) for 2 h at room temperature using hybridization bags cut to size. The membrane was then washed twice with 50–100 ml of TBS (0.01% Tween-20) for 15 min each and incubated with the secondary donkey anti-mouse IgGHRP conjugate (1:50,000 dilution in StartingBlock blocking buffer) for 1 h at room temperature. The membrane was washed twice with 50–100 ml of TBS (0.01% Tween-20), for 15 min each, incubated with 3 ml ECL peroxidase substrate solution (1:1 peroxidase substrate to luminol enhancer solution) for approximately 1 min, and the immunoblot was developed for 8 min using Hyperfilm ECL. LC3-II densitometry was accomplished using Image J software (Rasband, W.S., ImageJ, US National Institutes of Health, Bethesda, MD, http://rsb.info.nih.gov/ij/, 1997–2007.).
Lysotracker Red Kinetics and Fluorescent Microscopy
The Lysotracker Red kinetic assay is based on the method of Rodriguez-Enriquez et al. (2006). Modifications include substitution of a 96-well plating format for the 48-well, altered Lysotracker Red dye incubation conditions, elimination of the cell fixation step, and addition of Celltracker Green CMFDA dye for normalization to viable cell number. Lysotracker Red DND-99 is a cationic fluorescent dye that preferentially accumulates in acidic lysosomal compartments. The amount of dye taken up by cells in culture can be used as an indicator of lysosome content. Celltracker Green is deacetylated within viable cells to a thiol-reactive dye that remains in the cytosol and is used to normalize the lysotracker signal to viable cells. The LLC-PK1 cells were plated at a density of 1 × 105 cells/ml in a 96-well plate (1 × 104 cells per well) format. Cells were treated for 6, 24, or 48 h with 0.00004–10nM CdSe QD, or 0.0004–100nM InGaP QD, or media negative control. Following treatment for the designated time period, plates were washed twice with cell culture media, 100 μl of 50nM Lysotracker Red/10μM Celltracker Green co-staining solution in phenol-free RPMI-1640 media was added, and the plate was incubated for 1 h at 37°C. The plates were then washed twice with 200 μl of phenol-free RPMI-1640 media and then left in 200 μl phenol-free RPMI-1640 media for reading. Plate fluorescence was read at 544 nm/590 nm for Lysotracker Red and 492 nm/517 nm Celltracker Green.
For Lysotracker Red fluorescent microscopy studies, LLC-PK1 cells were plated in six-well plates at a density of 250,000 cells/ml. Cells were preincubated for 24 h prior to addition of test sample, reaching an approximate confluence of 80%. Cells were then treated with 10nM CdSe QD, 100nM InGaP QD, or control media for 48 h followed by staining with Lysotracker Red (50nM in media) for 1 h and then with Hoechst nuclear stain (5 μg/ml in media) for 30 min. Cells were washed two times with PBS to remove any cell-associated dye and then left in PBS for fluorescent microscopy imaging. Images were acquired using a Nikon Eclipse TE 2000-U (Nikon) fluorescent microscope equipped with an RT-KE mono CCD camera and SPOT version 4.6 imaging software (Diagnostic Instruments, Inc.). Lysotracker Red and Hoechst images were collected with bandpass excitation/emission filter sets of 528–553/590–650 for the Lysotracker Red stain and 340–380/435–485 for the Hoechst stain. The same microscope settings were used for the treated and control samples. (Note: under these treatment and imaging conditions, background QD fluorescence that might interfere with the Lysotracker Red signal was not observed.)
Statistical and IC50 Analyses
Statistical differences (n ≥ 3, p ≤ 0.05) were determined by Student's t-test, or ANOVA and Dunnett's post hoc test using Statistica 7.1 (Tulsa, OK). Determination of IC50 was accomplished by nonlinear fit of cytotoxicity data to a sigmoidal Hill equation, E = E0 + (Emax – E0) × (Cγ/(Cγ + EC50γ)), using WinNonlin version 4.1 software (Pharsight Corp., Cary, NC).
RESULTS
Quantum dot characterization (for details see Supplementary Data).
The absorption spectrum for the CdSe QD gradually decreased from 300 nm to ∼530 nm, with a peak at 597 nm. The fluorescence spectrum of the CdSe QD had a narrow emission, with maximum emission wavelength at 610 nm with a full width at half maximum (FWHM) of 40 nm. The absorption spectrum for the InGaP QD gradually decreased in absorbance from 300 nm to ∼700 nm, with no distinct peak. The fluorescence spectrum of the InGaP QD had a maximum emission wavelength at 642 nm and FWHM of 94 nm, with a front shoulder at 600 nm. These spectra suggest that the InGaP QD has a broader distribution than the CdSe QD, while the shoulder in the InGaP QD spectrum suggests a portion of the population has a smaller size.
The CdSe QD had a narrow size distribution, with an average transmission electron microscopy (TEM)-measured diameter of 5.1 ± 0.7 nm and an average atomic force microscopy (AFM)-measured height of 6.9 ± 0.8 nm (Figs. 1A and 1B). In contrast, the InGaP ZnS-PEG QD had a relatively broad distribution, with an average TEM-measured diameter of 3.7 ± 1.4 nm, with particle diameters ranging from 1.1 to 9.5 nm, and an average AFM-measured height of 4.2±1.1 nm, with particle heights ranging from 1.8 to 8.4 nm (Figs. 1C and 1D). This broad distribution is consistent with the broad FWHM observed for the InGaP QD.
FIG. 1.
AFM measurement of CdSe and InGaP QDs. Representative AFM images of CdSe QD (A) and InGaP (C), with corresponding size distribution histogram (B) and (D), respectively. The average AFM-measured height of the CdSe QD was 6.9 ± 0.8 nm, and the average AFM-measured height of the InGaP QD was 4.2 ± 1.1 nm.
QD-induced cytotoxity, morphological changes, and internalization.
The CdSe and InGaP QDs were highly toxic to LLC-PK1 cells as measured by MTT assay (Figs. 2A and 2B). The CdSe QD was found to be ∼10-fold more toxic than the InGaP QD, with 48 h IC50 values of 10 and 100nM, respectively. Treatment of cells with 4–1000nM of either QD resulted in a dose-responsive loss of cell viability, as measured by the MTT assay. In agreement with this cytotoxicity data, a progressive decrease in cell density relative to media-treated controls was observed by light microscopy in LLC-PK1 cells treated with cytotoxic concentrations of either CdSe or InGaP QD (data not shown). Morphological changes associated with both CdSe and InGaP QD treatment included marked vacuolization. For the CdSe QD, the vacuolization was observed as early as 6 h posttreatment, peaked at 24 h, and appeared to regress by the 48 h time point. Vacuolization that was mild relative to that observed in the CdSe QD–treated cells was observed at the 24 and 48 h time points for the InGaP QD–treated cells. Short-term treatment of the LLC-PK1 cells with high concentrations of either QD also resulted in cell uptake observed by fluorescent confocal microscopy (Fig. 3). The red fluorescence in the confocal images shows internalization and cytosolic localization of the QD, while Hoechst nuclear staining is displayed as blue fluorescence.
FIG. 2.
Cytotoxicity. LLC-PK1 cells were treated for 24 and 48 h with 4–1000nM CdSe QD (A) or InGaP QD (B). Cytotoxicity was determined at each time point by the MTT assay. Data are presented as the percentage of control viability, mean ± SD, N = 3.
FIG. 3.
Confocal QD images—30 min. LLC-PK1 cells were treated for 30 min with control media (Control), 10μM CdSe QD (CdSe), or 20μM InGaP QD (InGaP). Quantum dot fluorescence is displayed in red, and Hoechst nuclear stain is shown in blue. Magnification 500×.
The QD-induced morphological changes identified by light microscopy were further characterized by TEM (for materials and methods see Supplemental Data). CdSe QD treatment resulted in development of double-membrane autophagic vacuoles several microns in diameter with internalized electron-dense cellular debris (Figs. 4 and 5). In comparison to CdSe QD treatment, InGaP QD treatment resulted in development of smaller multilamellar vesicles and darkly staining lysosomal remnants (Figs. 4 and 5). Ultrastructural changes resulting from CdSe and InGaP QD treatment in the LLC-PK1 cells were consistent with lysosomal disorders, including autophagy and phospholipidosis. Morphological changes characteristic of necrosis (e.g., pyknosis, cytoplasmic swelling) or apoptosis (e.g., perinuclear chromatin, membrane blebbing) were not observed.
FIG. 4.
Low magnification TEM photomicrographs—6 h. LLC-PK1 cells were treated for 6 h with media (Control), 10nM CdSe QD (CdSe), or 100nM InGaP QD (InGaP). White arrows show autophagic vacuoles, consisting of double-layered membranes containing cellular debris. Black arrows show lysosomal remnants. Magnification 5000×.
FIG. 5.
High magnification TEM photomicrographs—24 h. LLC-PK1 cells were treated for 24 h with 10nM mg CdSe QD (A) or 100nM InGaP QD (B). White arrows show autophagic vacuoles, consisting of double-layered membranes containing cellular debris. Black arrows show lysosomal remnants, consisting of multilamellar vacuoles and electron-dense deposits. Magnification 5000–20,000×.
Caspase 3 activity.
In this study, caspase 3 activation was used as a measure of apoptosis. Apoptosis in mammalian cells is initiated by activation of the caspase family of cysteine proteases. This assay quantified caspase 3 activation in vitro by measuring the cleavage of the caspase 3 substrate DEVD-7-amino-4-trifluoromethyl coumarin (AFC) to free AFC, which emits yellow-green fluorescence (λmax = 505 nm). This free AFC is measured using a microtiter plate reader. Cells were treated for 24 and 48 h with 10nM CdSe QD, 100nM InGaP QD, 50μM cisplatin positive control (24 h only), or media negative control. The 10nM CdSe and 100nM InGaP QD concentrations correspond to the respective IC50 values for each QD. The caspase 3 activity of the 50μM cisplatin-treated positive control group was 1641 ± 200% (mean ± SD) of control at 24 h (Fig. 6). Treatment of the LLC-PK1 cells with CdSe QD resulted in a significant decrease in caspase 3 activity compared to control at 24 and 48 h, 21 ± 2 and 29 ± 7% (mean ± SD) of control, respectively (Fig. 6). Caspase 3 activity for the InGaP QD treatment group was similar to control at 24 h and decreased to 65 ± 3% (mean ± SD) of control at 48 h (Fig. 6). The caspase 3 activities were normalized to total cell lysate protein; thus, caspase 3 activity reflects the average cellular activity and should not be affected by changes in cell density. The decreased caspase 3 activity observed for the QD-treated cells may reflect decreased numbers of cells entering the caspase-dependent apoptosis pathway in response to the treatment. In this regard, the decreases in caspase 3 activity are consistent with the temporal pattern of cytotoxicity for the QD species, suggesting that the mechanism of cell death was not caspase-dependent apoptosis.
FIG. 6.
Caspase 3 activity in QD-treated cells. LLC-PK1 cells were treated for 24 and 48 h with 10nM CdSe QD, 100nM InGaP QD, 50μM cisplatin positive control, or media negative control. Data are presented as % media control caspase 3 activity normalized to total cellular protein. Bars correspond to the mean ± SE of three individual samples.*p ≤ 0.05, significantly different than media control by ANOVA, and Dunnett's post hoc test. The 50μM cisplatin positive control value was 1600 ± 116% of media control (mean ± SE) and was significantly different than media control, p ≤ 0.05, by ANOVA, and Dunnett's post hoc test.
Oxidative stress.
Oxidative stress was assessed in LLC-PK1 cells by measurement of lipid peroxides in media and cellular reduced glutathione content, at 3, 6, and 24 h posttreatment with cytotoxic concentrations of QD. The 5mM diethyl maleate positive control groups at 6 h, were 8.13 ± 3.94 and 5.69 ± 2.16 nmoles MDA/μg total protein (mean ± SD) compared to media control values of 0.82 ± 0.06 and 0.97 ± 0.21 nmoles MDA/μg total protein (mean ± SD), for the CdSe and InGaP QD treatment experiments, respectively (Figs. 7A and 7B). Treatment of cells with 10nM CdSe QD resulted in a small, yet statistically significant, increase in lipid peroxides over media control values at 24 h (Fig. 7), 0.35 ± 0.04 versus 0.54 ± 0.05 nmoles MDA/μg total protein (mean ± SD), for control versus CdSe QD–treated, respectively (Fig. 7A). Differences in lipid peroxides in comparison to media control were not observed for the earlier 10nM CdSe QD treatment periods or for any of the 100nM InGaP QD treatment periods (Figs. 7A and 7B).
FIG. 7.
Lipid peroxidation in QD-treated cells. LLC-PK1 cells were treated for 3, 6, or 24 h with 10nM CdSe QD (A), 100nM InGaP QD (B), 5mM DEM positive control, or media negative control. Lipid peroxides in media were determined by TBARS assay and are expressed in MDA equivalents normalized to total protein. Data represent the mean ± SD, N = 3. * p ≤ 0.05, significantly different than media control by Student's t-test. The 5mM DEM positive control value was 8.13 ± 2.32 nmoles MDA equivalent/μg protein (mean ± SD) and was significantly different than media control, p ≤ 0.05, by ANOVA, and Dunnett's post hoc test.
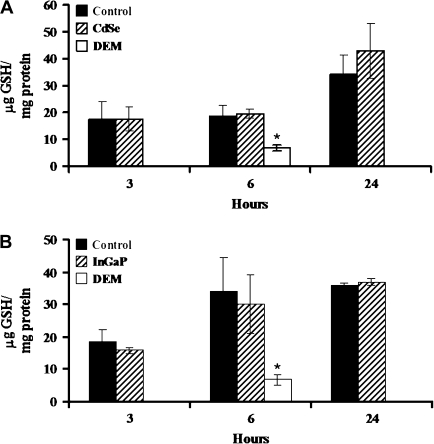
For the cellular reduced glutathione content analysis, 0.1mM diethyl maleate positive control values were 6.8 ± 1.0 and 7.0 ± 1.7 μg GSH/mg total protein (mean ± SD) compared to media control values of 18.6 ± 4.1 and 30.2 ± 10.4 μg GSH/mg total protein (mean ± SD), for the CdSe and InGaP QD treatment experiments, respectively (Figs. 8A and 8B). Treatment with 10nM CdSe QD or 100nM InGaP QD did not result in differences in reduced glutathione concentration in comparison to media control for the 3, 6 or 24 h treatment periods (Figs. 8A and 8B). The cytotoxicities of CdSe and InGaP QDs do not appear to be associated with increased generation of lipid peroxides or decreases in reduced glutathione content. These data do not support the involvement of metal-induced oxidative stress.
FIG. 8.
Reduced glutathione in QD-treated cells. LLC-PK1 cells were treated for 3, 6, or 24 h with 10nM CdSe QD (A), 100nM InGaP QD (B), 0.1mM DEM positive control, or media negative control. Reduced glutathione in media was determined by the DTNB assay and are expressed in μg glutathione/mg total protein. Data represent the mean ± SD, N = 3. * p ≤ 0.05 significantly different than media control by ANOVA, with Dunnett's post hoc test.
Metallothionein expression study.
Metallothionein gene expression was used as an indicator of metal-responsive gene induction. LLC-PK1 cells, in six-well plate format, were incubated for 8 and 24 h with either 2 or 10nM CdSe QD, 20 or 100nM InGaP QD, media negative control, or 10μM zinc sulfate positive control. The low and high concentrations of CdSe and InGaP QD correspond to nontoxic and toxic concentrations, respectively. Following the incubation periods, total RNA was prepared and MT-I mRNA expression was determined by RT-PCR analysis. Treatment of the LLC-PK1 cells with CdSe QD did not result in significant changes in MT-I mRNA levels for any treatment concentration or time point (Fig. 9). By contrast, treatment of LLC-PK cells with high concentrations of InGaP QD resulted in approximately 20- and 40-fold upregulation of MT-I mRNA in comparison to control at 8 and 24 h, respectively (Fig. 9). Treatment with similar dilutions of a 10-kDa cut-off filtrate of the InGaP QD solution did not result in any changes in MT-I transcription (data not shown), suggesting that residual metal in the suspending solution was not responsible for the observed changes in gene regulation resulting from treatment with the InGaP QD.
FIG. 9.
MT-IA gene regulation. LLC-PK1 cells were treated for 8 or 24 h with 2nM CdSe QD (CdSe-L), 10nM CdSe QD (CdSe-H), 20nM InGaP QD (InGaP-L), 100nM InGaP QD (InGaP-H), media negative control (Neg), or 10μM zinc sulfate positive control (Pos). For each treatment condition and time point, MT-IA mRNA expression relative to control media was determined. Data are expressed as the mean relative gene expression ± SD, of three independent determinations.
Autophagy induction.
Autophagy induction in QD-treated LLC-PK1 cells was determined by spectrophotometric measurement of Lysotracker Red dye uptake and LC3 western blot analysis. Lysotracker Red is a lysotropic dye that has been used previously to monitor autolysosome formation (Klionsky et al., 2007). For the spectrophotometry studies, cells were treated for 6, 24, or 48 h with 0.00004–10nM CdSe QD, or 0.0004–100nM InGaP QD, or media negative control in a 96-well microplate format. Following the treatment period, plates were stained with both Lysotracker Red and Celltracker Green, and fluorescence was determined using a microplate spectrophotometer. It should be noted that appropriate unstained control wells were included for all treatment concentrations and time points. Under these treatment conditions, QD fluorescence that might interfere with lysotracker signal was not observed (data not shown). The Lysotracker Red fluorescence was normalized to Celltracker Green fluorescence and expressed as percentage of control. The normalized Lysotracker Red fluorescence presented by treatment concentration and time point is displayed in Figure 10. The normalized Lysotracker Red fluorescence for the CdSe QD–treated cells was observed to increase in at the 24- and 48-h time points, with a maximum value of 519 ± 29% for the 2.5nM concentration at 24 h (Fig. 10A). Interestingly, at the highest CdSe QD concentration of 10nM, the Lysotracker Red fluorescence dropped back to control values, presumably due to rupture of the autolysosome subsequent to cytotoxicity. The normalized Lysotracker Red fluorescence for the InGaP QD–treated cells also increased at the 24- and 48-h time points, with a maximum value of 237 ± 56% (Fig. 10B).
FIG. 10.
Lysotracker kinetics. LLC-PK1 cells were treated for 6, 24, or 48 h with 0.00004–10nM CdSe QD or 0.0004–100nM InGaP QD. Following these incubation periods, cells were stained with Lysotracker Red and Celltracker Green. Data are presented as percent control Lysotacker Red fluorescence normalized to Celltracker Green fluorescence.
In agreement with this Lysotracker Red spectrophotometric data, treatment of cells with QD resulted in increased Lysotracker Red staining in comparison to media control, as determined by fluorescent microscopy (Fig. 11). Cells were treated for 48 h with 10nM CdSe QD, 100nM InGaP QD, or control media, followed by staining with Lysotracker Red and Hoechst nuclear stain. Appropriate unstained controls were included for all treatment groups, and QD fluorescence was undetectable under these treatment conditions (data not shown). The red fluorescence in the fluorescent microscopy images shows Lysotracker Red staining, while Hoechst nuclear staining is displayed as blue fluorescence (Fig. 11). Fluorescent microscopy images demonstrate increased Lysotracker Red staining of the CdSe- and InGaP QD–treated cells.
FIG. 11.
Lysotracker fluorescent microscopy images—48 h. LLC-PK1 cells were treated for 48 h with media (Control), 10nM CdSe QD (CdSe), or 100nM InGaP QD (InGaP), followed by staining with Lysotracker Red dye. Lysotracker stain is displayed in red and Hoescht stain is shown in blue. Magnification 100×.
The microtubule associated protein light chain 3 (MAP LC3)-II is an established marker of autophagy induction (Kabeya et al., 2000). This marker is a lipidated form of the cytosolic protein MAP LC3-I that is incorporated into the early autophagsome. In order to determine if Lysotracker Red staining corresponded with increased autophagy, cellular lysate from cells treated with QDs were analyzed by LC3 western blot. LLC-PK1 cells were treated in T-75 flasks with positive control Hank's balanced salt solution, negative control media, 10nM CdSe QD, or 100nM InGaP QD, for 6 h. Cell lysates were prepared from these treated cells and subjected to LC3 western blot (Fig. 12). The LC3 western shows the slower migrating LC3-I and the faster migrating LC3-II forms of the MAP protein (Fig. 12A). The densitometry values for the LC3-II form are displayed below the western (Fig. 12B). CdSe QD treatment resulted in greater LC3-II immunoblot staining than starvation positive control and negative media control treatments. The InGaP QD treatment resulted in LC3-II immunoblot staining of lesser intensity than the CdSe QD treatment and greater intensity than the negative control media treatment.
FIG. 12.
LC3 immunoblot. LLC-PK1 cells were treated for 6 h with media, starvation buffer, 10nM CdSe QD, or 100nM InGaP QD in duplicate. Cell lysate proteins were separated by SDS-PAGE, transferred to nitrocellulose membrane, and probed for LC3 immunoreactive proteins. LC3-I and -II protein bands are labeled on the immunoblot (A) and densitometry of the LC3-II band is displayed (B).
DISCUSSION
The CdSe QD and InGaP QDs were found to be highly toxic to the LLC-PK1 cells, with 48-h IC50 values of 10 and 100nM, respectively (Figs. 2, 4 and 5). The difference in the toxic potency of the two QD species could be due to physical characteristics or base chemical composition. Size-dependent differences in cytotoxicity have been observed previously for other QD species, with smaller QDs of identical composition being more toxic (Lovrić et al., 2005a; Zhang et al., 2007). In this study, the larger (5.1 ± 0.7 nm TEM, 6.9 ± 0.8 nm AFM) CdSe/ZnS core was more toxic than the smaller (3.7 ± 1.4 nm TEM, 4.2 ± 1.1 nm AFM) InGaP/ZnS core QD. Although the hydrodynamic size of these QDs could not be determined due to high laser absorption during dynamic light scattering, it is highly likely that this parameter was of similar value for the CdSe and InGaP QDs, as the same PEG chains were used in synthesis and the core sizes were comparable. Studies have suggested that certain QD-coating materials are cytotoxic and can be responsible for biological incompatibility (Duan and 2007; Shiohara et al., 2004). In the present case, identical surface lipids and PEG chains were incorporated into the resulting QDs, so toxicity differences resulting from the surface coatings are unlikely. However, due to differences in core composition, chemical and surface-effects unrelated to size are likely.
Several studies have suggested that a primary mechanism of QD cytotoxicity is release of toxic metals from the nanocrystal core, and point to the importance of the outermost capping/coating layers. Studies have correlated CdSe QD cytotoxicity in rat fibroblasts with the concentration of Cd-surface atoms, suggesting the involvement of free Cd ions (Kirchner et al., 2005). Capping of the CdSe QD with polymer, silica, or ZnS reduced cytotoxicity. In a study by Derfus et al. (2004), air and ultraviolet light exposure of uncapped, tri-n-octylphosphine oxide–coated CdSe QD resulted in enhanced cytotoxicity to primary rat hepatocytes that correlated with release of free Cd. Consistent with the loss of Cd from the CdSe core, air exposure also resulted in a blue-shift in the absorbance spectra and loss of fluorescence. Capping of the CdSe QD with ZnS partially prevented the release of free Cd and potentiation of cytotoxicity by either air exposure or ultraviolet irradiation. A study by Chan et al. (2006) also found that ZnS capping prevented generation of ROS and induction of apoptosis in human neuroblastoma cells by CdSe QD. As Cd has been shown to induce ROS generation and apoptosis (Oh and Lim, 2006), the investigators hypothesized that these responses could have been the result of exposure to free Cd, and ZnS capping may have prevented the Cd release. While these studies clearly support a role for free metal in CdSe QD cytotoxicity, other studies have been inconclusive. A recent study by Cho et al. (2007) found that intracellular free Cd ion concentrations did not correlate with CdTe QD cytotoxicity in MCF-7 cells. This lack of correlation suggested that Cd-dependent mechanisms alone were not responsible for the cytotoxicity and lead the researchers to speculate that other mechanisms involving the intact QD were also important.
In the present study, cytotoxic concentrations of CdSe QD did not result in upregulation of MT-1 mRNA (Fig. 9). This lack of a metal-associated stress gene response suggests that the ZnS cap and CdSe core remained intact or that free metals were below the threshold for MT-1 gene induction, which in LLC-PK1 cells has been shown to occur at low nontoxic concentrations of Zn and Cd (Alscher et al., 2005; Montine and Borch, 1990). This data is consistent with the fact that biochemical mechanisms previously associated with cadmium cytotoxicity in LLC-PK1 cells are absent in cells treated with cytotoxic concentrations of CdSe QD. For example, caspase 3-dependent apoptosis is associated with Cd-induced cytotoxicity in LLC-PK1 cells (Liu et al., 2007), and caspase 3 activation was not seen following CdSe QD treatment in the present study (Fig. 6). Caspase 3-depedent apoptosis has also been observed following treatment of human neuroblastoma cells with uncapped CdSe QD (Chan et al., 2006). In agreement with the results of the present study, ZnS capping prevented the caspase 3–dependent apoptosis. Lipid peroxidation is another Cd-associated mechanism that has been described in a variety of kidney cell lines treated with toxic concentrations of Cd (Pari et al., 2007), which was not observed for the CdSe QD–treated LLC-PK1 cells (Fig. 7). An additional oxidative stress response that has been characterized in Cd-treated LLC-PK1 cells is diminution of reduced glutathione levels (Wispriyono et al., 1998), which were unaffected by treatment with the CdSe QD, supporting the lack of free Cd involvement (Fig. 8). Together, these data do not support the primary involvement of free Cd in CdSe QD cytotoxicity.
In contrast to this lack of metal-associated responses for the CdSe QD, treatment of the LLC-PK1 cells with cytotoxic concentrations of InGaP QD resulted in a robust upregulation of MT-1 mRNA (Fig. 9). Pilot studies treating with similar dilutions of a 10-kDa cut-off filtrate of the InGaP QD solution, which removes the intact InGaP QD, did not result in any changes in MT-I transcription. This would suggest that residual dissociated metal ions in the suspending solution were not responsible for the increase in MT-1 gene expression and that the ZnS shell is either degraded in the media or within the cell, releasing free metal. We also cannot rule out the there also exists the possibility that the MT-1 induction was the result of cellular interactions with the uncoated metal core itself. Despite this upregulation of MT-I, evidence of metal-associated oxidative stress, such as loss of reduced glutathione and extensive lipid peroxidation, was not observed with InGaP QD treatment (Figs. 7 and 8). These results are in contrast to previous studies with other QD species that have demonstrated oxidative stress. For example, lipid peroxidation has been shown in mussels and neuroblastoma cells exposed the CdTe QD (Choi et al., 2007; Gagné et al., 2007). An additional finding was that InGaP QD cytotoxicity in LLC-PK1 cells did not involve caspase 3–dependent apoptosis that has been observed for other QD species (Fig. 6) (Chan et al., 2006).
A common finding in the CdSe QD– and InGaP QD–treated cells was evidence of autophagy induction. Autophagy is a lysosomal mechanism by which cellular organelles are recycled and long-lived proteins are degraded, complementing the breakdown of short-lived proteins by the proteosome (Yoshimori 2004). Autophagy has also been proposed as a pathway of programmed cell death (Type II) similar in homeostatic function to apoptosis (Type I) (Levine and Yuan, 2005). The TEM (Figs. 4 and 5), Lysotracker Red (Figs. 10 and 11), and MAP-LC3 (Fig. 12) data support an association between autophagy and CdSe and InGaP QD cytotoxicity. The TEM images depict an increase in autophagic vacuoles in the CdSe QD–treated cells (Fig. 5). Treatment of cells with the less toxic InGaP QD also resulted in increased lysosomal activity, though autophagosome formation was less pronounced (Fig. 5). Uptake of Lysotracker Red, a lysotropic dye that can be used to monitor autophagy (Klionsky et al., 2007), corresponded with the concentration-response and temporal pattern of the observed cytotoxicity for CdSe and InGaP QDs (Figs. 10 and 11). Treatment of cells with cytotoxic concentrations of CdSe and InGaP QD also resulted in increased MAP LC3-I to -II conversion (Fig. 12), a marker of autophagy (Mizushima and Yoshimori, 2007). Although autophagy has not been described previously as a response of LLC-PK1 to metal treatment, a recent study has demonstrated that toxic concentrations of cadmium can induce autophagy in human cord blood hemopoietic stem cells (Di Gioacchino et al., 2008). As discussed above, the absence of well-characterized Cd-associated responses in the present study casts doubt upon the involvement of free Cd in the observed autophagy development.
Recent studies have suggested that QDs may accumulate in organs, such as liver, kidney, and spleen, and remain for prolonged periods (Yang et al., 2007). Since this long-term exposure could result in chronic toxicities, it is very important to understand what the potential cellular responses to QDs are. The induction of autophagy by QDs may be a toxic mechanism responsible for the observed loss of cell viability, a protective cell stress response, or both. Preliminary attempts to prevent the QD-induced cell death by inhibition of autophagy were unsuccessful, as the inhibitors used (3-methyladenine, ammonium chloride, and bafilomycin A) were themselves highly toxic to the LLC-PK1 cells at effective concentrations (data not shown). Although the experiments in the present study do not demonstrate that autophagy is responsible for the loss of viability, the correlation of autophagy with the temporal pattern and concentration-response of QD-induced cytotoxicity is certainly supportive. It has been hypothesized by Zabinyk et al. (2007) that autophagy may be a common cellular response to nanomaterials. This theory certainly has merit, as many nanomaterials including protein-coated QDs, hydroxyl fullerene, carbon black, carbon nanotubes, and nanoscale neodymium oxide have all been shown to induce autophagy in vitro (Chen et al., 2005; Jia et al., 2005; Seleverstov et al., 2006; Yamawaki and Iwai, 2006a,b). In the case of nanocrystalline fullerene, inhibition of fullerene-induced autophagy by bafilomycin A1 treatment prevented cytotoxicity to human glioma cells (Harhaji et al., 2007), supporting a role for autophagy in nanoparticle-induced cell death.
The mechanism responsible for the induction of autophagy by CdSe and InGaP QD, and other nanomaterials, is presently unknown. Although overt oxidative stress was not observed in the present study, it is possible that there were localized areas of oxidative stress within the cell, as has been observed in other QD studies (Funnell and Maysinger, 2006). This may be significant, as oxidative stress has been shown to regulate autophagy (Scherz-Shouval et al., 2007). Alternatively, changes in gene regulation could be responsible for autophagy induction, as some QDs have been shown to localize to the nucleus (Lovrić et al., 2005a; Ryman-Rasmussen et al., 2007). A previous study in fibroblasts has shown that treatment with silica-coated CdSe/ZnS QDs can result in changes in expression of a variety of genes, including those involved in intracellular vesicle formation (Zhang et al., 2006). It is also quite possible that autophagy induction is the direct result of interaction with the endosome/lysosome itself, as QDs have been shown to localize to this compartment (Hanaki et al., 2003). The QD may be perceived by the cell as an endosomal pathogen and targeted to the autophagy pathway for destruction (Huang and Klionsky, 2007). Cytosolic QDs may interfere with the kinase-mediated autophagy regulatory cascades (Meijer and Codogno, 2006) or may be misidentified as an aggregation-prone protein, which are commonly degraded by autophagy (Williams et al., 2006). Future studies are needed to delineate the mechanisms responsible for and the consequences of nanomaterial-mediated autophagy induction.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
FUNDING
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health under contract N01-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US government.
Supplementary Material
Acknowledgments
The authors would like to thank Barry Neun for assistance with Lysotracker Red kinetics, Jaime Rodriguez for assistance with the LC3 Immunoblot, Timothy Potter for assistance with cell culture, David Parmiter for assistance with electron microscopy, and Evident Technologies, Inc. for supplying the quantum dots used in the studies
References
- Aldana J, Wang YA, Peng X. Photochemical instability of CdSe nanocrystals coated by hydrophilic thiols. J. Am. Chem. Soc. 2001;123:8844–8850. doi: 10.1021/ja016424q. [DOI] [PubMed] [Google Scholar]
- Aldana J, Lavelle N, Wang Y, Peng X. Size-dependent dissociation pH of thiolate ligands from cadmium chalcogenide nanocrystals. J. Am. Chem. Soc. 2005;127:2496–2504. doi: 10.1021/ja047000+. [DOI] [PubMed] [Google Scholar]
- Alley MC, Scudiero DA, Monks A, Hursey ML, Czerwinski MJ, Fine DL, Abbott BJ, Mayo JG, Shoemaker RH, Boyd MR. Feasibility of drug screening with panels of human tumor cell lines using a microculture tetrazolium assay. Cancer Res. 1988;48:589–601. [PubMed] [Google Scholar]
- Alscher DM, Braun N, Biegger D, Stuelten C, Gawronski K, Mürdter TE, Kuhlmann U, Fritz P. Induction of metallothionein in proximal tubular cells by zinc and its potential as an endogenous antioxidant. Kidney Blood Press. Res. 2005;28:127–133. doi: 10.1159/000084921. [DOI] [PubMed] [Google Scholar]
- Ballou B, Ernst LA, Andreko S, Harper T, Fitzpatrick JA, Waggoner AS, Bruchez MP. Sentinel lymph node imaging using quantum dots in mouse tumor models. Bioconjug. Chem. 2007;18:389–396. doi: 10.1021/bc060261j. [DOI] [PubMed] [Google Scholar]
- Bentzen EL, Tomlinson ID, Mason J, Gresch P, Warnement MR, Wright D, Sanders-Bush E, Blakely R, Rosenthal SJ. Surface modification to reduce nonspecific binding of quantum dots in live cell assays. Bioconjug. Chem. 2005;16:1488–1494. doi: 10.1021/bc0502006. [DOI] [PubMed] [Google Scholar]
- Bruchez M, Moronne M, Gin P, Weiss S, Alivisatos AP. Semiconductor nanocrystals as fluorescent biological labels. Science. 1998;281:2013–2016. doi: 10.1126/science.281.5385.2013. [DOI] [PubMed] [Google Scholar]
- Chen Y, Yang L, Feng C, Wen LP. Nano neodymium oxide induces massive vacuolization and autophagic cell death in non-small cell lung cancer NCI-H460 cells. Biochem. Biophys. Res. Commun. 2005;337:52–60. doi: 10.1016/j.bbrc.2005.09.018. [DOI] [PubMed] [Google Scholar]
- Chan WH, Shiao NH, Lu PZ. CdSe quantum dots induce apoptosis in human neuroblastoma cells via mitochondrial-dependent pathways and inhibition of survival signals. Toxicol Lett. 2006;167:191–200. doi: 10.1016/j.toxlet.2006.09.007. [DOI] [PubMed] [Google Scholar]
- Cho SJ, Maysinger D, Jain M, Röder B, Hackbarth S, Winnik FM. Long-term exposure to CdTe quantum dots causes functional impairments in live cells. Langmuir. 2007;23:1974–1980. doi: 10.1021/la060093j. [DOI] [PubMed] [Google Scholar]
- Choi AO, Cho SJ, Desbarats J, Lovrić J, Maysinger D. Quantum dot-induced cell death involves Fas upregulation and lipid peroxidation in human neuroblastoma cells. J. Nanobiotechnol. 2007;2007(5):1–13. doi: 10.1186/1477-3155-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derfus AM, Chan WCW, Bhatia SN. Probing the cytotoxicity of semiconductor quantum dots. Nano Lett. 2004;4:11–18. doi: 10.1021/nl0347334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derfus AM, Chen AA, Min DH, Ruoslahti E, Bhatia SN. Targeted quantum dot conjugates for siRNA delivery. Bioconjug. Chem. 2007;18:1391–1396. doi: 10.1021/bc060367e. [DOI] [PubMed] [Google Scholar]
- Duan H, Nie S. Cell-penetrating quantum dots based on multivalent and endosome-disrupting surface coatings. J. Am. Chem. Soc. 2007;129:3333–3338. doi: 10.1021/ja068158s. [DOI] [PubMed] [Google Scholar]
- Funnell WR, Maysinger D. Three-dimensional reconstruction of cell nuclei, internalized quantum dots and sites of lipid peroxidation. J. Nanobiotechnol. 2006;4:1–19. doi: 10.1186/1477-3155-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagné F, Auclair J, Turcotte P, Fournier M, Gagnon C, Sauvé S, Blaise C. Ecotoxicity of CdTe quantum dots to freshwater mussels: impacts on immune system, oxidative stress and genotoxicity. Aquat. Toxicol. 2007;86:333–340. doi: 10.1016/j.aquatox.2007.11.013. [DOI] [PubMed] [Google Scholar]
- Gennari A, Cortese E, Boveri M, Casado J, Prieto P. Sensitive endpoints for evaluating cadmium-induced acute toxicity in LLC-PK1 cells. Toxicology. 2003;183:211–220. doi: 10.1016/s0300-483x(02)00546-2. [DOI] [PubMed] [Google Scholar]
- Di Gioacchino M, Petrarca C, Perrone A, Farina M, Sabbioni E, Hartung T, Martino S, Esposito DL, Lotti LV, Mariani-Costantini R. Autophagy as an ultrastructural marker of heavy metal toxicity in human cord blood hematopoietic stem cells. Sci. Total Environ. 2008;392:39250–39258. doi: 10.1016/j.scitotenv.2007.11.009. [DOI] [PubMed] [Google Scholar]
- Hanaki K, Momo A, Oku T, Komoto A, Maenosono S, Yamaguchi Y, Yamamoto K. Semiconductor quantum dot/albumin complex is a long-life and highly photostable endosome marker. Biochem. Biophys. Res. Commun. 2003;302:496–501. doi: 10.1016/s0006-291x(03)00211-0. [DOI] [PubMed] [Google Scholar]
- Harhaji L, Isakovic A, Raicevic N, Markovic Z, Todorovic-Markovic B, Nikolic N, Vranjes-Djuric S, Markovic I, Trajkovic V. Multiple mechanisms underlying the anticancer action of nanocrystalline fullerene. Eur. J. Pharmacol. 2007;568:89–98. doi: 10.1016/j.ejphar.2007.04.041. [DOI] [PubMed] [Google Scholar]
- Huang J, Klionsky DJ. Autophagy and human disease. Cell Cycle. 2007;6:1837–1849. doi: 10.4161/cc.6.15.4511. [DOI] [PubMed] [Google Scholar]
- Ipe BI, Lehnig M, Niemeyer CM. On the generation of free radical species from quantum dots. Small. 2005;1:706–709. doi: 10.1002/smll.200500105. [DOI] [PubMed] [Google Scholar]
- Jia G, Wang H, Yan L, Wang X, Pei R, Yan T, Zhao Y, Guo X. Cytotoxicity of carbon nanomaterials: single-wall nanotube, multi-wall nanotube, and fullerene. Environ. Sci. Technol. 2005;39:1378–1383. doi: 10.1021/es048729l. [DOI] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchner C, Liedl T, Kudera S, Pellegrino T, Javier AM, Gaub HE, Stolzle S, Fertig N, Parak WJ. Cytotoxicity of colloidal CdSe and CdSe/ZnS nanoparticles. Nano Lett. 2005;5:331–338. doi: 10.1021/nl047996m. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Cuervo AM, Seglen PO. Methods for monitoring autophagy from yeast to human. Autophagy. 2007;2007(3):181–206. doi: 10.4161/auto.3678. [DOI] [PubMed] [Google Scholar]
- Levine B, Yuan J. Autophagy in cell death: an innocent convict? J. Clin. Invest. 2005;115:2679–2688. doi: 10.1172/JCI26390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zhang SP, Cai YQ. Cytoprotective effects of selenium on cadmium-induced LLC-PK1 cells apoptosis by activating JNK pathway. Toxicol. In Vitro. 2007;21:677–684. doi: 10.1016/j.tiv.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Lovrić J, Bazzi HS, Cuie Y, Fortin GR, Winnik FM, Maysinger D. Differences in subcellular distribution and toxicity of green and red emitting CdTe quantum dots. J. Mol. Med. 2005a;83:377–385. doi: 10.1007/s00109-004-0629-x. [DOI] [PubMed] [Google Scholar]
- Lovrić J, Cho SJ, Winnik FM, Maysinger D. Unmodified cadmium telluride quantum dots induce reactive oxygen species formation leading to multiple organelle damage and cell death. Chem. Biol. 2005b;12:1227–1234. doi: 10.1016/j.chembiol.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Meijer AJ, Codogno P. Signalling and autophagy regulation in health, aging and disease. Mol. Aspects Med. 2006;27:411–425. doi: 10.1016/j.mam.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T. How to interpret LC3 immunoblotting. Autophagy. 2007;3:542–545. doi: 10.4161/auto.4600. [DOI] [PubMed] [Google Scholar]
- Montine TJ, Borch RF. Role of endogenous sulfur-containing nucleophiles in an in vitro model of cis-diamminedichloroplatinum(II)-induced nephrotoxicity. Biochem. Pharmacol. 1990;39:1751–1757. doi: 10.1016/0006-2952(90)90121-z. [DOI] [PubMed] [Google Scholar]
- Oh SH, Lim SC. A rapid and transient ROS generation by cadmium triggers apoptosis via caspase-dependent pathway in HepG2 cells and this is inhibited through N-acetylcysteine-mediated catalase upregulation. Toxicol. Appl. Pharmacol. 2006;212:212–223. doi: 10.1016/j.taap.2005.07.018. [DOI] [PubMed] [Google Scholar]
- Pari L, Murugavel P, Sitasawad SL, Kumar KS. Cytoprotective and antioxidant role of diallyl tetrasulfide on cadmium induced renal injury: an in vivo and in vitro study. Life Sci. 2007;80:650–658. doi: 10.1016/j.lfs.2006.10.013. [DOI] [PubMed] [Google Scholar]
- Rhyner MN, Smith AM, Gao X, Mao H, Yang L, Nie S. Quantum dots and multifunctional nanoparticles: new contrast agents for tumor imaging. Nanomedicine. 2006;1:209–217. doi: 10.2217/17435889.1.2.209. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Enriquez S, Kim I, Currin RT, Lemasters JJ. Tracker dyes to probe mitochondrial autophagy (mitophagy) in rat hepatocytes. Autophagy. 2006;2:39–46. doi: 10.4161/auto.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryman-Rasmussen JP, Riviere JE, Monteiro-Riviere NA. Surface coatings determine cytotoxicity and irritation potential of quantum dot nanoparticles in epidermal keratinocytes. J. Invest. Dermatol. 2007;127:143–153. doi: 10.1038/sj.jid.5700508. [DOI] [PubMed] [Google Scholar]
- Seleverstov O, Zabirnyk O, Zscharnack M, Bulavina L, Nowicki M, Heinrich JM, Yezhelyev M, Emmrich F, O'Regan R, Bader A. Quantum dots for human mesenchymal stem cells labeling. A size-dependent autophagy activation. Nano Lett. 2006;6:2826–2832. doi: 10.1021/nl0619711. [DOI] [PubMed] [Google Scholar]
- Scherz-Shouval R, Shvets E, Elazar Z. Oxidation as a post-translational modification that regulates autophagy. Autophagy. 2007;3:371–373. doi: 10.4161/auto.4214. [DOI] [PubMed] [Google Scholar]
- Shaik IH, Mehvar R. Rapid determination of reduced and oxidized glutathione levels using a new thiol-masking reagent and the enzymatic recycling method: application to the rat liver and bile samples. Anal. Bioanal. Chem. 2006;385:105–113. doi: 10.1007/s00216-006-0375-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiohara A, Hoshino A, Hanaki K, Suzuki K, Yamamoto K. On the cyto-toxicity caused by quantum dots. Microbiol. Immunol. 2004;48:669–675. doi: 10.1111/j.1348-0421.2004.tb03478.x. [DOI] [PubMed] [Google Scholar]
- Uyeda HT, Medintz IL, Jaiswal JK, Simon SM, Mattoussi H. Synthesis of compact multidentate ligands to prepare stable hydrophilic quantum dot fluorophores. J. Am. Chem. Soc. 2005;127:3870–3878. doi: 10.1021/ja044031w. [DOI] [PubMed] [Google Scholar]
- Valko M, Morris H, Cronin MT. Metals, toxicity and oxidative stress. Curr. Med. Chem. 2005;12:1161–1208. doi: 10.2174/0929867053764635. [DOI] [PubMed] [Google Scholar]
- Wey HE, Pyron L, Woolery M. Essential fatty acid deficiency in cultured human keratinocytes attenuates toxicity due to lipid peroxidation. Toxicol. Appl. Pharmacol. 1993;120:72–79. doi: 10.1006/taap.1993.1088. [DOI] [PubMed] [Google Scholar]
- Williams A, Jahreiss L, Sarkar S, Saiki S, Menzies FM, Ravikumar B, Rubinsztein DC. Aggregate-prone proteins are cleared from the cytosol by autophagy: therapeutic implications. Curr. Top. Dev. Biol. 2006;76:89–101. doi: 10.1016/S0070-2153(06)76003-3. [DOI] [PubMed] [Google Scholar]
- Wispriyono B, Matsuoka M, Igisu H, Matsuno K. Protection from cadmium cytotoxicity by N-acetylcysteine in LLC-PK1 cells. J. Pharmacol. Exp. Ther. 1998;287:344–351. [PubMed] [Google Scholar]
- Yamawaki H, Iwai N. Cytotoxicity of water-soluble fullerene in vascular endothelial cells. Am. J. Physiol. Cell Physiol. 2006a;290:C1495–C1502. doi: 10.1152/ajpcell.00481.2005. [DOI] [PubMed] [Google Scholar]
- Yamawaki H, Iwai N. Mechanisms underlying nano-sized air-pollution-mediated progression of atherosclerosis: carbon black causes cytotoxic injury/inflammation and inhibits cell growth in vascular endothelial cells. Circ. J. 2006b;70:129–140. doi: 10.1253/circj.70.129. [DOI] [PubMed] [Google Scholar]
- Yang RS, Chang LW, Wu JP, Tsai MH, Wang HJ, Kuo YC, Yeh TK, Yang CS, Lin P. Persistent tissue kinetics and redistribution of nanoparticles, quantum dot 705, in mice: ICP-MS quantitative assessment. Environ. Health Perspect. 2007;115:1339–1343. doi: 10.1289/ehp.10290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimori T. Autophagy: a regulated bulk degradation process inside cells. Biochem. Biophys. Res. Commun. 2004;313:453–458. doi: 10.1016/j.bbrc.2003.07.023. [DOI] [PubMed] [Google Scholar]
- Zabirnyk O, Yezhelyev M, Seleverstov O. Nanoparticles as a novel class of autophagy activators. Autophagy. 2007;3:278–281. doi: 10.4161/auto.3916. [DOI] [PubMed] [Google Scholar]
- Zhang T, Stilwell JL, Gerion D, Ding L, Elboudwarej O, Cooke PA, Gray JW, Alivisatos AP, Chen FF. Cellular effect of high doses of silica-coated quantum dot profiled with high throughput gene expression analysis and high content cellomics measurements. Nano Lett. 2006;6:800–808. doi: 10.1021/nl0603350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen W, Zhang J, Liu J, Chen G, Pope C. In vitro and in vivo toxicity of CdTe nanoparticles. J. Nanosci. Nanotechnol. 2007;7:497–503. doi: 10.1166/jnn.2007.125. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.