Abstract
The aryl hydrocarbon receptor (AHR) is known for its role in the adaptive and toxic responses to a large number of environmental contaminants, as well as its role in hepatovascular development. The classical AHR pathway involves ligand binding, nuclear translocation, heterodimerization with the AHR nuclear translocator (ARNT), and binding of the heterodimer to dioxin response elements (DREs), thereby modulating the transcription of an array of genes. The AHR has also been implicated in signaling events independent of nuclear localization and DNA binding, and it has been suggested that such pathways may play important roles in the toxicity of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD). Here, we report the generation of a mouse model that expresses an AHR protein capable of ligand binding, interactions with chaperone proteins, functional heterodimerization with ARNT, and nuclear translocation, but is unable to bind DREs. Using this model, we provide evidence that DNA binding is required AHR-mediated liver development, as Ahrdbd/dbd mice exhibit a patent ductus venosus, similar to what is seen in Ahr−/− mice. Furthermore, Ahrdbd/dbd mice are resistant to TCDD-induced toxicity for all endpoints tested. These data suggest that DNA binding is necessary for AHR-mediated developmental and toxic signaling.
Keywords: aryl hydrocarbon receptor, dioxin, TCDD, ductus venosus
The aryl hydrocarbon receptor (AHR)1 is a basic helix-loop-helix (bHLH)-per-ARNT-sim (PAS) protein that mediates the toxic response to an array of lipophilic environmental toxicants, including 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD). Those responses include thymic involution, liver hypertrophy, tumor promotion, epithelial hyperplasia, and teratogenesis (Poland and Knutson, 1982). The generation of an Ahr-null allele in mice has also provided evidence that the receptor plays an important role in mammalian development (Gonzalez and Fernandez-Salguero, 1998; Schmidt et al., 1996). Characterization of the Ahr-null mouse revealed a transient microvesicular steatosis in perinatal hepatocytes, prolonged extramedullary hematopoiesis, and a reduced relative liver size throughout life. Recent evidence has shown that Ahr-null mice fail to resolve a fetal vascular structure, the ductus venosus (DV), which may be the underlying cause of liver atrophy. These outcomes are suggestive of a role for the AHR in vascular biology or hematopoiesis during mammalian development (Lahvis et al., 2000; Walisser et al., 2005).
In response to xenobiotic agonists, the AHR functions as a ligand-activated transcription factor. Upon binding agonists such as TCDD, the AHR translocates from the cytoplasm to the nucleus, where it dimerizes with another bHLH-PAS protein known as the AHR nuclear translocator (ARNT) (Hankinson, 1995). This heterodimeric complex recognizes dioxin response elements (DREs), which regulate the transcription of a battery of genes encoding xenobiotic metabolizing enzymes (XMEs). These XMEs include Phase I enzymes such as Cytochromes P450 1A1, 1A2, and 1B1, as well as the Phase II enzymes, GST-Ya, and UDPGT (reviewed in Hankinson, 1995; Schmidt et al., 1996).
Although the mechanism for AHR-mediated transcriptional activation of XMEs is well-established, it has been difficult to link specific target genes to most TCDD-induced toxic responses. Similarly, null alleles of three known transcriptional targets of AHR, Cytochromes P450 1a1, 1a2, and 1b1, have not been reported to possess any of the same phenotypes of Ahr−/− mice (Buters et al., 1999; Liang et al., 1996; Pineau et al., 1995). This latter observation suggests that none of the most responsive AHR target genes play an individual role in the developmental signaling of the AHR.
Our inability to link DRE-regulated genes to most aspects of TCDD toxicity or AHR developmental biology has led to the development of a number of models which propose that the AHR takes part in important signaling events that are independent of DRE binding or even ARNT dimerization. Included in this list of models is the hypothesis that the ligand-activated AHR signals through direct interactions with cellular proteins such as cSrc kinase, the retinoblastoma protein (Rb), and RelA (Blankenship and Matsumura, 1997; Enan and Matsumura, 1996; Ge and Elferink, 1998; Puga et al., 2000). Similarly, it has been proposed that TCDD toxicity may occur as a result of the activated AHR sequestering available ARNT in the cell. In in vitro and cell culture model systems, the capacity of an activated AHR to reduce ARNT participation in hypoxia signal transduction has been demonstrated and has also been challenged (Berghard et al., 1993; Chan et al., 1999; Gradin et al., 1996; Pollenz et al., 1999).
The various proposals suggesting that AHR may be involved in cellular signal transduction mechanisms independent of interactions with ARNT or DREs has led to a complicated picture of the mechanism of AHR-mediated development and TCDD toxicity. In an effort to test the role of DRE-independent signaling by the AHR in these processes, we have developed mouse models with deficiencies in specific signaling steps. We have shown previously that nuclear translocation of the AHR is required for normal liver development and TCDD-induced toxicity in mice (Bunger et al., 2003). Through the use of a similar gene-targeting approach, we have now generated a mouse line that expresses a mutant Ahr that is unable to bind DREs. We present evidence which suggests that the binding of AHR to DREs is required for developmental processes as well as AHR-mediated toxicity in vivo.
MATERIALS AND METHODS
Oligonucleotides and expression constructs.
Oligonucleotides were synthesized by Invitrogen (Carlsbad, CA) and are designated as follows:
OL659: 5′-ATCCAGAAGAGCTTATCAGTGGTTCTGC-3′
OL941: 5′-CTGAGGGGACGTTTTAATG-3′
OL942: 5′-AACATTTGCACTCATGGATAG-3′
OL1352: 5′-GGTACCTCTGAGTTCAAGTCTAGTCTG-3′
OL1353: 5′-GGTACCGCATGCTTACTAGTAGTTTTTCTAG-3′
OL1503:5′-GCCACCATGAGCAGCGGCGCCAACATCACCTATGCCAGCCGCAAGCGGCGCAAGCCGGTGCAGAAAACAGTAAAGCCCGGGCCCGCTGAA-3′
OL1793:5′-GTAAAGCCCGGGCCCGCTGAAGGAATTAAGTCAAATCCTTCTAAGCGACACAGAGGATCCGACCGGCTGAACACAGAGTTAGA-3′
OL2639:5′-ACTAGTCGACCTAACCCATTTGCTGTCCACCAGTCATGCTAGCCATACTCTGCACCTTGCTTAG-3′
PL65 and PL613 were described previously (Carver et al., 1998; Jain et al., 1994). To generate the pTgTAHRT7 (PL1550) construct, PL65 was used as a template for 22 rounds of PCR amplification using OL1503 (forward) and OL2639 (reverse). The oligo, OL1503, contains a consensus “Kozak start site” and a mutation creating an SrfI restriction site that replaces the isoleucine at position 25 (I25) with glycine in the AHR coding region. The oligonucleotide, OL2639, contains the region of AHR cDNA preceding the stop codon as well as a sequence for the T7-epitope and a translational stop. The pTgtAHRdbdT7 (PL1548) construct was generated by two-step PCR using PL65 as a template. In step 1, the forward oligo was OL1793, which contains the GGATCC insertion mutation, and the reverse oligo was OL2639. The product of the first step was then used as a template for 20 rounds of amplification using OL1503 and OL2639. Sequencing of all constructs was performed to ensure that no mutations were randomly generated. The plasmid, PL256, is a luciferase reporter driven by a DRE-containing promoter element from the upstream region of the CYP1A1 gene (DRE-luc) (Postlind et al., 1993).
Protein analysis.
All western blot analyses, gel-shift, and photoaffinity labeling experiments were performed essentially as described (Carber and Bradfield, 1997; Chan et al., 1999; Jain et al., 1994; Poland et al., 1991, 1986, 1994). In vitro protein expression was carried out using a transcription/translation system reticulocyte lysate system (Promega, Madison, WI). Microsomes were isolated from approximately 0.5 g of mouse liver which was homogenized in ice-cold MENG buffer (25mM 4-morpholinepropanesulfonic acid pH7.5, 0.025% wt/vol sodium azide, 1mM ethylene glycol bis(2-aminoethyl ether)tetraacetic acid, 10% glycerin vol/vol or glycerol) followed by two centrifugation steps at 10,000 × g and 100,000 × g. The microsomal pellet was resuspended in 250 μl of 15mM Tris-Cl pH8/250mM sucrose. Ethoxyresorufin O-deethylase (EROD) assays were performed in a 96-well format. In each well, 3 μl of 0.1mM ethoxyresorufin and 20 μl of 5mM NADPH were mixed with 5 μl of the total microsomal prep in 200 μl in MENG buffer. Following incubation at 25°C for 10 min, the production of hydroxyresorufin was measured using a fluorimeter (fMax, Molecular Devices, Sunnyvale, CA) at 510-nm excitation and 590-nm emission. Total protein concentrations were determined using the bicinchoninic acid assay (Pierce, Rockford, IL). Units are expressed as relative fluorescence/minute/mg protein (RFU/min/mg protein) as calculated using SoftMax Pro software (Molecular Devices).
Coimmunoprecipitation (Co-IP) experiments were performed by incubating approximately 10 fmol of reticulocyte lysate–expressed proteins with 5 μg antibody in 500 μl of cold MENG buffer supplemented with 15mM NaCl, 0.1mM dithiotreitol, and 0.1% NP-40. Bound protein-antibody complexes were precipitated with either protein A-sepharose (Sigma, St Louis, MO) or T7-antibody-coupled agarose (Novagen, La Jolla, CA) for 1.5 h at 4°C, washed four times with cold MENG buffer, eluted in 2× sodium dodecyl sulfate (SDS) sample buffer, and analyzed by polyacrylamide gel electrophoresis (PAGE).
Cell culture conditions and treatments.
Embryonic stem (ES) cells, designated GS-1, were purchased from Genome Systems (St Louis, MO). The ES cells were cultured on a confluent layer of mouse embryonic fibroblasts derived from PGK-NeoR transgenic mice (The Jackson Laboratory, Bar Harbor, ME) in Dulbecco's modified Eagle medium (DMEM)-high glucose supplemented with 20% fetal bovine serum (HyClone, Logan, UT), 0.1mM nonessential amino acids, 2mM L-glutamine, 10mM 4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES), 100 U/ml penicillin, 100 μg/ml streptomycin, and 1000 U/ml ESGRO (Invitrogen). To generate Ahr−/− fibroblasts, heterozygous Ahr−/− mice, which were previously backcrossed to C57BL/6J for 16 generations, were intercrossed to generate littermate +/+, +/−, and −/− littermate embryos. Following isolation of embryos from their yolk sacs, the heads and livers were removed by dissection. DNA was isolated from each individual embryo and was used for genotyping as described below. At passage 2, +/+ and −/− fibroblasts were placed on a 3T3 protocol and maintained on this protocol until passage 25 (Nilausen and Green, 1965). Cells were grown in DMEM-high glucose and supplemented with 10% fetal bovine serum, 0.1mM nonessential amino acids, 2mM L-glutamine, 10mM HEPES, 100 U/ml penicillin, and 100 μg/ml streptomycin. At passage 28, individual clones were isolated from each genotype, maintained in the same media and passaged regularly at subconfluence. Transient transfections were performed using Fugene-6 (Roche, Palo Alto, CA) with 1 μg total DNA and a 3:1 Fugene-6:DNA ratio. When TCDD was used, cells were allowed to recover for one day after transfection and 1nM TCDD in dimethyl sulfoxide (DMSO) was added directly to the media (at a final DMSO concentration of 0.1% vol/vol). Immunofluorescence was performed as described previously using a high-affinity rabbit polyclonal antibody raised against recombinant AHR (BEAR-3) and a flourescein isothiocyanate (FITC)–conjugated goat anti-rabbit secondary antibody (Jackson Immunochemicals, West Grove, PA) (Jain et al., 1994).
Mammalian 2-hybrid analysis.
The “bait” expression construct, PL283, contains a Gal DNA-binding domain fusion of ARNT, which is also deleted for the transactivation domain (Jain et al., 1994). The plasmid, PGL5 (Promega, Madison, WI), was used as a reporter and contains five Gal upstream activation sequences (UAS) upstream of an SV40 minimal promoter and the luciferase gene. A green fluorescence protein expression construct (Clontech, Mountain View, CA) was used as a control for transfection efficiency. Briefly, equal amounts of plasmids pTgTAHRdbd (PL1548) or pTgTAHRs (PL1550) were cotransfected with PL283 and the pGL5 reporter. These cells were treated with 1nM TCDD and luciferase assays were performed using The Luciferase Assay kit (Promega) and read on a luminometer.
Generation of Ahrdbd/dbd mice.
A 15-kb region of homology surrounding exon 2 of Ahr was isolated from a 129SvJ genomic library (Genome Systems) as described (Schmidt et al., 1993, 1996). A six nucleotide insertion (GAATTC) was introduced into exon 2 by megaprimer PCR using OL1793 and OL942. This product was used as a reverse megaprimer for PCR with OL659. An SphI fragment from this PCR product was used to replace exon 2 in an 8-kb BamHI genomic fragment. A 5.5-kb region containing the mutated exon 2 was amplified with OL1352 and OL1353 and cloned into the KpnI site of ploxPNT (Tybulewicz et al., 1991). A 7-kb SphI fragment from the 5′ region of exon 2 was cloned into the NotI/XhoI site of this construct to generate the final targeting construct, designated PL1238.
Ten micrograms of the targeting construct was electroporated into GS1 ES cells (Genome Systems) and selection was performed using 200 μg/ml G418 and 1mM Ganciclovir. Clones were screened by Southern blot on BamHI-digested genomic DNA using a probe 3′ to the end of the targeting construct (PL311). Correctly targeted clones were injected into 3.5-day postcoital C57BL/6J blastocysts, and the resulting chimeras were backcrossed to C57BL/6J to determine the contribution of the ES clones to the germline. Mice were genotyped using the PCR primers, OL941 and OL942. PCR was carried out for 40 cycles (95°C, 30″; 60°C, 30″; 72°C, 2′) in buffer containing 3.5mM MgCl2. A BamHI digest cuts the 380-bp PCR product from the targeted Ahrdbd allele into two fragments of 240 and 140 bp, which were detected on a 2% agarose gel. Removal of the neomycin cassetted inserted into the Ahr locus as part of the gene-targeting process was performed by breeding Ahrdbd/dbd mice at N6 to CMV-Cre/tg mice. The F2 generation was genotyped for the presence of the neomycin cassette by PCR. Animals where neomycin was successfully removed were then backcrossed three generations to C57BL/6 (Ahrdbd/dbd, Floxed).
Animals.
Animals were housed in a selective pathogen-free facility on corn cob bedding with food and water ad libitum according to the rules and guidelines set by the University of Wisconsin—Madison Animal Care and Use Committee. Where appropriate, animals were injected once i.p. with p-dioxane alone or with 100 μg/kg TCDD in p-dioxane. After 6 days, animals were weighed and sacrificed by cervical dislocation and organs were immediately removed and weighed. Tissues for histopathological analysis were fixed in 10% neutral-buffered formalin and embedded in paraffin wax. Five or 10-μm sections were stained with hematoxalin and eosin (H&E) or Oil Red-O and hematoxalin. For angiography, 1 ml of Omnipaque 300 (Nycomed, Inc., Princeton, NJ) was injected into the hepatic portal vein post mortem. Continuous X-ray images were obtained over a period of 10 s using an OEC 9800 Portable Vascular C-ARM (Medical Systems, Inc., Salt Lake City, UT). Patent DV was also scored by trypan blue perfusion. Statistical analyses were performed using Student's t-test. For induction of cleft palate, pregnant dams were i.p. injected with either DMSO or 128 μg TCDD/kg body weight on embryonic day 10 (ED10). Litters were scored for the presence of cleft palate and hydronephrosis on ED17–18.
RESULTS
Characterization of the AHRdbd Protein In Vitro
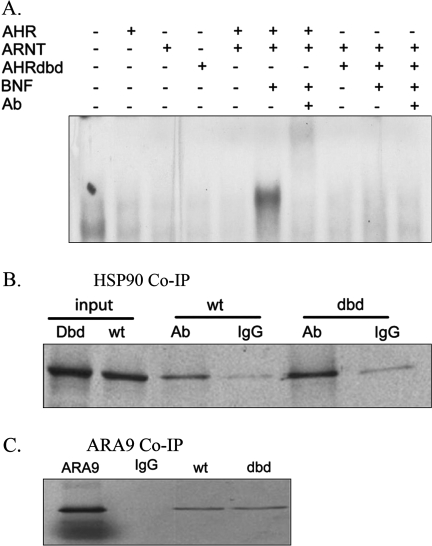
A mutant AHR cDNA, designated “AHRdbd,” was generated by the insertion of nucleotides GGTACC, coding for amino acids glycine and serine, between arginine-39 (R39) and aspartate-40 (D40) of the wild-type AHR cDNA (Fig. 1). Expression of the recombinant AHRdbd protein in rabbit reticulocyte lysate produced a protein approximately 95 kDa, in accordance with the known size of the wild-type protein. Photoaffinity labeling experiments indicated that the AHRdbd protein bound ligand with a capacity and affinity that was similar to its wild-type counterpart (data not shown). The DRE binding properties of AHRdbd were analyzed by a gel-shift protocol. Neither AHR nor AHRdbd proteins interacted significantly with a 32P-labeled DRE oligonucleotide in the absence of ARNT or in the presence of ARNT when an agonist was not present. The addition of the agonist, β-napthoflavone (BNF), induced formation of the AHR/ARNT/DRE complex, but not the AHRdbd/ARNT/DRE complex (Fig. 2A).
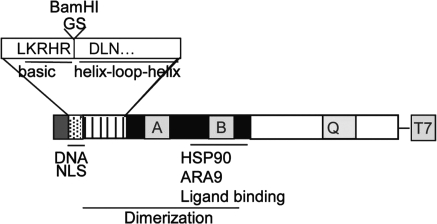
FIG. 1.
Schematic of the functional domains of the AHR protein. The enlarged portion depicts the GS insertion between the last residue of the basic domain and the first residue of the HLH domain, thereby introducing a BamHI restriction site.
FIG. 2.
Biochemical analysis of AHRdbd recombinant protein. (A) electromobility shift assay analysis of AHRdbd. AHR, ARNT, and AHRdbd proteins were expressed in reticulocyte lysate and equal quantities were incubated with a 32P-labeled, double-stranded oligo containing a single DRE consensus sequence. Shift of the AHR/ARNT heterodimer was induced by coincubation with 10μM BNF. An AHR-Ab was included to block the complex formation, controlling for specificity. (B,C) Co-IP of AHRdbd with HSP90. Wild-type or AHRdbd 35S-labeled in vitro–translated proteins were coincubated with reticulocyte lysate and HSP90-specific antibody (Ab) or preimmune IgG. Complexes were precipitated with Protein A-sepharose beads, separated on a 7.5% SDS-PAGE gel, and visualized with autoradiography. (D) Co-IP of AHRdbd with ARA9. T7-tagged AHR and AHRdbd were incubated with 35S-labeled ARA9. Complexes were precipitated using T7 antibody-coupled agarose beads and separated on a 7.5% SDS-PAGE gel.
Co-IP experiments were utilized to determine whether the AHRdbd protein retains the ability to interact with HSP90 and ARA9. An HSP90-specific antibody was capable of precipitating both 35S-labeled AHR and AHRdbd, and to the same degree (Fig. 2B). Furthermore, T7-peptide antibody-coupled agarose beads equally precipitated 35S-labeled ARA9 when incubated in the presence of T7-tagged AHR and T7-tagged AHRdbd (Fig. 2C). Together, these results indicate that AHRdbd is capable of interacting with ligand, HSP90, and ARA9 in a manner similar to the wild-type AHR, but is not capable of interacting with DREs.
Characterization of AHRdbd Signaling in Ahr−/− Fibroblasts
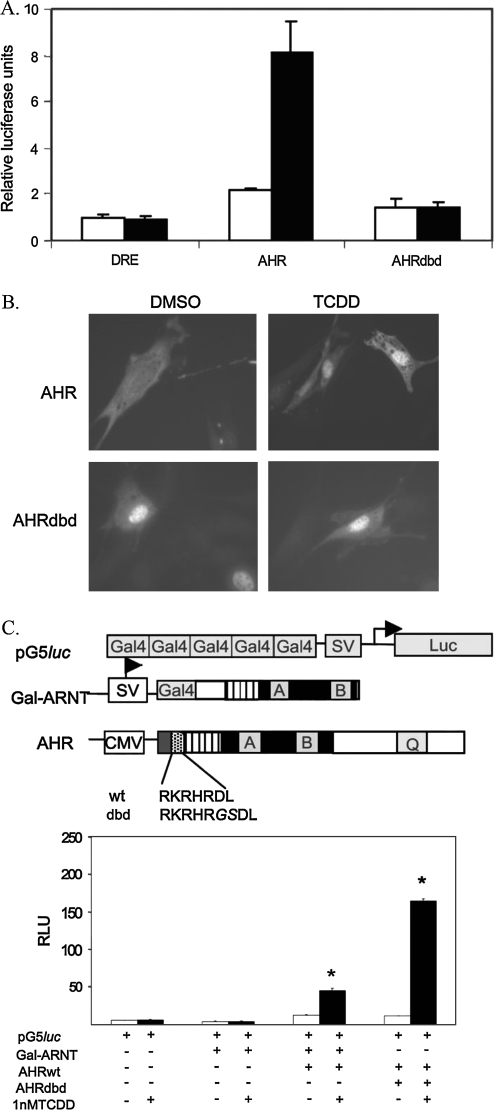
To determine whether the AHRdbd could signal effectively in cell culture, we performed transient transfections of AHR or AHRdbd with a DRE-driven luciferase reporter in immortalized Ahr−/− 3T3 fibroblasts. Upon transfection of wild-type AHR cDNA, luciferase activity increased relative to cells transfected with reporter alone. This response was enhanced 2.5-fold by exposure of the cells to 1nM TCDD. In comparison, luciferase activity in cells transfected with AHRdbd did not increase upon exposure of cells to TCDD (Fig. 3A).
FIG. 3.
Cellular characterization of the AHRdbd protein. (A) Luciferase assay for DRE-driven transcription. Ahr−/− 3T3 fibroblasts were transfected with equal amounts of DRE-Luc (PL256) and either AHR or AHRdbd recombinant cDNAs. Cells were then treated with 1nM TCDD (black bars) or 0.1% DMSO alone (white bars) for 24 h. Values represent relative luciferase units normalized to total protein levels. (B) Subcellular localization of AHRdbd. Indirect immunofluorescence was used to identify the subcellular localization of AHRdbd in Ahr−/− 3T3 fibroblasts transiently transfected with either Ahr+/+ or Ahrdbd/dbd. Prior to staining, nuclear translocation was induced by exposure of cells to 1nM TCDD for 2 h prior to staining. (C) Mammalian 2-hybrid analysis of AHRdbd interactions. The schematic diagram depicts the reporter construct (pG5luc), the “bait” construct (Gal-ARNT), and the “fish” construct (AHR), showing the amino acid sequence of the basic region in wild-type (wt) and AHRdbd (dbd) recombinant proteins. The two-hybrid analysis was carried out using equal amounts (0.33 μg) of transiently transfected Gal-ARNT and either wild-type AHR or AHRdbd, followed by incubation with 0.1% DMSO or 1nM TCDD. Values are expressed as relative luciferase units (*p < 0.001).
Indirect immunofluorescence analysis was used to determine the subcellular localization of AHRdbd. Ahr−/− 3T3 fibroblasts were transiently transfected with AHR or AHRdbd cDNAs and visualized using a BEAR3 primary antibody and a FITC-conjugated secondary antibody. The AHR was found to localize to the cytosol in untreated fibroblasts, but localized to the nucleus within 2 h of treatment with 1nM TCDD. Interestingly, the AHRdbd protein is constitutively nuclear in the absence of ligand, and localization was not altered by treatment with 1nM TCDD (Fig. 3B).
AHRdbd Interacts with ARNT in a Mammalian 2-Hybrid Assay
To determine whether AHRdbd functionally interacts with ARNT, we performed a mammalian two-hybrid analysis using Gal4-ARNT-ΔTAD as “bait” and the full-length AHRdbd as “fish.” Cotransfection of wild-type AHR with the Gal4-ARNT-ΔTAD along with a Gal4UAS-luciferase reporter showed a slight increase in RLU over reporter alone. Exposure to 1nM TCDD increased luciferase activity threefold. Cotransfection of the AHRdbd construct with the Gal4-ARNT-ΔTAD and reporter also slightly increased luciferase activity over cells with reporter alone and showed a 10-fold increase in luciferase activity after TCDD treatment (Fig. 3C).
Generation and Characterization of Ahrdbd/dbd Mice
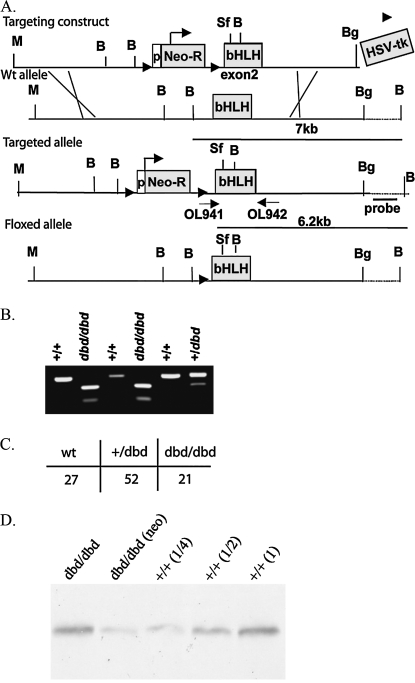
We used a megaprimer PCR approach to insert a GGATCC sequence (encoding Proline-Arginine) immediately downstream of the basic region of exon 2 in a 15-kb region of homologous genomic DNA derived from the Ahr locus (Fig. 4A). The final targeting construct, ploxPNT/AHRdbd, was electroporated into GS-1 ES cells (Genome Systems) and selected in both G418 and Ganciclovir (Roche). Double-selected clones (150 total) were screen by Southern blot and five correctly-targeted clones were identified. One clone gave rise to a chimera that transmitted the Ahrdbd allele to the germline. The resulting Ahrdbd/dbd Targeted allele mice were genotyped by PCR and restriction digest (Fig. 4B). To generate the Floxed allele mice, the neomycin cassette was excised by breeding to CMV-Cre mice, and the resulting offspring were genotyped by PCR. All Cre-positive mice were negative for neo. The N3F1 mice were born at the expected frequency (Fig. 4C) and were fertile. Western blot analysis of liver protein extracts from Ahrdbd/dbd (Targeted and Floxed) mice and wild-type littermates was used to quantify the relative in vivo expression levels of the AHRdbd protein, and showed that although the Targeted allele produced hypomorphic expression of the Ahrdbd protein as compared with wild-type littermates, the amount of protein produced in the Floxed mice was equivalent to wild-type littermates (Fig. 4D).
FIG. 4.
Generation of Ahrdbd mice. (A) Schematic diagram depicting the targeting construct used to generate the Ahrdbd allele in mice. Restriction enzyme sites shown are Mlu (M), BglII (Bg), BamHI (B), and SrfI (S). Arrowheads flanking the neomycin resistance cassette (Neo-R) indicate the location of LoxP sites. Shown in gray boxes are the locations of the Neo-R cassette, including the phosphoglycerate kinase promoter (p), the bHLH domain, and the Herpes Simplex Virus thymidine kinase gene cassette (HSV-tk). Primers used for PCR genotyping are shown (OL941 and OL942) as well as the location of the Southern probe used to genotype ES cells for recombination. The Floxed allele was generated by crossing the Ahrdbd Targeted allele animals to an animal expressing the Cre-recombinase protein driven by the CMV promoter, and the subsequent outcrossing to C57BL/6J to eliminate the Cre transgene. (B) PCR-based genotyping of Ahr+/+, dbd/dbd, and +/dbd mice. The amplified product from OL941 and OL942 is cut by BamHI only when the targeted allele is present. (C) Western blot analysis. AHR protein expression in liver extracts from wild-type and Ahrdbd/dbd mice. Lane 1: Floxed allele (Neo excised); lane 2: Targeted allele (Neo present); lane 3: Ahr+/+ (1/4 protein concentration); lane 4: Ahr+/+ (1/2 protein concentration); lane 5: Ahr+/+.
Characterization of AHRdbd Signaling In Vivo
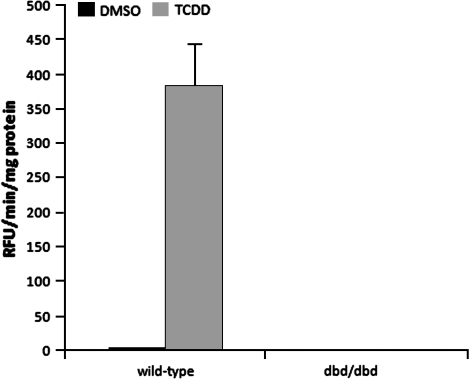
To determine whether AHRdbd signals effectively in vivo, Ahrdbd/dbd (Floxed) and wild-type mice were injected intraperitoneally with 100 μg/kg TCDD. After 6 days, liver microsomes were isolated and analyzed for EROD activity. Microsomes from wild-type mice showed a low basal EROD activity that was induced 10-fold by TCDD. In contrast, microsomes from Ahrdbd/dbd mice showed extremely low basal EROD activity, which was unaltered by TCDD treatment, indicating that AHRdbd lacks the ability to activate gene transcription from DRE elements in vivo (Fig. 5).
FIG. 5.
EROD analysis of liver microsomes from Ahrdbd/dbd mice. Wild-type 129SV/J and Ahdbd/dbd mice were administered a single injection of p-dioxane (-) or 32 μg/kg TCDD in p-dioxane (+) and sacrificed after 24 h. Microsomes were isolated from 0.5 g of liver, and EROD activity was quantified.
Ahrdbd/dbd (Floxed) Mice Exhibit Developmental Defects Similar to Ahr−/− Mice
Wild-type and Ahrdbd/dbd mice were examined for the developmental phenotypes found previously in Ahr−/− mice (Fernandez-Salguero et al., 1996, 1997; Gonzalez and Fernandez-Salguero, 1998; McDonnell et al., 1996; Schmidt et al., 1996; Lahvis and Bradfield, 1998; Lahvis et al., 2000; Peters et al., 1999; Zaher et al., 1998). Tissue wet weights were determined for liver, spleen, heart, thymus, and testis of 8-week-old male Ahrdbd/dbd and wild-type littermates. Similar to Ahr-null mice, the Ahrdbd/dbd mice were found to exhibit 25% smaller livers than wild-type littermate controls. Conversely, the hearts and spleens were 25 and 58% larger, respectively, in these animals (p < 0.005, Fig. 6A). Histopathological analysis of livers taken from Ahrdbd/dbd mice at postnatal days (PND) 7, 14, and 21 revealed a transient microvesicular steatosis around PND 7, which resolved by PND 14, and appeared identical to livers from age-matched Ahr−/− mice (Fig. 6B and data not shown). Histopathological analyses were also performed on adult spleen, heart, thymus, testis, lung, colon, kidney, eye, and brain, but revealed no significant differences between Ahrdbd/dbd and wild-type mice (data not shown). These findings are consistent with those reported in our previous work characterizing the Ahr−/− mouse (Schmidt et al., 1996).
FIG. 6.
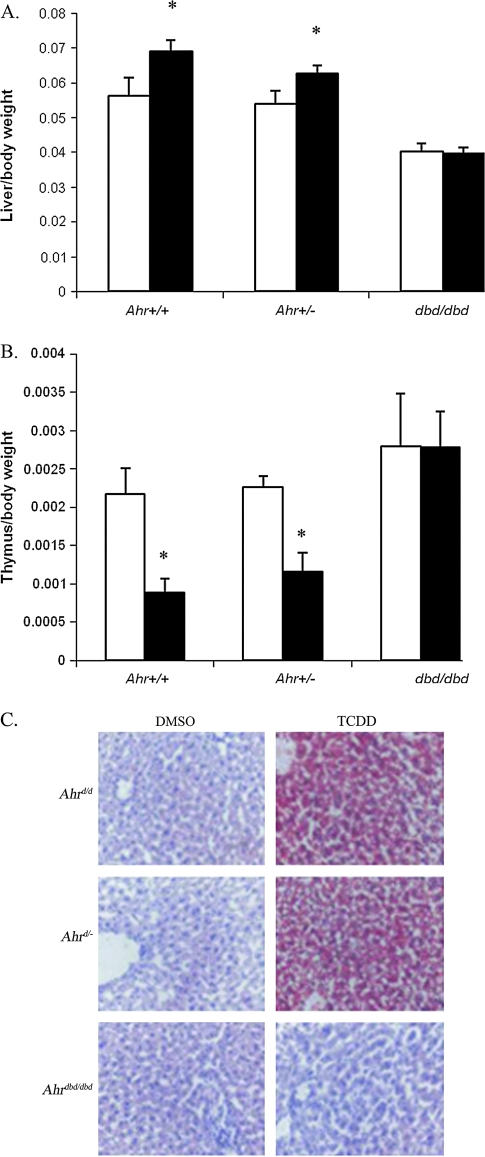
Developmental phenotype of Ahrdbd/dbd mice. (A) Relative organ wet weights of Ahrdbd/dbd mice (white bars) and wild-type littermates (black bars) sacrificed at 8 weeks of age (n = 5). *Indicates p < 0.01 by Student's t-test (wild-type versus Ahrdbd/dbd). (B) Representative H&E sections of livers from 7-day-old wild-type (littermate), Ahrdbd/dbd, and Ahr−/− mice (40× magnification). (C) Time-lapse angiography of wild-type (top row) and Ahrdbd/dbd (bottom row) littermates. Arrows identify key features as follows: BV, branching vessel; PV, portal vein; shIVC, suprahepatic inferior vena cava; ihIVC, infrahepatic inferior vena cava. Total time elapsed from the first panel to the last is approximately 10 s. (D) Incidence of patent DV in wild-type and Ahrdbd/dbd male mice as measured by trypan blue perfusion.
A consistent phenotype found in all Ahr−/− mice is the presence of a ductous venosus (DV) throughout life (Lahvis et al., 2005). To determine whether Ahrdbd/dbd mice exhibit a patent DV, the flow of contrast medium through the perfused liver was observed with the use of serial angiograms. In a wild-type littermate, contrast medium flowed into the portal vein and immediately into the portal branches of the liver (Fig. 6C). After filling the major branching veins of the liver, contrast entered the suprahepatic inferior vena cava (IVC) and then flowed retrograde, filling the infrahepatic IVC. However, in the Ahrdbd/dbd mice, contrast flowed directly from the portal vein to the IVC. The DV in the Ahrdbd/dbd mouse was clearly visible as a short segment that runs perpendicular to both the portal vein and the IVC. Trypan blue perfusion was also used to score for a patent DV. Whereas 0% of wild-type mice (0/6) showed a patent DV, 100% (6/6) of Ahrdbd/dbd mice scored positive for this structure, a vascular pattern consistent with the frequency of patent DV seen in Ahr−/− mice (Fig. 6D) (Lahvis et al., 2000).
Ahrdbd/dbd Mice are Resistant to TCDD-Induced Toxicity
To determine the importance of DNA binding in the AHR-mediated physiologic response to TCDD, 4-week-old male Ahrdbd/dbd mice, wild-type littermates, and Ahr+/− mice were treated with 100 μg/kg TCDD. Mice were sacrificed 6 days later and assayed for hepatomegaly and thymic involution, two classic endpoints associated with TCDD toxicity in these animals. In response to TCDD, the Ahr+/+ mice (n = 10) showed a 23% increase and 59% decrease in liver and thymus weights, respectively (p < 0.001; Figs. 7A and 7B). The Ahr+/− (n = 11) mice showed a 16% increase in liver weight (p < 0.001) and a 51% decrease in thymus weight 6 days after TCDD exposure (p < 0.001). In contrast, the Ahrdbd/dbd mice (n = 10) showed no significant difference in liver or thymus weights (p = 0.52, and p = 0.97, respectively), mimicking the response seen in Ahr−/− mice (Schmidt et al., 1996).
FIG. 7.
TCDD-induced phenotypic changes in Ahrdbd/dbd mice. (A) Hepatomegaly (expressed as relative liver weight) and (B) thymic involution (expressed as relative thymus weight) of DMSO- or TCDD-treated Ahr+/+ (n = 10), Ahr+/− (n = 11), and Ahrdbd/dbd (n = 10) mice as quantified 6 days after a single i.p. injection of p-dioxane (white bars) or 100 μg/kg TCDD (black bars). *Indicates p < 0.001. (C) Intrahepatic lipid accumulation. Frozen sections from DMSO- or TCDD-treated Ahr+/+, Ahr+/−, and Ahrdbd/dbd mice were stained with Oil Red-O (lipids, red) and hematoxylin (nuclei, blue).
TCDD is known to cause intrahepatic lipid accumulation in Ahr wild-type but not Ahr−/− mice (Poland and Knutson, 1982; data not shown). We therefore qualitatively examined the presence of lipids in Ahrdbd/dbd mice by Oil Red-O staining. As expected, TCDD caused a significant increase in hepatic lipid content in Ahr+/+ mice, and Ahr+/− mice accumulated lipid to the same degree. However, similar to Ahr−/− mice, Ahrdbd/dbd mice were resistant to the effects of TCDD and did not accumulate lipid (data not shown and Fig. 7C).
As TCDD is a potent teratogen, we sought to determine whether Ahrdbd/dbd mice were resistant to TCDD-induced cleft palate and hydronephrosis. Whereas 29% (10/35) of wild-type embryos (ED17–18) exhibited cleft palate upon TCDD treatment, Ahrdbd/dbd embryos were completely resistant (0/52). Similarly, 100% (35/35) of wild-type mice exhibited TCDD-induced hydronephrosis, but Ahrdbd/dbd mice were entirely resistant (0/52; Table 1).
TABLE 1.
TCDD-Induced Developmental Toxicity in Wild-Type and Ahrdbd/dbd Mice
| Ahr-wt | Ahrdbd/dbd | ||
| Cleft palate | DMSO | 0% (n = 6) | 0% (n = 8) |
| TCDD | 29% (n = 35) | 0% (n = 52) | |
| Hydronephrosis | DMSO | 0% (n = 6) | 0% (n = 8) |
| TCDD | 100% (n = 35) | 0% (n = 52) |
Note. Pregnant dams were injected with DMSO or TCDD on ED10, and litters were scored for the presence of cleft palate and hydronephrosis on ED17–18. Incidence is expressed as a percentage.
DISCUSSION
Several reports have suggested that AHR-DRE binding may not be a requirement for TCDD signaling and toxicity (Blankenship and Matsumura, 1997; Enan and Matsumura, 1996; Ge and Elferink, 1998; Puga et al., 2000). The implication of these models is that the AHR may signal in a toxicologically relevant manner through protein interactions in the cytosolic or nuclear compartment. In this regard, it has been reported that the cytosolic cSrc protein tyrosine kinase becomes activated in response to TCDD in cell-free extracts of guinea pig adipose tissue and mouse NIH3T3 cells (Enan and Matsumura, 1996). This interaction was shown to be dependent on AHR, as activity was lower in AHR-immunodepleted extracts. An interaction of AHR with the retinoblastoma protein (pRB) has also been proposed. The AHR was shown to be immunoprecipitated in rat hepatoma 5L cells by antibodies to pRB, but in yeast and cell-free interaction analysis, only truncated forms of AHR showed significant interactions. In a second study, this interaction was reported to be important in G1 cell-cycle arrest and occurred only after ligand-bound AHR translocated to the nucleus (Ge and Elferink, 1998; Puga et al., 2000). A third proposed mechanism through which AHR may mediate toxicity is through a repression of NF-κB. Experiments in vitro and in cell culture have shown that AHR-NF-κB interactions may occur through direct binding of AHR and the RelA subunit (Tian et al., 1999). We and others have considered the notion that cross-talk occurs between the AHR and HIF-1α signaling pathways via their common dimerization partner, ARNT, the underlying idea being that TCDD toxicity may be the result of ARNT sequestration rather than AHR-ARNT-DRE interactions (Berghard et al., 1993; Chan et al., 1999; Gradin et al., 1996; Pollenz et al., 1999).
We hypothesized that if any of these models were correct, then related toxic responses to TCDD should occur in animal models harboring a correctly folded AHR protein with a mutation that prevents its binding to the DRE. Such a mutant should be capable of sequestering ARNT, as well as interacting with cSrc, pRB, and NF-κB, and yet be unable to activate transcription of DRE-driven genes. We therefore used homologous recombination to replace the endogenous bHLH region of the Ahr locus with a bHLH region carrying both an I25G mutation (SrfI site) and a GS insertion (BamHI site) at amino acid residue 39. This type of insertion mutation was generated with the idea that it would effectively shift the basic region out of the major groove of DNA without disrupting the dimerization capability of the HLH domain or the overlapping nuclear localization signal (Bacsi and Hankinson, 1996; Ikuta et al., 1998). Upon construction of the corresponding mutant cDNA, we found that the AHRdbd protein does in fact form a robust, ligand-inducible interaction with the ARNT protein in a ligand dependent manner using mammalian two-hybrid analysis. Moreover, the resulting protein binds ligand and interacts with its known cellular chaperones, Hsp90 and ARA9. In keeping with our predicted impact on function, the AHRdbd protein was incapable of forming an AHRdbd/ARNT/DRE complex in a gel-shift analysis. Surprisingly, we found that although this mutation did not change amino acids thought to be directly involved in nuclear localization, it did appear to target the protein constitutively to the nuclear compartment. Although this finding implicates a disruption in ARA9- and HSP90-AHR interactions, Co-IP experiments demonstrate that these interactions were similar to those seen with wild-type AHR. Moreover, our two-hybrid experiments indicated that the AHRdbd still bound similar amounts of agonist upon exposure in cell culture.
In order to test the ability of Ahrdbd/dbd mice to signal in classical xenobiotic adaptation pathways, we quantified EROD activity following TCDD exposure. We found that Ahrdbd/dbd mice failed to show an increase in EROD activity in response to TCDD, indicating that DRE-mediated transcriptional events are eliminated in these mice. We also assayed for several of the known developmental defects observed in Ahr−/− mice and found that Ahrdbd/dbd mice are identical to Ahr−/− mice in all of these aspects, including a patent DV. The DV is a portal-systemic shunt that connects the umbilical cord blood with blood from both the portal vein and inferior vena cava (Schermerhorn et al., 1996). This structure normally resolves shortly after birth, yet remains open in Ahr−/− mice. Similar to Ahr−/− mice, Ahrdbd/dbd mice display a patent DV in addition to a transient perinatal microvesicular steatosis in hepatocytes and a 25% reduction in liver weight.
To determine whether Ahrdbd/dbd mice are sensitive to TCDD-induced toxicity, a number of classical toxic responses to TCDD were also quantified. We observed that Ahrdbd/dbd mice failed to exhibit the obvious liver and thymic toxicity normally associated with this dosing regimen. We also found that similar to the Ahr-null, TCDD-induced cleft palate, hydronephrosis, and intrahepatic lipid accumulation were nonexistent in these mice.
Although the AHRdbd protein appears to be constitutively nuclear in the absence of ligand, we showed previously that a mutation introduced at the nuclear localization sequence also causes abnormal liver development and renders mice resistant to TCDD-induced toxicity (Bunger et al., 2003). Therefore, it is unlikely that the cytosolic interactions lost due to localization are important in the developmental or toxic phenotypes of these animals, including those of cSrc, RelA, and pRB. Furthermore, in combination with our previously reported observations on Ahrnls/nls mice, these results convincingly show that nuclear localization alone is necessary but not sufficient for Ahr to function in development and toxicity. Therefore, sequestration of ARNT within the nucleus as a mechanism of toxicity is unlikely. However, we do not rule out the possibility that Ahrdbd/dbd mice may still show responsiveness to TCDD at other endpoints not tested, such as tumor promotion.
We show here that AHR-mediated XME induction may be directly related to a toxic response. Ahrdbd/dbd mice express an AHR that can be ligand-activated to form a heterodimer with the ARNT protein in a similar manner to wild-type AHR, but cannot bind to DREs and activate XME expression. The fact that these mice fail to exhibit a toxic response suggests that DRE-driven gene expression is indeed upstream of the physiologic effects of TCDD and that ARNT sequestration may in fact not play a significant role in the TCDD-induced toxic endpoints assessed here. Furthermore, a developmental phenotype of the Ahrdbd/dbd mice consistent with that of Ahr−/− mice suggests that DRE-driven genes are also involved in early liver development and vascular remodeling.
FUNDING
National Institutes of Health grant numbers (R01-ES-013566-01, P01-CA022484, T32-CA009135, and P30-CA014520).
References
- Bacsi SG, Hankinson O. Functional characterization of DNA-binding domains of the subunits of the heterodimeric aryl hydrocarbon receptor complex imputing novel and canonical basic helix-loop-helix protein-DNA interactions. J. Biol. Chem. 1996;271:8843–8850. doi: 10.1074/jbc.271.15.8843. [DOI] [PubMed] [Google Scholar]
- Berghard A, Gradin K, Pongratz I, Whitelaw M, Poellinger L. Cross-coupling of signal transduction pathways: The dioxin receptor mediates induction of cytochrome P-450IA1 expression via a protein kinase C-dependent mechanism. Mol. Cell. Biol. 1993;13:677–689. doi: 10.1128/mcb.13.1.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenship A, Matsumura F. 2,3,7,8-Tetrachlorodibenzo-p-dioxin-induced activation of a protein tyrosine kinase, pp60src, in murine hepatic cytosol using a cell-free system. Mol. Pharmacol. 1997;52:667–675. doi: 10.1124/mol.52.4.667. [DOI] [PubMed] [Google Scholar]
- Bunger MK, Moran SM, Glover E, Thomae TL, Lahvis GP, Lin BC, Bradfield CA. Resistance to 2,3,7,8-tetrachlorodibenzo-p-dioxin toxicity and abnormal liver development in mice carrying a mutation in the nuclear localization sequence of the aryl hydrocarbon receptor. J. Biol. Chem. 2003;278:17767–17774. doi: 10.1074/jbc.M209594200. [DOI] [PubMed] [Google Scholar]
- Buters JT, Doehmer J, Gonzalez FJ. Cytochrome P450-null mice. Drug Metab. Rev. 1999;31:437–447. doi: 10.1081/dmr-100101929. [DOI] [PubMed] [Google Scholar]
- Carver LA, Bradfield CA. Ligand-dependent interaction of the aryl hydrocarbon receptor with a novel immunophilin homolog in vivo. J. Biol. Chem. 1997;272:11452–11456. doi: 10.1074/jbc.272.17.11452. [DOI] [PubMed] [Google Scholar]
- Carver LA, LaPres JJ, Jain S, Dunham EE, Bradfield CA. Characterization of the Ah receptor-associated protein, ARA9. J. Biol. Chem. 1998;273:33580–33587. doi: 10.1074/jbc.273.50.33580. [DOI] [PubMed] [Google Scholar]
- Chan WK, Yao G, Gu YZ, Bradfield CA. Cross-talk between the aryl hydrocarbon receptor and hypoxia inducible factor signaling pathways. Demonstration of competition and compensation. J. Biol. Chem. 1999;274:12115–12123. doi: 10.1074/jbc.274.17.12115. [DOI] [PubMed] [Google Scholar]
- Enan E, Matsumura F. Identification of c-Src as the integral component of the cytosolic Ah receptor complex, transducing the signal of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) through the protein phosphorylation pathway. Biochem. Pharmacol. 1996;52:1599–1612. doi: 10.1016/s0006-2952(96)00566-7. [DOI] [PubMed] [Google Scholar]
- Fernandez-Salguero PM, Hilbert DM, Rudikoff S, Ward JM, Gonzalez FJ. Aryl-hydrocarbon receptor-deficient mice are resistant to 2,3,7,8-tetrachlorodibenzo-p-dioxin-induced toxicity. Toxicol. Appl. Pharmacol. 1996;140:173–179. doi: 10.1006/taap.1996.0210. [DOI] [PubMed] [Google Scholar]
- Fernandez-Salguero PM, Ward JM, Sundberg JP, Gonzalez FJ. Lesions of aryl-hydrocarbon receptor-deficient mice. Vet. Pathol. 1997;34:605–614. doi: 10.1177/030098589703400609. [DOI] [PubMed] [Google Scholar]
- Ge NL, Elferink CJ. A direct interaction between the aryl hydrocarbon receptor and retinoblastoma protein. Linking dioxin signaling to the cell cycle. J. Biol. Chem. 1998;273:22708–22713. doi: 10.1074/jbc.273.35.22708. [DOI] [PubMed] [Google Scholar]
- Gonzalez FJ, Fernandez-Salguero P. The aryl hydrocarbon receptor: Studies using the AHR-null mice. Drug Metab. Dispos. 1998;26:1194–1198. [PubMed] [Google Scholar]
- Gradin K, McGuire J, Wenger RH, Kvietikova I, Fhitelaw ML, Toftgard R, Tora L, Gassmann M, Poellinger L. Functional interference between hypoxia and dioxin signal transduction pathways: Competition for recruitment of the Arnt transcription factor. Mol. Cell. Biol. 1996;16:5221–5231. doi: 10.1128/mcb.16.10.5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hankinson O. The aryl hydrocarbon receptor complex. Annu. Rev. Pharmacol. Toxicol. 1995;35:307–340. doi: 10.1146/annurev.pa.35.040195.001515. [DOI] [PubMed] [Google Scholar]
- Ikuta T, Eguchi H, Tachibana T, Yoneda Y, Kawajiri K. Nuclear localization and export signals of the human aryl hydrocarbon receptor. J. Biol. Chem. 1998;273:2895–2904. doi: 10.1074/jbc.273.5.2895. [DOI] [PubMed] [Google Scholar]
- Jain S, Dolwick KM, Schmidt JV, Bradfield CA. Potent transactivation domains of the Ah receptor and the Ah receptor nuclear translocator map to their carboxyl termini. J. Biol. Chem. 1994;269:31518–31524. [PubMed] [Google Scholar]
- Lahvis GP, Bradfield CA. Ahr null alleles: Distinctive or different? Biochem. Pharmacol. 1998;56:781–787. doi: 10.1016/s0006-2952(98)00134-8. [DOI] [PubMed] [Google Scholar]
- Lahvis GP, Lindell SL, Thomas RS, McCuskey RS, Murphy C, Glover E, Bentz M, Southard J, Bradfield CA. Portosystemic shunting and persistent fetal vascular structures in aryl hydrocarbon receptor-deficient mice. Proc. Natl. Acad. Sci. U. S. A. 2000;97:10442–10447. doi: 10.1073/pnas.190256997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahvis G, Pyzalski RW, Glover E, Pitot HC, McElwee MK, Bradfield C. The aryl hydrocarbon receptor is required for developmental closure of the ductus venosus in the neonatal mouse. Mol. Pharmacol. 2005;67:714–720. doi: 10.1124/mol.104.008888. [DOI] [PubMed] [Google Scholar]
- Liang HC, Li H, McKinnon RA, Duffy JJ, Potter SS, Puga A, Nebert DW. Cyp1a2(-/-) null mutant mice develop normally but show deficient drug metabolism. Proc. Natl. Acad. Sci. U. S. A. 1996;93:1671–1676. doi: 10.1073/pnas.93.4.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonnell WM, Chensue SW, Askari FK, Moseley RH. Hepatic fibrosis in Ahr-/- mice. Science. 1996;271:223–224. [PubMed] [Google Scholar]
- Nilausen K, Green H. Reversible arrest of growth in G1 of an established fibroblast line (3T3) Exp. Cell. Res. 1965;40:166–168. doi: 10.1016/0014-4827(65)90306-x. [DOI] [PubMed] [Google Scholar]
- Peters JM, Narotsky MG, Elizondo G, Fernandez-Salguero PM, Gonzalez FJ, Abbott BD. Amelioration of TCDD-induced teratogenesis in aryl hydrocarbon receptor (AhR)-null mice. Toxicol. Sci. 1999;47:86–92. doi: 10.1093/toxsci/47.1.86. [DOI] [PubMed] [Google Scholar]
- Pineau T, Fernandez-Salguero P, Lee SS, McPhail T, Ward JM, Gonzalez FJ. Neonatal lethality associated with respiratory distress in mice lacking cytochrome P450 1A2. Proc. Natl. Acad. Sci. U. S. A. 1995;92:5134–5138. doi: 10.1073/pnas.92.11.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland A, Glover E, Bradfield CA. Characterization of polyclonal antibodies to the Ah receptor prepared by immunization with a synthetic peptide hapten. Mol. Pharmacol. 1991;39:20–26. [PubMed] [Google Scholar]
- Poland A, Glover E, Ebetino FH, Kende AS. Photoaffinity labeling of the Ah receptor. J. Biol. Chem. 1986;261:6352–6365. [PubMed] [Google Scholar]
- Poland A, Knutson JC. 2,3,7,8-tetrachlorodibenzo-p-dioxin and related halogenated aromatic hydrocarbons: Examination of the mechanism of toxicity. Annu. Rev. Pharmacol. Toxicol. 1982;22:517–554. doi: 10.1146/annurev.pa.22.040182.002505. [DOI] [PubMed] [Google Scholar]
- Poland A, Palen D, Glover E. Analysis of the four alleles of the murine aryl hydrocarbon receptor. Mol. Pharmacol. 1994;46:915–921. [PubMed] [Google Scholar]
- Pollenz RS, Davarinos NA, Shearer TP. Analysis of aryl hydrocarbon receptor-mediated signaling during physiological hypoxia reveals lack of competition for the aryl hydrocarbon nuclear translocator transcription factor. Mol. Pharmacol. 1999;56:1127–1137. doi: 10.1124/mol.56.6.1127. [DOI] [PubMed] [Google Scholar]
- Postlind H, Vu TP, Tukey RH, Quattrochi LC. Response of human CYP1-luciferase plasmids to 2,3,7,8-tetrachlorodibenzo-p-dioxin and polycyclic aromatic hydrocarbons. Toxicol. Appl. Pharmacol. 1993;118:255–262. doi: 10.1006/taap.1993.1031. [DOI] [PubMed] [Google Scholar]
- Puga A, Barnes SJ, Dalton TP, Chang C, Knudsen ES, Maier MA. Aromatic hydrocarbon receptor interaction with the retinoblastoma protein potentiates repression of E2F-dependent transcription and cell cycle arrest. J. Biol. Chem. 2000;275:2943–2950. doi: 10.1074/jbc.275.4.2943. [DOI] [PubMed] [Google Scholar]
- Schermerhorn T, Center SA, Dykes NL, Rowland PH, Yeager AE, Erb HN, Oberhansley K, Bonda M. Characterization of hepatoportal microvascular dysplasia in a kindred of cairn terriers. J. Vet. Intern. Med. 1996;10:219–230. doi: 10.1111/j.1939-1676.1996.tb02053.x. [DOI] [PubMed] [Google Scholar]
- Schmidt JV, Carver LA, Bradfield CA. Molecular characterization of the murine Ahr gene. Organization, promoter analysis, and chromosomal assignment. J. Biol. Chem. 1993;268:22203–22209. [PubMed] [Google Scholar]
- Schmidt JV, Su GH, Reddy JK, Simon MC, Bradfield CA. Characterization of a murine Ahr null allele: Involvement of the Ah receptor in hepatic growth and development. Proc. Natl. Acad. Sci. U. S. A. 1996;93:6731–6736. doi: 10.1073/pnas.93.13.6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Ke S, Denison MS, Rabson AB, Gallo MA. Ah receptor and NF-kappaB interactions, a potential mechanism for dioxin toxicity. J. Biol. Chem. 1999;274:510–515. doi: 10.1074/jbc.274.1.510. [DOI] [PubMed] [Google Scholar]
- Tybulewicz VL, Crawford CE, Jackson PK, Bronson RT, Mulligan RC. Neonatal lethality and lymphopenia in mice with a homozygous disruption of the c-abl proto-oncogene. Cell. 1991;65:1153–1163. doi: 10.1016/0092-8674(91)90011-m. [DOI] [PubMed] [Google Scholar]
- Walisser JA, Glover E, Pande K, Liss AL, Bradfield CA. Aryl hydrocarbon receptor-dependent liver development and hepatotoxicity are mediated by different cell types. Proc. Natl. Acad. Sci. U. S. A. 2005;102:17858–17863. doi: 10.1073/pnas.0504757102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaher H, Fernandez-Salguero PM, Letterio J, Sheikh MS, Fornace AJ, Jr, Roberts AB, Gonzalez FJ. The involvement of aryl hydrocarbon receptor in the activation of transforming growth factor-beta and apoptosis. Mol. Pharmacol. 1998;54:313–321. doi: 10.1124/mol.54.2.313. [DOI] [PubMed] [Google Scholar]